Identification of Disease Resistance Parents and Genome-Wide Association Mapping of Resistance in Spring Wheat
Abstract
:1. Introduction
2. Results
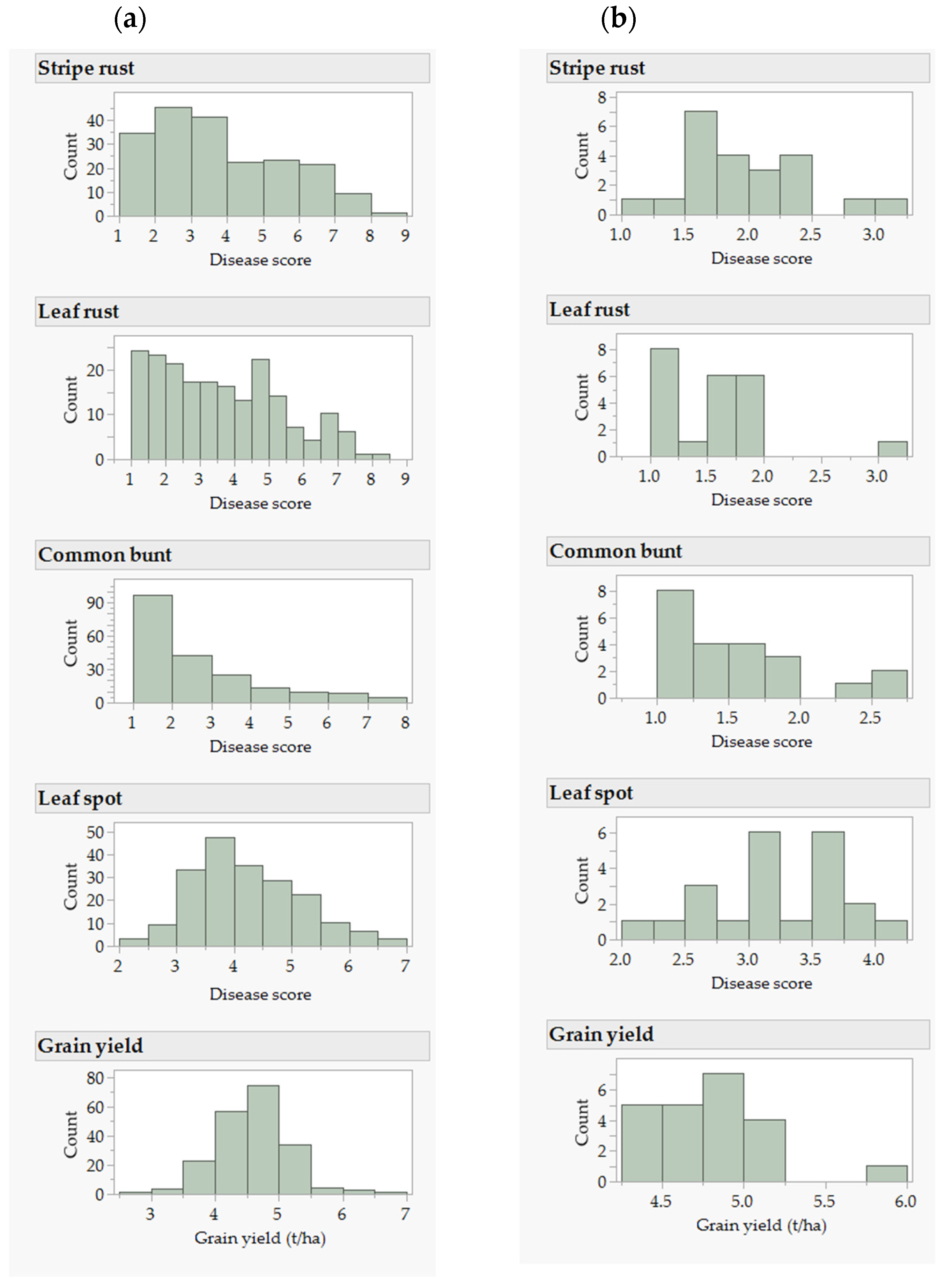
2.1. Phenotypic and Genetic Variation
2.2. Comparison of Economic Weights for Multi-Trait Selection
2.3. Disease Resistant Parents Selected Using Single-Trait and Multi-Trait RLPSI
2.4. Genome-Wide Association Mapping
3. Discussion
4. Materials and Methods
4.1. Genotyping and Phenotyping
4.2. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lehermeier, C.; Teyssèdre, S.; Schön, C.-C. Genetic gain increases by applying the usefulness criterion with improved variance prediction in selection of crosses. Genetics 2017, 207, 1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, L.; Jacobson, A.; Zhong, S.; Bernardo, R. Prediction of genetic variance in biparental maize populations: Genomewide marker effects versus mean genetic variance in prior populations. Crop Sci. 2015, 55, 1181–1188. [Google Scholar] [CrossRef]
- Zhong, S.; Jannink, J.L. Using quantitative trait loci results to discriminate among crosses on the basis of their progeny mean and variance. Genetics 2007, 177, 567–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkhamer, R.L.; Lanning, S.P.; Martens, R.J.; Martin, J.M.; Talbert, L.E. Predicting progeny variance from parental divergence in hard red spring wheat. Crop Sci. 1998, 38, 243–248. [Google Scholar] [CrossRef]
- Longin, F.H.; Liu, W.; Ranc, N.; Reif, J.C. Association of progeny variance and genetic distances among parents and implications for the design of elite maize breeding programs. Maydica 2011, 56, 19–23. [Google Scholar]
- Daetwyler, H.D.; Hayden, M.J.; Spangenberg, G.C.; Hayes, B.J. Selection on optimal haploid value increases genetic gain and preserves more genetic diversity relative to genomic selection. Genetics 2015, 200, 1341–1348. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, M.; Tiede, T.; Smith, K.P. Popvar: A genome-wide procedure for predicting genetic variance and correlated response in biparental breeding populations. Crop Sci. 2015, 55, 2068–2077. [Google Scholar] [CrossRef]
- Han, Y.; Cameron, J.N.; Wang, L.; Beavis, W.D. The predicted cross value for genetic introgression of multiple alleles. Genetics 2017, 205, 1409–1423. [Google Scholar] [CrossRef] [Green Version]
