Conventional and Omics Approaches for Understanding the Abiotic Stress Response in Cereal Crops—An Updated Overview
Abstract
1. Introduction
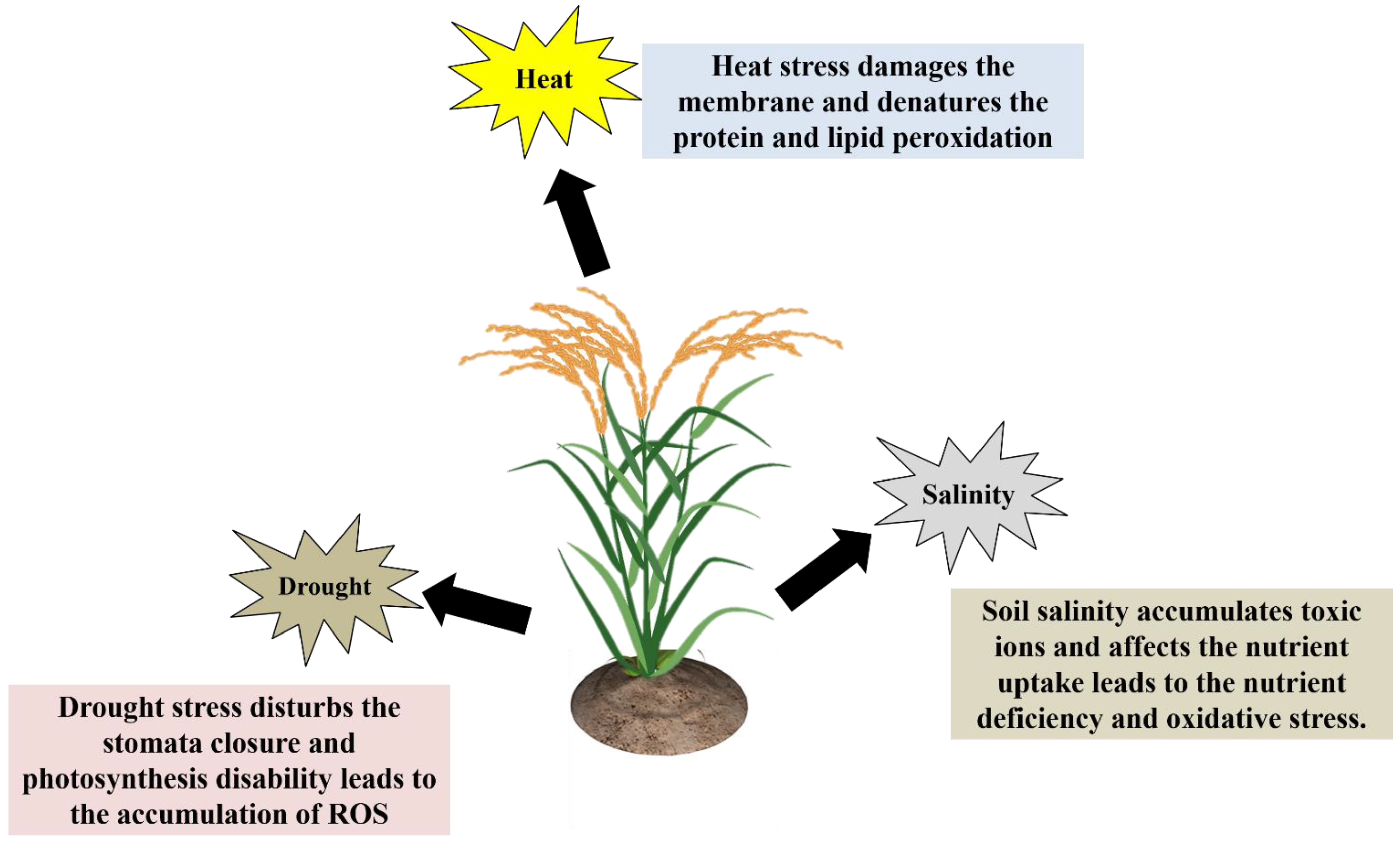
2. General Effects of Drought, Heat, and Salt Stress on Plant Growth and Development
2.1. Drought Stress
2.2. Temperature Stress (Heat)
2.3. Salinity Stress
2.4. Oxidative Damage and Antioxidant Enzyme Defense System in Plants under Abiotic Stress
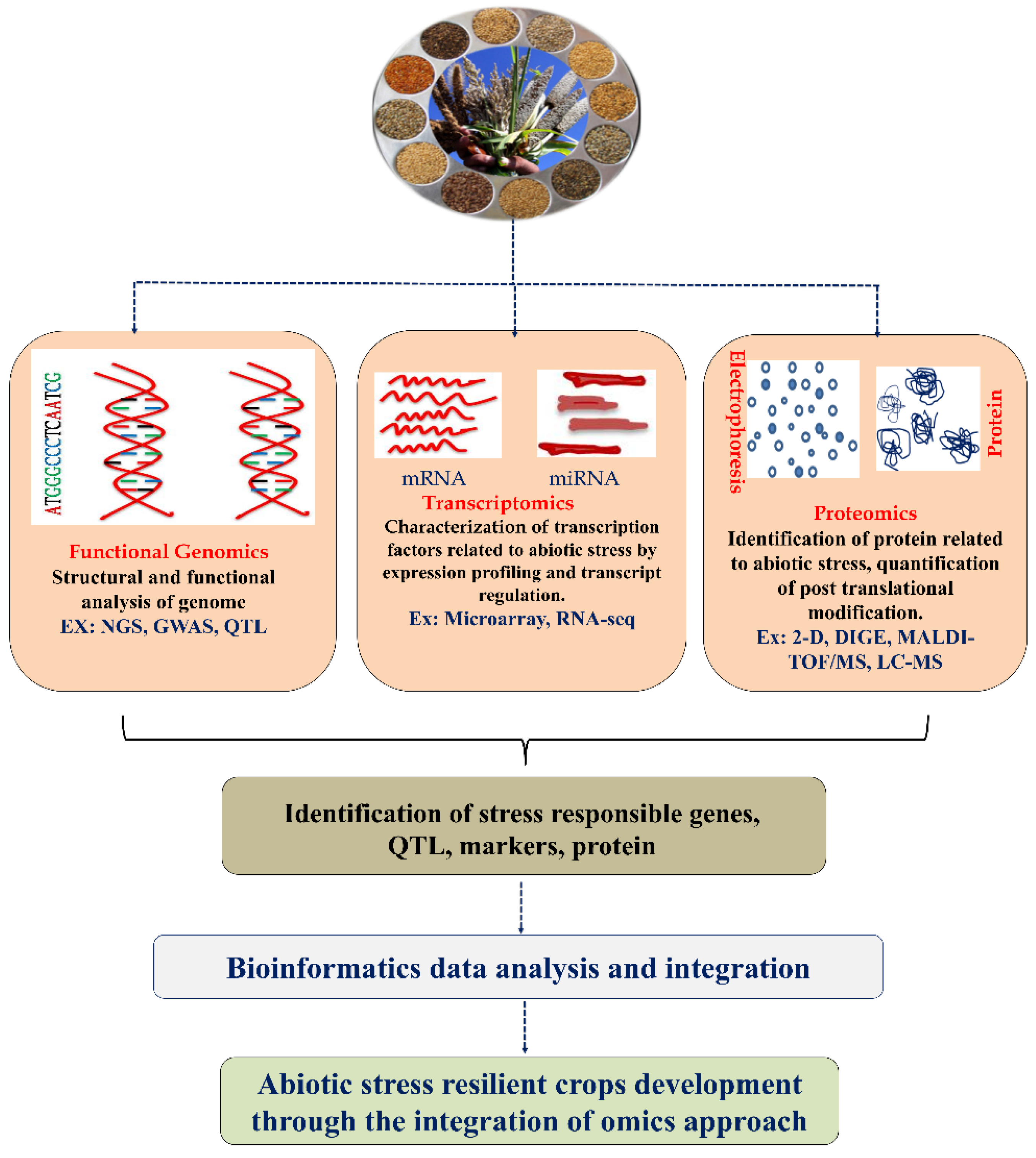
3. Functional Genomic Approaches to Identify Stress-Responsive Genes in Cereals
3.1. Genome Sequencing
| S. No. | Crop (Common Name) | Botanical Name | Genome Size (Mb) | Sequencing Methods | Reference |
|---|---|---|---|---|---|
| 1 | Rice | Oryza sativa ssp. japonica (Nipponbare) | 420 | Sanger, WGS | [78] |
| Oryza sativa ssp. japonica (Nipponbare) | 389 | Sanger, BAC-by-BAC | [79] | ||
| Oryza sativa ssp. indica | 466 | Sanger, WGS | [77] | ||
| 2 | Maize | Zea mays (Palomero Toluqueno) (popcorn) | 2100 | Sanger, WGS | [126] |
| Zea mays (B73) | 2300 | Sanger, BAC-by-BAC | [80] | ||
| 3 | Sorghum | Sorghum bicolor (L.) Moench | 730 | Sanger, WGS | [81] |
| 4 | Foxtail millet | Setaria italica | 515 | Illumina, WGS | [82] |
| 5 | Bread wheat | Triticum aestivum | 17,000 | 454, WGS | [84] |
| 6 | Barley | Hordeum vulgare | 5100 | 454, BAC-by-BAC | [85] |
| 7 | Finger millet | Eleusine coracana | 1200 | Illumina, WGS | [86,104] |
| 8 | Pearl millet | Cenchrus americanus | 1800 | Illumina, WGS | [108] |
| 9 | Proso millet | Panicum miliaceum | 923 | Illumina short-read coupled with Pac-Bio long-read sequencing | [106] |
| 10 | Barnyard millet | Echinochloa esculenta | 1.27 | Illumina HiSeq platform | [107] |
3.2. Molecular Markers
3.3. Genome-Wide Association Studies (GWAS)
4. Transcriptome and Gene Expression Analysis of Abiotic Stress Responses in Cereals
4.1. Drought Stress
4.2. Heat Stress
4.3. Salinity Stress
5. Proteomics Approaches to Identify the Functional and Structural Characteristics of the Abiotic Stress-Responsive Proteins in Cereal Crops
5.1. Drought Stress
5.2. Heat Stress
5.3. Salinity Stress
6. Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salse, J.; Feuillet, C. Comparative Genomics of Cereals. Genom.-Assist. Crop Improv. 2007, 177–205. [Google Scholar]
- Lata, C.; Shivhare, R. Engineering Cereal Crops for Enhanced Abiotic Stress Tolerance. Proc. Indian Natl. Sci. Acad. 2021, 87, 63–83. [Google Scholar] [CrossRef]
- TAŞĞIN, E. Macronutrients and Micronutrients in Nutrition. Int. J. Innov. Res. Rev. 2017, 1, 10–15. [Google Scholar]
- Ahmad, P.; Abdel Latef, A.A.H.; Rasool, S.; Akram, N.A.; Ashraf, M.; Gucel, S. Role of Proteomics in Crop Stress Tolerance. Front. Plant Sci. 2016, 7, 1336. [Google Scholar] [CrossRef]
- Raza, A.; Su, W.; Gao, A.; Mehmood, S.S.; Hussain, M.A.; Nie, W.; Lv, Y.; Zou, X.; Zhang, X. Catalase (CAT) Gene Family in Rapeseed (Brassica Napus L.): Genome-Wide Analysis, Identification, and Expression Pattern in Response to Multiple Hormones and Abiotic Stress Conditions. Int. J. Mol. Sci. 2021, 22, 4281. [Google Scholar] [CrossRef]
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-Morphological Traits. Front. Plant Sci. 2017, 8, 537. [Google Scholar] [CrossRef]
- Rani, S.; Kumar, P.; Suneja, P. Biotechnological Interventions for Inducing Abiotic Stress Tolerance in Crops. Plant Gene 2021, 27, 100315. [Google Scholar] [CrossRef]
- Giordano, M.; Petropoulos, S.A.; Rouphael, Y. Response and Defence Mechanisms of Vegetable Crops against Drought, Heat and Salinity Stress. Agriculture 2021, 11, 463. [Google Scholar] [CrossRef]
- Roy, S.J.; Tucker, E.J.; Tester, M. Genetic Analysis of Abiotic Stress Tolerance in Crops. Curr. Opin. Plant Biol. 2011, 14, 232–239. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef]
- Comas, L.H.; Trout, T.J.; DeJonge, K.C.; Zhang, H.; Gleason, S.M. Water Productivity under Strategic Growth Stage-Based Deficit Irrigation in Maize. Agric. Water Manag. 2019, 212, 433–440. [Google Scholar] [CrossRef]
- World Health Organization. The State of Food Security and Nutrition in the World 2021: Transforming Food Systems for Food Security, Improved Nutrition and Affordable Healthy Diets for All; Food & Agriculture Org.: Rome, Italy, 2021; Volume 2021, ISBN 925134325X. [Google Scholar]
- Wang, H.; Qin, F. Genome-Wide Association Study Reveals Natural Variations Contributing to Drought Resistance in Crops. Front. Plant Sci. 2017, 8, 1110. [Google Scholar] [CrossRef] [PubMed]
- Pandian, S.; Rakkammal, K.; Rency, A.S.; Muthuramalingam, P.; Pandian, S.K.; Ramesh, M. Abiotic Stress and Applications of Omics Approaches to Develop Stress Tolerance in Agronomic Crops. In Agronomic Crops; Springer: Singapore, 2020; pp. 557–578. [Google Scholar]
- Zhan, X.; Lu, Y.; Zhu, J.; Botella, J.R. Genome Editing for Plant Research and Crop Improvement. J. Integr. Plant Biol. 2021, 63, 3–33. [Google Scholar] [CrossRef] [PubMed]
- Paes de Melo, B.; de Carpinetti, P.A.; Fraga, O.T.; Rodrigues-Silva, P.L.; Fioresi, V.S.; de Camargos, L.F.; da Silva Ferreira, M.F. Abiotic Stresses in Plants and Their Markers: A Practice View of Plant Stress Responses and Programmed Cell Death Mechanisms. Plants 2022, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Yaqoob, U.; Jan, N.; Raman, P.V.; Siddique, K.H.M.; John, R. Crosstalk between Brassinosteroid Signaling, ROS Signaling and Phenylpropanoid Pathway during Abiotic Stress in Plants: Does It Exist? Plant Stress 2022, 4, 100075. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of Abiotic Stress on Plants: A Systems Biology Perspective. BMC Plant Biol. 2011, 11, 1–14. [Google Scholar] [CrossRef]
- Pandian, S.; Rakkammal, K.; Rathinapriya, P.; Rency, A.S.; Satish, L.; Ramesh, M. Physiological and Biochemical Changes in Sorghum under Combined Heavy Metal Stress: An Adaptive Defence against Oxidative Stress. Biocatal. Agric. Biotechnol. 2020, 29, 101830. [Google Scholar] [CrossRef]
- Rathinapriya, P.; Pandian, S.; Rakkammal, K.; Balasangeetha, M.; Alexpandi, R.; Satish, L.; Rameshkumar, R.; Ramesh, M. The Protective Effects of Polyamines on Salinity Stress Tolerance in Foxtail Millet (Setaria Italica L.), an Important C4 Model Crop. Physiol. Mol. Biol. Plants 2020, 26, 1815–1829. [Google Scholar] [CrossRef]
- He, M.; He, C.-Q.; Ding, N.-Z. Abiotic Stresses: General Defenses of Land Plants and Chances for Engineering Multistress Tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef]
- Chaudhry, S.; Sidhu, G.P.S. Climate Change Regulated Abiotic Stress Mechanisms in Plants: A Comprehensive Review. Plant Cell Rep. 2021, 41, 1–31. [Google Scholar] [CrossRef]
- Kamara, A.Y.; Menkir, A.; Badu-Apraku, B.; Ibikunle, O. The Influence of Water Deficit on Growth, Yield and Yield Components of Some Maize Genotypes. J. Agric. Sci. 2003, 141, 43–50. [Google Scholar] [CrossRef]
- Balla, K.; Rakszegi, M.; Li, Z.; Bekes, F.; Bencze, S.; Veisz, O. Quality of Winter Wheat in Relation to Heat and Drought Shock after Anthesis. Czech J. Food Sci. 2011, 29, 117–128. [Google Scholar] [CrossRef]
- Lafitte, H.R.; Yongsheng, G.; Yan, S.; Li, Z.K. Whole Plant Responses, Key Processes, and Adaptation to Drought Stress: The Case of Rice. J. Exp. Bot. 2007, 58, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Zeid, I.M.; Shedeed, Z.A. Response of Alfalfa to Putrescine Treatment under Drought Stress. Biol. Plant. 2006, 50, 635–640. [Google Scholar] [CrossRef]
- Shi, J.-F.; Mao, X.-G.; Jing, R.-L.; Pang, X.-B.; Wang, Y.-G.; Chang, X.-P. Gene Expression Profiles of Response to Water Stress at the Jointing Stage in Wheat. Agric. Sci. China 2010, 9, 325–330. [Google Scholar] [CrossRef]
- Devnarain, N.; Crampton, B.G.; Chikwamba, R.; Becker, J.V.W.; O’Kennedy, M.M. Physiological Responses of Selected African Sorghum Landraces to Progressive Water Stress and Re-Watering. S. Afr. J. Bot. 2016, 103, 61–69. [Google Scholar] [CrossRef]
- Tang, S.; Li, L.; Wang, Y.; Chen, Q.; Zhang, W.; Jia, G.; Zhi, H.; Zhao, B.; Diao, X. Genotype-Specific Physiological and Transcriptomic Responses to Drought Stress in Setaria Italica (an Emerging Model for Panicoideae Grasses). Sci. Rep. 2017, 7, 10009. [Google Scholar] [CrossRef]
- Satish, L.; Rency, A.S.; Ramesh, M. Spermidine Sprays Alleviate the Water Deficit-Induced Oxidative Stress in Finger Millet (Eleusine Coracana L. Gaertn.) Plants. 3 Biotech 2018, 8, 63. [Google Scholar] [CrossRef]
- Barzana, G.; Rios, J.J.; Lopez-Zaplana, A.; Nicolas-Espinosa, J.; Yepes-Molina, L.; Garcia-Ibañez, P.; Carvajal, M. Interrelations of Nutrient and Water Transporters in Plants under Abiotic Stress. Physiol. Plant. 2021, 171, 595–619. [Google Scholar] [CrossRef]
- Carmody, M.; Waszczak, C.; Idänheimo, N.; Saarinen, T.; Kangasjärvi, J. ROS Signalling in a Destabilised World: A Molecular Understanding of Climate Change. J. Plant Physiol. 2016, 203, 69–83. [Google Scholar] [CrossRef]
- Wang, J.; Li, C.; Li, L.; Reynolds, M.; Mao, X.; Jing, R. Exploitation of Drought Tolerance-Related Genes for Crop Improvement. Int. J. Mol. Sci. 2021, 22, 10265. [Google Scholar] [CrossRef] [PubMed]
- Alhaithloul, H.A.S. Impact of Combined Heat and Drought Stress on the Potential Growth Responses of the Desert Grass Artemisia Sieberi Alba: Relation to Biochemical and Molecular Adaptation. Plants 2019, 8, 416. [Google Scholar] [CrossRef] [PubMed]
- Recchia, G.H.; Caldas, D.G.G.; Beraldo, A.L.A.; Da Silva, M.J.; Tsai, S.M. Transcriptional Analysis of Drought-Induced Genes in the Roots of a Tolerant Genotype of the Common Bean (Phaseolus Vulgaris L.). Int. J. Mol. Sci. 2013, 14, 7155–7179. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, J.L.; Prueger, J.H. Temperature Extremes: Effect on Plant Growth and Development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Rai, A.; Rai, G.K.; Dubey, R.S. Heat Stress and Its Effects on Plant Growth and Metabolism. In Abiotic Stress Tolerance Mechanisms in Plants; CRC Press: Boca Raton, FL, USA, 2021; pp. 203–265. ISBN 1003163831. [Google Scholar]
- Jajoo, A.; Allakhverdiev, S.I. High-Temperature Stress in Plants: Consequences and Strategies for Protecting Photosynthetic Machinery. Plant Stress Physiol. 2017, 2017, 138–154. [Google Scholar]
- Chalanika De Silva, H.C.; Asaeda, T. Effects of Heat Stress on Growth, Photosynthetic Pigments, Oxidative Damage and Competitive Capacity of Three Submerged Macrophytes. J. Plant Interact. 2017, 12, 228–236. [Google Scholar] [CrossRef]
- Yang, H.; Gu, X.; Ding, M.; Lu, W.; Lu, D. Heat Stress during Grain Filling Affects Activities of Enzymes Involved in Grain Protein and Starch Synthesis in Waxy Maize. Sci. Rep. 2018, 8, 15665. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant Tolerance to High Temperature in a Changing Environment: Scientific Fundamentals and Production of Heat Stress-Tolerant Crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef]
- Roy, S.J.; Negrão, S.; Tester, M. Salt Resistant Crop Plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of Salinity Stress on Plants and Its Tolerance Strategies: A Review. Environ. Sci. Pollut. Res. 2015, 22, 4056–4075. [Google Scholar] [CrossRef] [PubMed]
- Muneer, S.; Park, Y.G.; Manivannan, A.; Soundararajan, P.; Jeong, B.R. Physiological and Proteomic Analysis in Chloroplasts of Solanum Lycopersicum L. under Silicon Efficiency and Salinity Stress. Int. J. Mol. Sci. 2014, 15, 21803–21824. [Google Scholar] [CrossRef] [PubMed]
- Qados, A.M.S.A. Effect of Salt Stress on Plant Growth and Metabolism of Bean Plant Vicia Faba (L.). J. Saudi Soc. Agric. Sci. 2011, 10, 7–15. [Google Scholar]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating Physiological Responses of Plants to Salinity Stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nxele, X.; Klein, A.; Ndimba, B.K. Drought and Salinity Stress Alters ROS Accumulation, Water Retention, and Osmolyte Content in Sorghum Plants. S. Afr. J. Bot. 2017, 108, 261–266. [Google Scholar] [CrossRef]
- Jha, Y.; Subramanian, R.B. Regulation of Plant Physiology and Antioxidant Enzymes for Alleviating Salinity Stress by Potassium-Mobilizing Bacteria. In Potassium Solubilizing Microorganisms for Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2016; pp. 149–162. [Google Scholar]
- Suriyan, C.-U.; Supaibulwattana, K.; Kirdmanee, C. Comparative Effects of Salt Stress and Extreme PH Stress Combined on Glycinebetaine Accumulation, Photosynthetic Abilities and Growth Characters of Two Rice Genotypes. Rice Sci. 2009, 16, 274–282. [Google Scholar]
- Sahi, C.; Singh, A.; Kumar, K.; Blumwald, E.; Grover, A. Salt Stress Response in Rice: Genetics, Molecular Biology, and Comparative Genomics. Funct. Integr. Genom. 2006, 6, 263–284. [Google Scholar] [CrossRef]
- Swami, A.K.; Alam, S.I.; Sengupta, N.; Sarin, R. Differential Proteomic Analysis of Salt Stress Response in Sorghum Bicolor Leaves. Environ. Exp. Bot. 2011, 71, 321–328. [Google Scholar] [CrossRef]
- Almodares, A.; Hadi, M.R.; Dosti, B. Effects of Salt Stress on Germination Percentage and Seedling Growth in Sweet Sorghum Cultivars. J. Biol. Sci. 2007, 7, 1492–1495. [Google Scholar] [CrossRef]
- Gill, P.K.; Sharma, A.D.; Singh, P.; Bhullar, S.S. Changes in Germination, Growth and Soluble Sugar Contents of Sorghum Bicolor (L.) Moench Seeds under Various Abiotic Stresses. Plant Growth Regul. 2003, 40, 157–162. [Google Scholar] [CrossRef]
- Sidhu, G.P.S.; Bali, A.S.; Singh, H.P.; Batish, D.R.; Kohli, R.K. Insights into the Tolerance and Phytoremediation Potential of Coronopus Didymus L.(Sm) Grown under Zinc Stress. Chemosphere 2020, 244, 125350. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive Oxygen Species, Abiotic Stress and Stress Combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Sandalio, L.M.; Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Luis, A. Role of Peroxisomes as a Source of Reactive Oxygen Species (ROS) Signaling Molecules. Peroxisomes Key Role Cell. Signal. Metab. 2013, 69, 231–255. [Google Scholar]
- Pei, Z.-M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium Channels Activated by Hydrogen Peroxide Mediate Abscisic Acid Signalling in Guard Cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Al Mahmud, J.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, MA, USA, 2015; ISBN 0198717482. [Google Scholar]
- Kerchev, P.; Waszczak, C.; Lewandowska, A.; Willems, P.; Shapiguzov, A.; Li, Z.; Alseekh, S.; Mühlenbock, P.; Hoeberichts, F.A.; Huang, J. Lack of GLYCOLATE OXIDASE1, but Not GLYCOLATE OXIDASE2, Attenuates the Photorespiratory Phenotype of CATALASE2-Deficient Arabidopsis. Plant Physiol. 2016, 171, 1704–1719. [Google Scholar] [CrossRef]
- Caverzan, A.; Casassola, A.; Brammer, S.P. Antioxidant Responses of Wheat Plants under Stress. Genet. Mol. Biol. 2016, 39, 1–6. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Al Mahmud, J.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants under Abiotic Stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef]
- Gill, S.S.; Anjum, N.A.; Gill, R.; Yadav, S.; Hasanuzzaman, M.; Fujita, M.; Mishra, P.; Sabat, S.C.; Tuteja, N. Superoxide Dismutase—Mentor of Abiotic Stress Tolerance in Crop Plants. Environ. Sci. Pollut. Res. 2015, 22, 10375–10394. [Google Scholar] [CrossRef] [PubMed]
- Pradedova, E.V.; Isheeva, O.D.; Salyaev, R.K. Superoxide Dismutase of Plant Cell Vacuoles. Biochem. Suppl. Ser. Membr. Cell Biol. 2009, 3, 24–32. [Google Scholar] [CrossRef]
- Krönoiger, W.; Rennenberg, H.; Polle, A. Purification of Two Superoxide Dismutase Isozymes and Their Subcellular Localization in Needles and Roots of Norway Spruce (Picea Abies L.) Trees. Plant Physiol. 1992, 100, 334–340. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reddy, C.D.; Venkaiah, B. Isoenzymes of Superoxide Dismutase from Mung Bean (Phaseolus Aureus) Seedlings. Curr. Sci. 1982, 51, 987–988. [Google Scholar]
- Sharma, P.; Dubey, R.S. Drought Induces Oxidative Stress and Enhances the Activities of Antioxidant Enzymes in Growing Rice Seedlings. Plant Growth Regul. 2005, 46, 209–221. [Google Scholar] [CrossRef]
- Khanna-Chopra, R.; Selote, D.S. Acclimation to Drought Stress Generates Oxidative Stress Tolerance in Drought-Resistant than-Susceptible Wheat Cultivar under Field Conditions. Environ. Exp. Bot. 2007, 60, 276–283. [Google Scholar] [CrossRef]
- Zhang, J.; Kirkham, M.B. Antioxidant Responses to Drought in Sunflower and Sorghum Seedlings. New Phytol. 1996, 132, 361–373. [Google Scholar] [CrossRef]
- Boeckx, T.; Winters, A.L.; Webb, K.J.; Kingston-Smith, A.H. Polyphenol Oxidase in Leaves: Is There Any Significance to the Chloroplastic Localization? J. Exp. Bot. 2015, 66, 3571–3579. [Google Scholar] [CrossRef]
- Pandey, S.; Fartyal, D.; Agarwal, A.; Shukla, T.; James, D.; Kaul, T.; Negi, Y.K.; Arora, S.; Reddy, M.K. Abiotic Stress Tolerance in Plants: Myriad Roles of Ascorbate Peroxidase. Front. Plant Sci. 2017, 8, 581. [Google Scholar] [CrossRef]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of Crop Wild Relatives: Expanding the Gene Pool for Crop Improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef]
- D’Agostino, N.; Tripodi, P. NGS-Based Genotyping, High-Throughput Phenotyping and Genome-Wide Association Studies Laid the Foundations for next-Generation Breeding in Horticultural Crops. Diversity 2017, 9, 38. [Google Scholar] [CrossRef]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.-S.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X. A Draft Sequence of the Rice Genome (Oryza Sativa L. Ssp. Indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.A.; Ricke, D.; Lan, T.-H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H. A Draft Sequence of the Rice Genome (Oryza Sativa L. Ssp. Japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Project, I.R.G.S. The Map-Based Sequence of the Rice Genome. Nature 2005, 436, 793–800. [Google Scholar]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A. The B73 Maize Genome: Complexity, Diversity, and Dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A. The Sorghum Bicolor Genome and the Diversification of Grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, X.; Quan, Z.; Cheng, S.; Xu, X.; Pan, S.; Xie, M.; Zeng, P.; Yue, Z.; Wang, W. Genome Sequence of Foxtail Millet (Setaria Italica) Provides Insights into Grass Evolution and Biofuel Potential. Nat. Biotechnol. 2012, 30, 549–554. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J. Reference Genome Sequence of the Model Plant Setaria. Nat. Biotechnol. 2012, 30, 555–561. [Google Scholar] [CrossRef]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D. Analysis of the Bread Wheat Genome Using Whole-Genome Shotgun Sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef]
- Mayer, K.F.; Waugh, R.; Langridge, P.; Close, T.J.; Wise, R.P.; Graner, A.; Matsumoto, T.; Sato, K.; Schulman, A.; Muehlbauer, G.J. A Physical, Genetic and Functional Sequence Assembly of the Barley Genome. Int. Barley Genome Seq. Consort. 2012, 491, 711–716. [Google Scholar]
- Hatakeyama, M.; Aluri, S.; Balachadran, M.T.; Sivarajan, S.R.; Patrignani, A.; Grüter, S.; Poveda, L.; Shimizu-Inatsugi, R.; Baeten, J.; Francoijs, K.-J. Multiple Hybrid de Novo Genome Assembly of Finger Millet, an Orphan Allotetraploid Crop. DNA Res. 2018, 25, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Braslavsky, I.; Hebert, B.; Kartalov, E.; Quake, S.R. Sequence Information Can Be Obtained from Single DNA Molecules. Proc. Natl. Acad. Sci. USA 2003, 100, 3960–3964. [Google Scholar] [CrossRef] [PubMed]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Levene, M.J.; Korlach, J.; Turner, S.W.; Foquet, M.; Craighead, H.G.; Webb, W.W. Zero-Mode Waveguides for Single-Molecule Analysis at High Concentrations. Science 2003, 299, 682–686. [Google Scholar] [CrossRef] [PubMed]
- Greenleaf, W.J.; Block, S.M. Single-Molecule, Motion-Based DNA Sequencing Using RNA Polymerase. Science 2006, 313, 801. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F. Systems Biology Approach in Plant Abiotic Stresses. Plant Physiol. Biochem. 2017, 121, 58–73. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, H.; Li, H.; Tang, H.; Zhang, L.; Xu, H.; Jiao, F.; Wang, N.; Yang, L. Short Tandem Repeats in Plants: Genomic Distribution and Function Prediction. Electron. J. Biotechnol. 2021, 50, 37–44. [Google Scholar] [CrossRef]
- Feuillet, C.; Stein, N.; Rossini, L.; Praud, S.; Mayer, K.; Schulman, A.; Eversole, K.; Appels, R. Integrating Cereal Genomics to Support Innovation in the Triticeae. Funct. Integr. Genom. 2012, 12, 573–583. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Mao, L.; Chen, M.; Chu, Q.; Jia, L.; Sultana, M.H.; Wu, D.; Kong, X.; Qiu, J.; Ye, C.-Y.; Zhu, Q.-H. RiceRelativesGD: A Genomic Database of Rice Relatives for Rice Research. Database 2019, 2019, baz110. [Google Scholar] [CrossRef]
- McLaren, C.G.; Bruskiewich, R.M.; Portugal, A.M.; Cosico, A.B. The International Rice Information System. A Platform for Meta-Analysis of Rice Crop Data. Plant Physiol. 2005, 139, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, J.; Chen, L.; Huang, X.; Cheng, Z.; Han, B.; Zhang, Q.; Wu, C. Rice Functional Genomics Research: Past Decade and Future. Mol. Plant 2018, 11, 359–380. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, M.R.; Cannon, E.K.; Portwood, J.L.; Harper, L.C.; Gardiner, J.M.; Schaeffer, M.L.; Andorf, C.M. A Pan-Genomic Approach to Genome Databases Using Maize as a Model System. BMC Plant Biol. 2021, 21, 385. [Google Scholar] [CrossRef]
- Lawrence, C.J.; Dong, Q.; Polacco, M.L.; Seigfried, T.E.; Brendel, V. MaizeGDB, the Community Database for Maize Genetics and Genomics. Nucleic Acids Res. 2004, 32, D393–D397. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.T.; Portwood, J.L.; Gardiner, J.M.; Harper, L.C.; Lawrence-Dill, C.J.; Friedberg, I.; Andorf, C.M. MaizeDIG: Maize Database of Images and Genomes. Front. Plant Sci. 2019, 10, 1050. [Google Scholar] [CrossRef]
- Lawrence, C.J.; Harper, L.C.; Schaeffer, M.L.; Sen, T.Z.; Seigfried, T.E.; Campbell, D.A. MaizeGDB: The Maize Model Organism Database for Basic, Translational, and Applied Research. Int. J. Plant Genom. 2008, 2008, 496957. [Google Scholar] [CrossRef][Green Version]
- Shamimuzzaman, M.; Gardiner, J.M.; Walsh, A.T.; Triant, D.A.; Le Tourneau, J.J.; Tayal, A.; Unni, D.R.; Nguyen, H.N.; Portwood, J.L.; Cannon, E.K.S. MaizeMine: A Data Mining Warehouse for the Maize Genetics and Genomics Database. Front. Plant Sci. 2020, 11, 592730. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; Liu, A.; Zou, J.; Zhou, X.; Xiang, J.; Rerksiri, W.; Peng, Y.; Xiong, X.; Chen, X. Expression Profile in Rice Panicle: Insights into Heat Response Mechanism at Reproductive Stage. PLoS ONE 2012, 7, e49652. [Google Scholar] [CrossRef]
- Hittalmani, S.; Mahesh, H.B.; Shirke, M.D.; Biradar, H.; Uday, G.; Aruna, Y.R.; Lohithaswa, H.C.; Mohanrao, A. Genome and Transcriptome Sequence of Finger Millet (Eleusine Coracana (L.) Gaertn.) Provides Insights into Drought Tolerance and Nutraceutical Properties. BMC Genom. 2017, 18, 465. [Google Scholar] [CrossRef]
- Xiao, C.-L.; Chen, Y.; Xie, S.-Q.; Chen, K.-N.; Wang, Y.; Han, Y.; Luo, F.; Xie, Z. MECAT: Fast Mapping, Error Correction, and de Novo Assembly for Single-Molecule Sequencing Reads. Nat. Methods 2017, 14, 1072–1074. [Google Scholar] [CrossRef]
- Zou, C.; Li, L.; Miki, D.; Li, D.; Tang, Q.; Xiao, L.; Rajput, S.; Deng, P.; Peng, L.; Jia, W. The Genome of Broomcorn Millet. Nat. Commun. 2019, 10, 436. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Qiu, J.; Ye, C.; Jin, G.; Mao, L.; Zhang, H.; Yang, X.; Peng, Q.; Wang, Y.; Jia, L. Echinochloa Crus-Galli Genome Analysis Provides Insight into Its Adaptation and Invasiveness as a Weed. Nat. Commun. 2017, 8, 1031. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Shi, C.; Thudi, M.; Mariac, C.; Wallace, J.; Qi, P.; Zhang, H.; Zhao, Y.; Wang, X.; Rathore, A. Pearl Millet Genome Sequence Provides a Resource to Improve Agronomic Traits in Arid Environments. Nat. Biotechnol. 2017, 35, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Sutton, G.G.; White, O.; Adams, M.D.; Kerlavage, A.R. TIGR Assembler: A New Tool for Assembling Large Shotgun Sequencing Projects. Genome Sci. Technol. 1995, 1, 9–19. [Google Scholar] [CrossRef]
- Myers, E.W. The Fragment Assembly String Graph. Bioinformatics 2005, 21, ii79–ii85. [Google Scholar] [CrossRef]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z. Genome Sequencing in Microfabricated High-Density Picolitre Reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef]
- Warren, R.L.; Sutton, G.G.; Jones, S.J.M.; Holt, R.A. Assembling Millions of Short DNA Sequences Using SSAKE. Bioinformatics 2007, 23, 500–501. [Google Scholar] [CrossRef]
- Jeck, W.R.; Reinhardt, J.A.; Baltrus, D.A.; Hickenbotham, M.T.; Magrini, V.; Mardis, E.R.; Dangl, J.L.; Jones, C.D. Extending Assembly of Short DNA Sequences to Handle Error. Bioinformatics 2007, 23, 2942–2944. [Google Scholar] [CrossRef]
- Dohm, J.C.; Lottaz, C.; Borodina, T.; Himmelbauer, H. SHARCGS, a Fast and Highly Accurate Short-Read Assembly Algorithm for de Novo Genomic Sequencing. Genome Res. 2007, 17, 1697–1706. [Google Scholar] [CrossRef]
- Butler, J.; MacCallum, I.; Kleber, M.; Shlyakhter, I.A.; Belmonte, M.K.; Lander, E.S.; Nusbaum, C.; Jaffe, D.B. ALLPATHS: De Novo Assembly of Whole-Genome Shotgun Microreads. Genome Res. 2008, 18, 810–820. [Google Scholar] [CrossRef]
- Hernandez, D.; François, P.; Farinelli, L.; Østerås, M.; Schrenzel, J. De Novo Bacterial Genome Sequencing: Millions of Very Short Reads Assembled on a Desktop Computer. Genome Res. 2008, 18, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.R.; Delcher, A.L.; Koren, S.; Venter, E.; Walenz, B.P.; Brownley, A.; Johnson, J.; Li, K.; Mobarry, C.; Sutton, G. Aggressive Assembly of Pyrosequencing Reads with Mates. Bioinformatics 2008, 24, 2818–2824. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A Parallel Assembler for Short Read Sequence Data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y. Erratum: SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler. Gigascience 2015, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Duan, C.; Argout, X.; Gébelin, V.; Summo, M.; Dufayard, J.-F.; Leclercq, J.; Piyatrakul, P.; Pirrello, J.; Rio, M.; Champion, A. Identification of the Hevea BrasiliensisAP2/ERF Superfamily by RNA Sequencing. BMC Genom. 2013, 14, 30. [Google Scholar] [CrossRef]
- Yang, Y.; Smith, S.A. Optimizing de Novo Assembly of Short-Read RNA-Seq Data for Phylogenomics. BMC Genom. 2013, 14, 328. [Google Scholar] [CrossRef]
- Wang, W.; Ding, G.-D.; White, P.J.; Wang, X.-H.; Jin, K.-M.; Xu, F.-S.; Shi, L. Mapping and Cloning of Quantitative Trait Loci for Phosphorus Efficiency in Crops: Opportunities and Challenges. Plant Soil 2019, 439, 91–112. [Google Scholar] [CrossRef]
- Vielle-Calzada, J.-P.; Martínez de la Vega, O.; Hernández-Guzmán, G.; Ibarra-Laclette, E.; Alvarez-Mejía, C.; Vega-Arreguín, J.C.; Jiménez-Moraila, B.; Fernández-Cortés, A.; Corona-Armenta, G.; Herrera-Estrella, L. The Palomero Genome Suggests Metal Effects on Domestication. Science 2009, 326, 1078. [Google Scholar] [CrossRef] [PubMed]
- Rashid, B.; Husnain, T.; Riazuddin, S. Genomic Approaches and Abiotic Stress Tolerance in Plants. In Emerging Technologies and Management of Crop Stress Tolerance; Elsevier: Amsterdam, The Netherlands, 2014; pp. 1–37. [Google Scholar]
- Priyadarshan, P.M. Breeding for Abiotic Stress Adaptation. In Plant Breeding: Classical to Modern; Springer: Berlin/Heidelberg, Germany, 2019; pp. 413–455. [Google Scholar]
- Kumar, S.; Bhati, J.; Saha, A.; Lal, S.B.; Pandey, P.K.; Mishra, D.C.; Farooqi, M.S.; Kumar, A.; Chaturvedi, K.K.; Rai, A. CerealESTDb: A Comprehensive Resource for Abiotic Stress-Responsive Annotated ESTs with Predicted Genes, Gene Ontology, and Metabolic Pathways in Major Cereal Crops. Front. Genet. 2022, 13, 842868. [Google Scholar] [CrossRef] [PubMed]
- Riaz, A.; Kanwal, F.; Börner, A.; Pillen, K.; Dai, F.; Alqudah, A.M. Advances in Genomics-Based Breeding of Barley: Molecular Tools and Genomic Databases. Agronomy 2021, 11, 894. [Google Scholar] [CrossRef]
- Udoh, L.I.; Obaseojei, W.P.; Uzoebo, C. Single Nucleotide Polymorphisms: A Modern Tool to Screen Plants for Desirable Traits. In Plant Breeding-Current and Future Views; IntechOpen: London, UK, 2021; ISBN 1839683104. [Google Scholar]
- Bhattarai, G.; Shi, A.; Kandel, D.R.; Solís-Gracia, N.; da Silva, J.A.; Avila, C.A. Genome-Wide Simple Sequence Repeats (SSR) Markers Discovered from Whole-Genome Sequence Comparisons of Multiple Spinach Accessions. Sci. Rep. 2021, 11, 9999. [Google Scholar] [CrossRef] [PubMed]
- Yonemaru, J.; Yamamoto, T.; Fukuoka, S.; Uga, Y.; Hori, K.; Yano, M. Q-TARO: QTL Annotation Rice Online Database. Rice 2010, 3, 194–203. [Google Scholar] [CrossRef]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y. Development and Mapping of 2240 New SSR Markers for Rice (Oryza Sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef]
- Ganie, S.A.; Borgohain, M.J.; Kritika, K.; Talukdar, A.; Pani, D.R.; Mondal, T.K. Assessment of Genetic Diversity of Saltol QTL among the Rice (Oryza Sativa L.) Genotypes. Physiol. Mol. Biol. Plants 2016, 22, 107–114. [Google Scholar] [CrossRef]
- Ghimire, K.H.; Quiatchon, L.A.; Vikram, P.; Swamy, B.P.M.; Dixit, S.; Ahmed, H.; Hernandez, J.E.; Borromeo, T.H.; Kumar, A. Identification and Mapping of a QTL (QDTY1. 1) with a Consistent Effect on Grain Yield under Drought. Field Crops Res. 2012, 131, 88–96. [Google Scholar] [CrossRef]
- Tang, W.; Wu, T.; Ye, J.; Sun, J.; Jiang, Y.; Yu, J.; Tang, J.; Chen, G.; Wang, C.; Wan, J. SNP-Based Analysis of Genetic Diversity Reveals Important Alleles Associated with Seed Size in Rice. BMC Plant Biol. 2016, 16, 93. [Google Scholar]
- Rauf, S.; Al-Khayri, J.M.; Zaharieva, M.; Monneveux, P.; Khalil, F. Breeding Strategies to Enhance Drought Tolerance in Crops. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Springer: Berlin/Heidelberg, Germany, 2016; pp. 397–445. [Google Scholar]
- Wilkinson, P.A.; Winfield, M.O.; Barker, G.L.A.; Allen, A.M.; Burridge, A.; Coghill, J.A.; Edwards, K.J. CerealsDB 2.0: An Integrated Resource for Plant Breeders and Scientists. BMC Bioinform. 2012, 13, 219. [Google Scholar] [CrossRef]
- Kayıhan, C.; Eyidoğan, F. Omics in Oxidative Stress Tolerance in Crops. React. Oxyg. Nitrogen Sulfur Species Plants Prod. Metab. Signal. Def. Mech. 2019, 195–224. [Google Scholar] [CrossRef]
- McNally, K.L.; Childs, K.L.; Bohnert, R.; Davidson, R.M.; Zhao, K.; Ulat, V.J.; Zeller, G.; Clark, R.M.; Hoen, D.R.; Bureau, T.E. Genomewide SNP Variation Reveals Relationships among Landraces and Modern Varieties of Rice. Proc. Natl. Acad. Sci. USA 2009, 106, 12273–12278. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.B.; Jing, R.L.; Yuan, H.Y.; Wei, B.; Chang, X.P. Single Nucleotide Polymorphism of TaDREB1 Gene in Wheat Germplasm. Sci. Agric. Sin. 2005, 38, 2387–2394. [Google Scholar]
- Garg, D.; Sareen, S.; Dalal, S.; Tiwari, R.; Singh, R. Heat Shock Protein Based SNP Marker for Terminal Heat Stress in Wheat (Triticum Aestivum L.). Aust. J. Crop Sci. 2012, 6, 1516–1521. [Google Scholar]
- Setter, T.L.; Yan, J.; Warburton, M.; Ribaut, J.-M.; Xu, Y.; Sawkins, M.; Buckler, E.S.; Zhang, Z.; Gore, M.A. Genetic Association Mapping Identifies Single Nucleotide Polymorphisms in Genes That Affect Abscisic Acid Levels in Maize Floral Tissues during Drought. J. Exp. Bot. 2011, 62, 701–716. [Google Scholar] [CrossRef] [PubMed]
- Assenov, B.; Andjelkovic, V.; Ignjatovic-Micic, D.; Vancetovic, J.; Nikolic, A.; Christov, N.K.; Tsonev, S.; Abu-Mhadi, N.; Vassilev, D.; Muhovski, Y. Identification of SNP Mutations in MYBE-1 Gene Involved in Drought Stress Tolerance in Maize. Bulg. J. Agric. Sci 2013, 19, 181–185. [Google Scholar]
- Close, T.J.; Wanamaker, S.I.; Caldo, R.A.; Turner, S.M.; Ashlock, D.A.; Dickerson, J.A.; Wing, R.A.; Muehlbauer, G.J.; Kleinhofs, A.; Wise, R.P. A New Resource for Cereal Genomics: 22K Barley GeneChip Comes of Age. Plant Physiol. 2004, 134, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Stein, N.; Prasad, M.; Scholz, U.; Thiel, T.; Zhang, H.; Wolf, M.; Kota, R.; Varshney, R.K.; Perovic, D.; Grosse, I. A 1000-Loci Transcript Map of the Barley Genome: New Anchoring Points for Integrative Grass Genomics. Theor. Appl. Genet. 2007, 114, 823–839. [Google Scholar] [CrossRef]
- Sato, K.; Nankaku, N.; Takeda, K. A High-Density Transcript Linkage Map of Barley Derived from a Single Population. Heredity 2009, 103, 110–117. [Google Scholar] [CrossRef]
- Caicedo, A.L.; Purugganan, M.D. Comparative Plant Genomics. Frontiers and Prospects. Plant Physiol. 2005, 138, 545–547. [Google Scholar] [CrossRef][Green Version]
- Gaut, B.S.; Wright, S.I.; Rizzon, C.; Dvorak, J.; Anderson, L.K. Recombination: An Underappreciated Factor in the Evolution of Plant Genomes. Nat. Rev. Genet. 2007, 8, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Feuillet, C.; Keller, B. Comparative Genomics in the Grass Family: Molecular Characterization of Grass Genome Structure and Evolution. Ann. Bot. 2002, 89, 3–10. [Google Scholar] [CrossRef]
- Iqbal, M.; Shahzad, R.; Shahzad, R.; Bilal, K.; Qaisar, R.; Nisar, A.; Kanwal, S.; Bhatti, M. DNA Fingerprinting of Crops and Its Applications in the Field of Plant Breeding. J. Agric. Res 2021, 59, 13–28. [Google Scholar]
- Mace, E.; Innes, D.; Hunt, C.; Wang, X.; Tao, Y.; Baxter, J.; Hassall, M.; Hathorn, A.; Jordan, D. The Sorghum QTL Atlas: A Powerful Tool for Trait Dissection, Comparative Genomics and Crop Improvement. Theor. Appl. Genet. 2019, 132, 751–766. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Batra, R.; Sharma, S.; Saripalli, G.; Gautam, T.; Singh, R.; Pal, S.; Malik, P.; Kumar, M.; Jan, I. WheatQTLdb: A QTL Database for Wheat. Mol. Genet. Genom. 2021, 296, 1051–1056. [Google Scholar] [CrossRef]
- Steele, K.A.; Ogden, R.; McEwing, R.; Briggs, H.; Gorham, J. InDel Markers Distinguish Basmatis from Other Fragrant Rice Varieties. Field Crops Res. 2008, 105, 81–87. [Google Scholar] [CrossRef]
- Steele, K.A.; Virk, D.S.; Kumar, R.; Prasad, S.C.; Witcombe, J.R. Field Evaluation of Upland Rice Lines Selected for QTLs Controlling Root Traits. Field Crops Res. 2007, 101, 180–186. [Google Scholar] [CrossRef]
- Bernier, J.; Kumar, A.; Venuprasad, R.; Spaner, D.; Verulkar, S.; Mandal, N.P.; Sinha, P.K.; Peeraju, P.; Dongre, P.R.; Mahto, R.N. Characterization of the Effect of a QTL for Drought Resistance in Rice, Qtl12. 1, over a Range of Environments in the Philippines and Eastern India. Euphytica 2009, 166, 207–217. [Google Scholar] [CrossRef]
- Welcker, C.; Boussuge, B.; Bencivenni, C.; Ribaut, J.M.; Tardieu, F. Are Source and Sink Strengths Genetically Linked in Maize Plants Subjected to Water Deficit? A QTL Study of the Responses of Leaf Growth and of Anthesis-Silking Interval to Water Deficit. J. Exp. Bot. 2007, 58, 339–349. [Google Scholar] [CrossRef]
- Sadok, W.; Naudin, P.; Boussuge, B.; Muller, B.; Welcker, C.; Tardieu, F. Leaf Growth Rate per Unit Thermal Time Follows QTL-dependent Daily Patterns in Hundreds of Maize Lines under Naturally Fluctuating Conditions. Plant. Cell Environ. 2007, 30, 135–146. [Google Scholar] [CrossRef]
- Reymond, M.; Muller, B.; Leonardi, A.; Charcosset, A.; Tardieu, F. Combining Quantitative Trait Loci Analysis and an Ecophysiological Model to Analyze the Genetic Variability of the Responses of Maize Leaf Growth to Temperature and Water Deficit. Plant Physiol. 2003, 131, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Osthoff, A.; Donà Dalle Rose, P.; Baldauf, J.A.; Piepho, H.P.; Hochholdinger, F. Transcriptomic Reprogramming of Barley Seminal Roots by Combined Water Deficit and Salt Stress. BMC Genom. 2019, 20, 325. [Google Scholar] [CrossRef] [PubMed]
- Serraj, R.; Hash, T.C.; Buhariwalla, H.K.; Bidinger, F.R.; Folkertsma, R.T.; Chandra, S.; Gaur, P.M.; Kashiwagi, J.; Nigam, S.N.; Rupakula, A. Marker-Assisted Breeding for Crop Drought Tolerance at ICRISAT: Achievements and Prospects. In Proceedings of the International Congress “In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution”, Avenue Media, Bologna, Italy, 27–31 May 2005; pp. 217–238. [Google Scholar]
- Yang, D.-L.; Jing, R.-L.; Chang, X.-P.; Li, W. Identification of Quantitative Trait Loci and Environmental Interactions for Accumulation and Remobilization of Water-Soluble Carbohydrates in Wheat (Triticum Aestivum L.) Stems. Genetics 2007, 176, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Båga, M.; Chodaparambil, S.V.; Limin, A.E.; Pecar, M.; Fowler, D.B.; Chibbar, R.N. Identification of Quantitative Trait Loci and Associated Candidate Genes for Low-Temperature Tolerance in Cold-Hardy Winter Wheat. Funct. Integr. Genom. 2007, 7, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Bálint, A.F.; Szira, F.; Röder, M.S.; Galiba, G.; Börner, A. Mapping of Loci Affecting Copper Tolerance in Wheat—The Possible Impact of the Vernalization Gene Vrn-A1. Environ. Exp. Bot. 2009, 65, 369–375. [Google Scholar] [CrossRef]
- Ma, Z.; Zhao, D.; Zhang, C.; Zhang, Z.; Xue, S.; Lin, F.; Kong, Z.; Tian, D.; Luo, Q. Molecular Genetic Analysis of Five Spike-Related Traits in Wheat Using RIL and Immortalized F2 Populations. Mol. Genet. Genom. 2007, 277, 31–42. [Google Scholar] [CrossRef]
- Navakode, S.; Weidner, A.; Varshney, R.; Lohwasser, U.; Scholz, U.; Börner, A. A QTL Analysis of Aluminium Tolerance in Barley, Using Gene-Based Markers. Cereal Res. Commun. 2009, 37, 531–540. [Google Scholar] [CrossRef]
- Xue, D.; Huang, Y.; Zhang, X.; Wei, K.; Westcott, S.; Li, C.; Chen, M.; Zhang, G.; Lance, R. Identification of QTLs Associated with Salinity Tolerance at Late Growth Stage in Barley. Euphytica 2009, 169, 187–196. [Google Scholar] [CrossRef]
- Francia, E.; Barabaschi, D.; Tondelli, A.; Laidò, G.; Rizza, F.; Stanca, A.M.; Busconi, M.; Fogher, C.; Stockinger, E.J.; Pecchioni, N. Fine Mapping of a HvCBF Gene Cluster at the Frost Resistance Locus Fr-H2 in Barley. Theor. Appl. Genet. 2007, 115, 1083–1091. [Google Scholar] [CrossRef]
- Elango, D.; Sandoya, G.; Chopra, S. Techniques and Tools of Modern Plant Breeding. In Plant Biotechnology; Springer: Berlin/Heidelberg, Germany, 2021; pp. 17–26. [Google Scholar]
- Chang, J.; Tian, J.; Yang, Y.; Zhong, R.; Li, J.; Zhai, K.; Ke, J.; Lou, J.; Chen, W.; Zhu, B. A Rare Missense Variant in TCF7L2 Associates with Colorectal Cancer Risk by Interacting with a GWAS-Identified Regulatory Variant in the MYC EnhancerAn Exome-Wide Association Study of Colorectal Cancer. Cancer Res. 2018, 78, 5164–5172. [Google Scholar] [CrossRef]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A Tutorial on Conducting Genome-wide Association Studies: Quality Control and Statistical Analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; Li, M. Genome-Wide Association Studies of 14 Agronomic Traits in Rice Landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.P.; Ramu, P.; Deshpande, S.P.; Hash, C.T.; Shah, T.; Upadhyaya, H.D.; Riera-Lizarazu, O.; Brown, P.J.; Acharya, C.B.; Mitchell, S.E. Population Genomic and Genome-Wide Association Studies of Agroclimatic Traits in Sorghum. Proc. Natl. Acad. Sci. USA 2013, 110, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Huang, X.; Zhi, H.; Zhao, Y.; Zhao, Q.; Li, W.; Chai, Y.; Yang, L.; Liu, K.; Lu, H. A Haplotype Map of Genomic Variations and Genome-Wide Association Studies of Agronomic Traits in Foxtail Millet (Setaria Italica). Nat. Genet. 2013, 45, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Kamali, S.; Singh, A. Genomics and Transcriptomics Approaches to Understand Abiotic Stress Response in Rice. In Omics Approach to Manage Abiotic Stress in Cereals; Springer: Berlin/Heidelberg, Germany, 2022; pp. 405–433. [Google Scholar]
- Imran, Q.M.; Falak, N.; Hussain, A.; Mun, B.-G.; Yun, B.-W. Abiotic Stress in Plants; Stress Perception to Molecular Response and Role of Biotechnological Tools in Stress Resistance. Agronomy 2021, 11, 1579. [Google Scholar] [CrossRef]
- Munaweera, T.I.K.; Jayawardana, N.U.; Rajaratnam, R.; Dissanayake, N. Modern Plant Biotechnology as a Strategy in Addressing Climate Change and Attaining Food Security. Agric. Food Secur. 2022, 11, 26. [Google Scholar] [CrossRef]
- Rane, J.; Singh, A.K.; Tiwari, M.; Prasad, P.V.V.; Jagadish, S.V.K. Effective Use of Water in Crop Plants in Dryland Agriculture: Implications of Reactive Oxygen Species and Antioxidative System. Front. Plant Sci. 2021, 12, 778270. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Palakolanu, S.R.; Chopra, P.; Rajurkar, A.B.; Gupta, R.; Iqbal, N.; Maheshwari, C. Improving Drought Tolerance in Rice: Ensuring Food Security through Multi-dimensional Approaches. Physiol. Plant. 2021, 172, 645–668. [Google Scholar] [CrossRef]
- Hu, W.; Hu, G.; Han, B. Genome-Wide Survey and Expression Profiling of Heat Shock Proteins and Heat Shock Factors Revealed Overlapped and Stress Specific Response under Abiotic Stresses in Rice. Plant Sci. 2009, 176, 583–590. [Google Scholar] [CrossRef]
- Liu, A.-L.; Zou, J.; Zhang, X.-W.; Zhou, X.-Y.; Wang, W.-F.; Xiong, X.-Y.; Chen, L.-Y.; Chen, X.-B. Expression Profiles of Class A Rice Heat Shock Transcription Factor Genes under Abiotic Stresses. J. Plant Biol. 2010, 53, 142–149. [Google Scholar] [CrossRef]
- Jung, H.-S.; Crisp, P.A.; Estavillo, G.M.; Cole, B.; Hong, F.; Mockler, T.C.; Pogson, B.J.; Chory, J. Subset of Heat-Shock Transcription Factors Required for the Early Response of Arabidopsis to Excess Light. Proc. Natl. Acad. Sci. USA 2013, 110, 14474–14479. [Google Scholar] [CrossRef] [PubMed]
- González-Schain, N.; Dreni, L.; Lawas, L.M.F.; Galbiati, M.; Colombo, L.; Heuer, S.; Jagadish, K.S.V.; Kater, M.M. Genome-Wide Transcriptome Analysis during Anthesis Reveals New Insights into the Molecular Basis of Heat Stress Responses in Tolerant and Sensitive Rice Varieties. Plant Cell Physiol. 2016, 57, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Wu, H.; Peng, H.; Yao, Y.; Ni, Z.; Li, Z.; Zhou, C.; Sun, Q. Heat Stress-Responsive Transcriptome Analysis in Heat Susceptible and Tolerant Wheat (Triticum Aestivum L.) by Using Wheat Genome Array. BMC Genom. 2008, 9, 432. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Khurana, N.; Tyagi, A.K.; Khurana, J.P.; Khurana, P. Identification and Characterization of High Temperature Stress Responsive Genes in Bread Wheat (Triticum Aestivum L.) and Their Regulation at Various Stages of Development. Plant Mol. Biol. 2011, 75, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Khurana, P. Use of Doubled Haploid Technology for Development of Stable Drought Tolerant Bread Wheat (Triticum Aestivum L.) Transgenics. Plant Biotechnol. J. 2011, 9, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Comastri, A.; Janni, M.; Simmonds, J.; Uauy, C.; Pignone, D.; Nguyen, H.T.; Marmiroli, N. Heat in Wheat: Exploit Reverse Genetic Techniques to Discover New Alleles within the Triticum Durum SHsp26 Family. Front. Plant Sci. 2018, 9, 1337. [Google Scholar] [CrossRef]
- Lin, Y.-X.; Jiang, H.-Y.; Chu, Z.-X.; Tang, X.-L.; Zhu, S.-W.; Cheng, B.-J. Genome-Wide Identification, Classification and Analysis of Heat Shock Transcription Factor Family in Maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef]
- Frey, F.P.; Urbany, C.; Hüttel, B.; Reinhardt, R.; Stich, B. Genome-Wide Expression Profiling and Phenotypic Evaluation of European Maize Inbreds at Seedling Stage in Response to Heat Stress. BMC Genom. 2015, 16, 123. [Google Scholar] [CrossRef]
- Johnson, S.M.; Lim, F.-L.; Finkler, A.; Fromm, H.; Slabas, A.R.; Knight, M.R. Transcriptomic Analysis of Sorghum Bicolor Responding to Combined Heat and Drought Stress. BMC Genom. 2014, 15, 456. [Google Scholar] [CrossRef]
- Mangelsen, E.; Kilian, J.; Harter, K.; Jansson, C.; Wanke, D.; Sundberg, E. Transcriptome Analysis of High-Temperature Stress in Developing Barley Caryopses: Early Stress Responses and Effects on Storage Compound Biosynthesis. Mol. Plant 2011, 4, 97–115. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; García-Pereira, M.J.; Gracia, M.P.; Igartua, E.; Casas, A.M.; Contreras-Moreira, B. Large Differences in Gene Expression Responses to Drought and Heat Stress between Elite Barley Cultivar Scarlett and a Spanish Landrace. Front. Plant Sci. 2017, 8, 647. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene Networks Involved in Drought Stress Response and Tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis Thaliana CBF1 Encodes an AP2 Domain-Containing Transcriptional Activator That Binds to the C-Repeat/DRE, a Cis-Acting DNA Regulatory Element That Stimulates Transcription in Response to Low Temperature and Water Deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-Binding Specificity of the ERF/AP2 Domain of Arabidopsis DREBs, Transcription Factors Involved in Dehydration-and Cold-Inducible Gene Expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB Genes in Rice, Oryza Sativa L., Encode Transcription Activators That Function in Drought-, High-salt-and Cold-responsive Gene Expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Chen, J.-Q.; Meng, X.-P.; Zhang, Y.; Xia, M.; Wang, X.-P. Over-Expression of OsDREB Genes Lead to Enhanced Drought Tolerance in Rice. Biotechnol. Lett. 2008, 30, 2191–2198. [Google Scholar] [CrossRef]
- Lenka, S.K.; Katiyar, A.; Chinnusamy, V.; Bansal, K.C. Comparative Analysis of Drought-responsive Transcriptome in Indica Rice Genotypes with Contrasting Drought Tolerance. Plant Biotechnol. J. 2011, 9, 315–327. [Google Scholar] [CrossRef]
- Yoo, Y.-H.; Nalini Chandran, A.K.; Park, J.-C.; Gho, Y.-S.; Lee, S.-W.; An, G.; Jung, K.-H. OsPhyB-Mediating Novel Regulatory Pathway for Drought Tolerance in Rice Root Identified by a Global RNA-Seq Transcriptome Analysis of Rice Genes in Response to Water Deficiencies. Front. Plant Sci. 2017, 8, 580. [Google Scholar] [CrossRef]
- Ma, J.; Li, R.; Wang, H.; Li, D.; Wang, X.; Zhang, Y.; Zhen, W.; Duan, H.; Yan, G.; Li, Y. Transcriptomics Analyses Reveal Wheat Responses to Drought Stress during Reproductive Stages under Field Conditions. Front. Plant Sci. 2017, 8, 592. [Google Scholar] [CrossRef]
- Li, P.; Cao, W.; Fang, H.; Xu, S.; Yin, S.; Zhang, Y.; Lin, D.; Wang, J.; Chen, Y.; Xu, C. Transcriptomic Profiling of the Maize (Zea Mays L.) Leaf Response to Abiotic Stresses at the Seedling Stage. Front. Plant Sci. 2017, 8, 290. [Google Scholar] [CrossRef]
- Zenda, T.; Liu, S.; Wang, X.; Liu, G.; Jin, H.; Dong, A.; Yang, Y.; Duan, H. Key Maize Drought-Responsive Genes and Pathways Revealed by Comparative Transcriptome and Physiological Analyses of Contrasting Inbred Lines. Int. J. Mol. Sci. 2019, 20, 1268. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Bai, L.; Wei, Z.; Yuan, H.; Wang, Y.; Xu, Q.; Tang, Y.; Nyima, T. Transcriptome Analysis Revealed the Drought-Responsive Genes in Tibetan Hulless Barley. BMC Genomi. 2016, 17, 386. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Cheng, J.; Wen, X.; Wang, J.; Shi, G.; Yao, J.; Hou, L.; Sun, Q.; Xiang, P.; Yuan, X. Transcriptomic Studies Reveal a Key Metabolic Pathway Contributing to a Well-Maintained Photosynthetic System under Drought Stress in Foxtail Millet (Setaria Italica L.). PeerJ 2018, 6, e4752. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Iqbal, J.; Naseer, S.; Yaseen, T.; Shaukat, M.; Bibi, H.; Ahmad, Y.; Daud, H.; Abbasi, N.L.; Mahmood, T. Molecular Mechanisms of Plant Tolerance to Heat Stress: Current Landscape and Future Perspectives. Plant Cell Rep. 2021, 40, 2247–2271. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.A.D.; Mittler, R.O.N. Could Heat Shock Transcription Factors Function as Hydrogen Peroxide Sensors in Plants? Ann. Bot. 2006, 98, 279–288. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b Act as Repressors of the Expression of Heat-Inducible Hsfs but Positively Regulate the Acquired Thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; Von Koskull-Döring, P. A Cascade of Transcription Factor DREB2A and Heat Stress Transcription Factor HsfA3 Regulates the Heat Stress Response of Arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar] [CrossRef]
- Chauhan, H.; Khurana, N.; Agarwal, P.; Khurana, P. Heat Shock Factors in Rice (Oryza Sativa L.): Genome-Wide Expression Analysis during Reproductive Development and Abiotic Stress. Mol. Genet. Genom. 2011, 286, 171–187. [Google Scholar] [CrossRef]
- Xue, G.-P.; Drenth, J.; McIntyre, C.L. TaHsfA6f Is a Transcriptional Activator That Regulates a Suite of Heat Stress Protection Genes in Wheat (Triticum Aestivum L.) Including Previously Unknown Hsf Targets. J. Exp. Bot. 2015, 66, 1025–1039. [Google Scholar] [CrossRef]
- Fang, C.; Dou, L.; Liu, Y.; Yu, J.; Tu, J. Heat Stress-Responsive Transcriptome Analysis in Heat Susceptible and Tolerant Rice by High-Throughput Sequencing. Ecol. Genet. Genom. 2018, 6, 33–40. [Google Scholar] [CrossRef]
- Kong, W.; Zhong, H.; Gong, Z.; Fang, X.; Sun, T.; Deng, X.; Li, Y. Meta-Analysis of Salt Stress Transcriptome Responses in Different Rice Genotypes at the Seedling Stage. Plants 2019, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Amirbakhtiar, N.; Ismaili, A.; Ghaffari, M.R.; Firouzabadi, F.N.; Shobbar, Z.S. Transcriptome Response of Roots to Salt Stress in a Salinity-Tolerant Bread Wheat Cultivar. PLoS ONE 2019, 14, e0213305. [Google Scholar] [CrossRef] [PubMed]
- Chandran, A.K.N.; Kim, J.-W.; Yoo, Y.-H.; Park, H.L.; Kim, Y.-J.; Cho, M.-H.; Jung, K.-H. Transcriptome Analysis of Rice-Seedling Roots under Soil–Salt Stress Using RNA-Seq Method. Plant Biotechnol. Rep. 2019, 13, 567–578. [Google Scholar] [CrossRef]
- Li, Q.; Ma, C.; Tai, H.; Qiu, H.; Yang, A. Comparative Transcriptome Analysis of Two Rice Genotypes Differing in Their Tolerance to Saline-Alkaline Stress. PLoS ONE 2020, 15, e0243112. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Zhou, H.; Sui, X.; Su, C.; Yu, Y.; Yang, H.; Dong, C.-H. Generation of New Salt-Tolerant Wheat Lines and Transcriptomic Exploration of the Responsive Genes to Ethylene and Salt Stress. Plant Growth Regul. 2021, 94, 33–48. [Google Scholar] [CrossRef]
- Mahajan, M.M.; Goyal, E.; Singh, A.K.; Gaikwad, K.; Kanika, K. Shedding Light on Response of Triticum Aestivum Cv. Kharchia Local Roots to Long-Term Salinity Stress through Transcriptome Profiling. Plant Growth Regul. 2020, 90, 369–381. [Google Scholar] [CrossRef]
- Bhanbhro, N.; Xiao, B.; Han, L.; Lu, H.; Wang, H.; Yang, C. Adaptive Strategy of Allohexaploid Wheat to Long-Term Salinity Stress. BMC Plant Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, P.; Qing, C.; Yang, C.; Shen, Y.; Ma, L. Comparative Transcriptome Analyses of Maize Seedling Root Responses to Salt Stress. PeerJ 2021, 9, e10765. [Google Scholar] [CrossRef]
- Chen, T.; Shabala, S.; Niu, Y.; Chen, Z.-H.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.; Zhou, M. Molecular Mechanisms of Salinity Tolerance in Rice. Crop J. 2021, 9, 506–520. [Google Scholar] [CrossRef]
- Singh, R.K.; Sood, P.; Prasad, A.; Prasad, M. Advances in Omics Technology for Improving Crop Yield and Stress Resilience. Plant Breed. 2021, 140, 719–731. [Google Scholar] [CrossRef]
- Plunk, E.C.; Chambers, W.S.; Richards, S.M. System Biology. In Metabolomics Perspectives; Elsevier: Amsterdam, The Netherlands, 2022; pp. 3–25. [Google Scholar]
- Agregán, R.; Echegaray, N.; López-Pedrouso, M.; Aadil, R.M.; Hano, C.; Franco, D.; Lorenzo, J.M. Proteomic Advances in Cereal and Vegetable Crops. Molecules 2021, 26, 4924. [Google Scholar] [CrossRef] [PubMed]
- Jorrin Novo, J.V. Proteomics and Plant Biology: Contributions to Date and a Look towards the next Decade. Expert Rev. Proteom. 2021, 18, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Holman, J.D.; Dasari, S.; Tabb, D.L. Informatics of Protein and Posttranslational Modification Detection via Shotgun Proteomics. In Proteomics for Biomarker Discovery; Springer: Berlin/Heidelberg, Germany, 2013; pp. 167–179. [Google Scholar]
- Que, S.; Li, K.; Chen, M.; Wang, Y.; Yang, Q.; Zhang, W.; Zhang, B.; Xiong, B.; He, H. PhosphoRice: A Meta-Predictor of Rice-Specific Phosphorylation Sites. Plant Methods 2012, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Helmy, M.; Tomita, M.; Ishihama, Y. OryzaPG-DB: Rice Proteome Database Based on Shotgun Proteogenomics. BMC Plant Biol. 2011, 11, 63. [Google Scholar] [CrossRef]
- Gu, H.; Zhu, P.; Jiao, Y.; Meng, Y.; Chen, M. PRIN: A Predicted Rice Interactome Network. BMC Bioinform. 2011, 12, 161. [Google Scholar] [CrossRef]
- Sun, Q.; Zybailov, B.; Majeran, W.; Friso, G.; Olinares, P.D.B.; van Wijk, K.J. PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 2009, 37, D969–D974. [Google Scholar] [CrossRef]
- Duncan, O.; Trösch, J.; Fenske, R.; Taylor, N.L.; Millar, A.H. Resource: Mapping the Triticum Aestivum Proteome. Plant J. 2017, 89, 601–616. [Google Scholar] [CrossRef]
- Mirzaei, M.; Pascovici, D.; Atwell, B.J.; Haynes, P.A. Differential Regulation of Aquaporins, Small GTP Ases and V-ATP Ases Proteins in Rice Leaves Subjected to Drought Stress and Recovery. Proteomics 2012, 12, 864–877. [Google Scholar] [CrossRef]
- Ji, K.; Wang, Y.; Sun, W.; Lou, Q.; Mei, H.; Shen, S.; Chen, H. Drought-Responsive Mechanisms in Rice Genotypes with Contrasting Drought Tolerance during Reproductive Stage. J. Plant Physiol. 2012, 169, 336–344. [Google Scholar] [CrossRef]
- Maksup, S.; Roytrakul, S.; Supaibulwatana, K. Physiological and Comparative Proteomic Analyses of Thai Jasmine Rice and Two Check Cultivars in Response to Drought Stress. J. Plant Interact. 2014, 9, 43–55. [Google Scholar] [CrossRef]
- Ford, K.L.; Cassin, A.; Bacic, A. Quantitative Proteomic Analysis of Wheat Cultivars with Differing Drought Stress Tolerance. Front. Plant Sci. 2011, 2, 44. [Google Scholar] [CrossRef]
- Alvarez, S.; Roy Choudhury, S.; Pandey, S. Comparative Quantitative Proteomics Analysis of the ABA Response of Roots of Drought-Sensitive and Drought-Tolerant Wheat Varieties Identifies Proteomic Signatures of Drought Adaptability. J. Proteome Res. 2014, 13, 1688–1701. [Google Scholar] [CrossRef]
- Hao, P.; Zhu, J.; Gu, A.; Lv, D.; Ge, P.; Chen, G.; Li, X.; Yan, Y. An Integrative Proteome Analysis of Different Seedling Organs in Tolerant and Sensitive Wheat Cultivars under Drought Stress and Recovery. Proteomics 2015, 15, 1544–1563. [Google Scholar] [CrossRef]
- Deng, X.; Liu, Y.; Xu, X.; Liu, D.; Zhu, G.; Yan, X.; Wang, Z.; Yan, Y. Comparative Proteome Analysis of Wheat Flag Leaves and Developing Grains under Water Deficit. Front. Plant Sci. 2018, 9, 425. [Google Scholar] [CrossRef]
- Benešová, M.; Hola, D.; Fischer, L.; Jedelský, P.L.; Hnilička, F.; Wilhelmová, N.; Rothova, O.; Kočová, M.; Prochazkova, D.; Honnerova, J. The Physiology and Proteomics of Drought Tolerance in Maize: Early Stomatal Closure as a Cause of Lower Tolerance to Short-Term Dehydration? PLoS ONE 2012, 7, e38017. [Google Scholar] [CrossRef]
- Zhao, F.; Zhang, D.; Zhao, Y.; Wang, W.; Yang, H.; Ta, F.; Li, C.; Hu, X. The Difference of Physiological and Proteomic Changes in Maize Leaves Adaptation to Drought, Heat, and Combined Both Stresses. Front. Plant Sci. 2016, 7, 1471. [Google Scholar] [CrossRef]
- Jedmowski, C.; Ashoub, A.; Beckhaus, T.; Berberich, T.; Karas, M.; Brüggemann, W. Comparative Analysis of Sorghum Bicolor Proteome in Response to Drought Stress and Following Recovery. Int. J. Proteom. 2014, 2014, 395905. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, J.; He, X.; Sun, H.; Zhang, G.; Wu, F. Comparative Proteomic Analysis of Drought Tolerance in the Two Contrasting Tibetan Wild Genotypes and Cultivated Genotype. BMC Genom. 2015, 16, 432. [Google Scholar] [CrossRef]
- Pan, J.; Li, Z.; Wang, Q.; Garrell, A.K.; Liu, M.; Guan, Y.; Zhou, W.; Liu, W. Comparative Proteomic Investigation of Drought Responses in Foxtail Millet. BMC Plant Biol. 2018, 18, 315. [Google Scholar] [CrossRef]
- Liao, H.J.; Qian, Q.; Liu, X.D. Heat Shock Suppresses Mating and Sperm Transfer in the Rice Leaf Folder Cnaphalocrocis Medinalis. Bull. Entomol. Res. 2014, 104, 383–392. [Google Scholar] [CrossRef]
- Mu, Q.; Zhang, W.; Zhang, Y.; Yan, H.; Liu, K.; Matsui, T.; Tian, X.; Yang, P. ITRAQ-Based Quantitative Proteomics Analysis on Rice Anther Responding to High Temperature. Int. J. Mol. Sci. 2017, 18, 1811. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, J.; Huang, X.; Guo, D.; Lou, H.; Hou, Z.; Su, M.; Liang, R.; Xie, C.; You, M. Differential Effects of a Post-Anthesis Heat Stress on Wheat (Triticum Aestivum L.) Grain Proteome Determined by ITRAQ. Sci. Rep. 2017, 7, 3468. [Google Scholar] [CrossRef]
- Kumar, R.R.; Singh, K.; Ahuja, S.; Tasleem, M.; Singh, I.; Kumar, S.; Grover, M.; Mishra, D.; Rai, G.K.; Goswami, S. Quantitative Proteomic Analysis Reveals Novel Stress-Associated Active Proteins (SAAPs) and Pathways Involved in Modulating Tolerance of Wheat under Terminal Heat. Funct. Integr. Genom. 2019, 19, 329–348. [Google Scholar] [CrossRef]
- Abou-Deif, M.H.; Rashed, M.A.-S.; Khalil, K.M.; Mahmoud, F.E.-S. Proteomic Analysis of Heat Shock Proteins in Maize (Zea Mays L.). Bull. Natl. Res. Cent. 2019, 43, 199. [Google Scholar] [CrossRef]
- Xu, J.; Lan, H.; Fang, H.; Huang, X.; Zhang, H.; Huang, J. Quantitative Proteomic Analysis of the Rice (Oryza Sativa L.) Salt Response. PLoS ONE 2015, 10, e0120978. [Google Scholar] [CrossRef]
- Lakra, N.; Kaur, C.; Singla-Pareek, S.L.; Pareek, A. Mapping the ‘Early Salinity Response’Triggered Proteome Adaptation in Contrasting Rice Genotypes Using ITRAQ Approach. Rice 2019, 12, 3. [Google Scholar] [CrossRef]
- Chen, F.; Fang, P.; Peng, Y.; Zeng, W.; Zhao, X.; Ding, Y.; Zhuang, Z.; Gao, Q.; Ren, B. Comparative Proteomics of Salt-Tolerant and Salt-Sensitive Maize Inbred Lines to Reveal the Molecular Mechanism of Salt Tolerance. Int. J. Mol. Sci. 2019, 20, 4725. [Google Scholar] [CrossRef]
- Rasoulnia, A.; Bihamta, M.R.; Peyghambari, S.A.; Alizadeh, H.; Rahnama, A. Proteomic Response of Barley Leaves to Salinity. Mol. Biol. Rep. 2011, 38, 5055–5063. [Google Scholar] [CrossRef]
- Fatehi, F.; Hosseinzadeh, A.; Alizadeh, H.; Brimavandi, T.; Struik, P.C. The Proteome Response of Salt-Resistant and Salt-Sensitive Barley Genotypes to Long-Term Salinity Stress. Mol. Biol. Rep. 2012, 39, 6387–6397. [Google Scholar] [CrossRef]
- Mostek, A.; Börner, A.; Badowiec, A.; Weidner, S. Alterations in Root Proteome of Salt-Sensitive and Tolerant Barley Lines under Salt Stress Conditions. J. Plant Physiol. 2015, 174, 166–176. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, J.; Wang, W.; Wang, Y.; Xu, J.; Li, Z.; Zhao, X.; Fu, B. Integrated Analysis of the Transcriptome and Metabolome Revealed the Molecular Mechanisms Underlying the Enhanced Salt Tolerance of Rice Due to the Application of Exogenous Melatonin. Front. Plant Sci. 2021, 11, 618680. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.Y.; Wang, L.H.; Dou, X.T.; Wang, Y.J.; Wang, H.Z. Comparative Metabolomic Profiling in the Roots of Salt-Tolerant and Salt-Intolerant Maize Cultivars Treated with NaCl Stress. Biol. Plant 2020, 64, 569–577. [Google Scholar] [CrossRef]

| Crop (Common Name) | QTL Trait Identified | Stress | Trait Improved | References |
|---|---|---|---|---|
| Rice | Seed weight and grain yield, osmotic potential, water potential, cell membrane stability | Drought | Yield, grain, plant biomass, root length | [25,155,156,157] |
| Maize | Leaf growth, ionic balance, osmotic adjustment | Drought | Yield and grain | [158,159,160] |
| Sorghum | Water-related attributes, ionic balance, osmotic adjustment | Drought | Delayed leaf senescence, yield and grain | [161] |
| Pearl millet | Osmotic potential, water-related attributes, cell membrane stability | Drought | Yield and grain | [162] |
| Wheat | Osmotic potential, seed weight and grain yield, salt tolerance | Drought, cold, salinity, metal toxicity, nutrient deficiency, heat | Plant biomass, early flower and maturity, yield | [163,164,165,166] |
| Barley | Water potential, osmotic potential, freezing tolerance | Drought, salinity, water logging, metal toxicity | Grain | [167,168,169] |
| Plant Species | Approach Used | Differentially Regulated Genes | Reference |
|---|---|---|---|
| Rice | Microarray | HsfA2a, HsfA2d, HsfA2f, HsfA3, HsfB2a, Hsfb, Hsfc, DREB, ERF, and members of HSP70, HSP90, and HSP100 | [103] |
| Microarray | Microarray Hsfs, sHSPs, members of HSP70, HSP90, and HSP100 gene families | [181] | |
| RT-PCR | OsHsfA4b, OsHsfA5, OsHsfA7, OsHsfA4d, OsHsf A2a, OsHsfA2c, and OsHsfA2d | [182] | |
| Microarray | Hsfs, bZIP TFs, HSP10s, and HSP20s | [183] | |
| RNA-seq, qRT–PCR | sHSP genes, HSP101 or heat shock factor (HSF) genes, TFs- WRKY, MYB, AP2/ERF | [184] | |
| RNA-seq, qRT–PCR | HSP 20 and HSP70 family), heat shock protein binding protein 1 (HSPBP1, HSP70-interacting protein), DREB, RAB, and late embryogenesis abundant (LEA) proteins | [184] | |
| Wheat | Microarray | Hsfs, HSPs, DREB2B and DREB6A, ERETC, and member of MBF1 | [185] |
| Microarray | Wheat microarray b-ZIP transcription factors andTaCAM3-1(zinc finger with calmodulin) | [186] | |
| Microarray | HSPs, transporters, protein modifiers, and signaling molecules | [187] | |
| qRT-PCR | Chloroplast-localized small heat shock proteins (sHSP) encoded by the Hsp26 gene | [188] | |
| Maize | qRT-PCR, RNA Sequence | ZmHsf-01, ZmHsf-03, ZmHsf-04, ZmHsf-06, ZmHsf-10, ZmHsf-11, ZmHsf-14, ZmHsf-15, ZmHsf-19, ZmHsf-20, ZmHsf-21, ZmHsf-22, ZmHsf-23, ZmHsf-24, and ZmHsf-25 | [189] |
| HSP 26, HSP 70, sHSP Cytokinin, signal transduction and DNA synthesis/chromatin structure | [190] | ||
| Photosynthesis, oxidation-reduction process, peptidase inhibitor activity, peptidase regulator activity and inositol tetrakisphosphate kinase activity | [190] | ||
| Sorghum | Microarray | TFs-MYB78 and ATAF1, chaperones. | [191] |
| Barley | Microarray, RNA Sequence | Raffinose synthase 1, UDPd-glucose 4-epimerase 1, UDP-d-glucose 4-epimerase 3, rehalose-6-phosphate synthase, trehalose-6-phosphate phosphatase, invertase inhibitor, heat shock transcription factor A2d, hexokinase 2, and SNF1-related protein kinases | [192] |
| Starch phosphorylation, chorismate biosynthesis, L-ascorbate biosynthesis and recycling, | [193] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rakkammal, K.; Priya, A.; Pandian, S.; Maharajan, T.; Rathinapriya, P.; Satish, L.; Ceasar, S.A.; Sohn, S.-I.; Ramesh, M. Conventional and Omics Approaches for Understanding the Abiotic Stress Response in Cereal Crops—An Updated Overview. Plants 2022, 11, 2852. https://doi.org/10.3390/plants11212852
Rakkammal K, Priya A, Pandian S, Maharajan T, Rathinapriya P, Satish L, Ceasar SA, Sohn S-I, Ramesh M. Conventional and Omics Approaches for Understanding the Abiotic Stress Response in Cereal Crops—An Updated Overview. Plants. 2022; 11(21):2852. https://doi.org/10.3390/plants11212852
Chicago/Turabian StyleRakkammal, Kasinathan, Arumugam Priya, Subramani Pandian, Theivanayagam Maharajan, Periyasamy Rathinapriya, Lakkakula Satish, Stanislaus Antony Ceasar, Soo-In Sohn, and Manikandan Ramesh. 2022. "Conventional and Omics Approaches for Understanding the Abiotic Stress Response in Cereal Crops—An Updated Overview" Plants 11, no. 21: 2852. https://doi.org/10.3390/plants11212852
APA StyleRakkammal, K., Priya, A., Pandian, S., Maharajan, T., Rathinapriya, P., Satish, L., Ceasar, S. A., Sohn, S.-I., & Ramesh, M. (2022). Conventional and Omics Approaches for Understanding the Abiotic Stress Response in Cereal Crops—An Updated Overview. Plants, 11(21), 2852. https://doi.org/10.3390/plants11212852