- Moeinizade, S.; Han, Y.; Pham, H.; Hu, G.; Wang, L. A look-ahead Monte Carlo simulation method for improving parental selection in trait introgression. Sci. Rep. 2021, 11, 3918. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.-Y.; Liao, C.-T. Identification of superior parental lines for biparental crossing via genomic prediction. PLoS ONE 2020, 15, e0243159. [Google Scholar] [CrossRef]
- Singh, R.P.; Juliana, P.; Huerta-Espino, J.; Govindan, V.; Crespo-Herrera, L.A.; Mondal, S.; Bhavani, S.; Singh, P.K.; He, X.; Ibba, M.I.; et al. Achieving genetic gains in practice. In Wheat Improvement: Food Security in a Changing Climate; Reynolds, M.P., Braun, H.-J., Eds.; Springer International Publishing: Cham, UK, 2022; pp. 97–123. [Google Scholar]
- Singh, R.P.; Rajaram, S.; Miranda, A.; Huerta-Espino, J.; Autrique, E. Comparison of two crossing and four selection schemes for yield, yield traits, and slow rusting resistance to leaf rust in wheat. Euphytica 1998, 100, 35–43. [Google Scholar] [CrossRef]
- McCallum, B.D.; Reimer, E.; McNabb, W.; Foster, A.; Rosa, S.; Xue, A. Physiologic specialization of Puccinia triticina, the causal agent of wheat leaf rust, in Canada in 2015–2019. Can. J. Plant Pathol. 2021, 33, 541–549. [Google Scholar] [CrossRef]
- Aboukhaddour, R.; Fetch, T.; McCallum, B.D.; Harding, M.W.; Beres, B.L.; Graf, R.J. Wheat diseases on the prairies: A Canadian story. Plant Pathol. 2020, 69, 418–432. [Google Scholar] [CrossRef]
- Kutcher, H.R.; Turkington, T.K.; McLaren, D.L.; Irvine, R.B.; Brar, G.S. Fungicide and cultivar management of leaf spot diseases of winter wheat in western Canada. Plant Dis. 2018, 102, 1828–1833. [Google Scholar] [CrossRef] [Green Version]
- Sabouri, H.; Rabiei, B.; Fazlalipour, M. Use of selection indices based on multivariate analysis for improving grain yield in rice. Rice Sci. 2008, 15, 303–310. [Google Scholar] [CrossRef]
- Cerón-Rojas, J.J.; Sahagún-Castellanos, J.; Castillo-González, F.; Santacruz-Varela, A.; Crossa, J. A restricted selection index method based on eigenanalysis. J. Agric. Biol. Environ. Stat. 2008, 13, 440–457. [Google Scholar] [CrossRef]
- Rapp, M.; Lein, V.; Lacoudre, F.; Lafferty, J.; Müller, E.; Vida, G.; Bozhanova, V.; Ibraliu, A.; Thorwarth, P.; Piepho, H.P.; et al. Simultaneous improvement of grain yield and protein content in durum wheat by different phenotypic indices and genomic selection. Theor. Appl. Genet. 2018, 131, 1315–1329. [Google Scholar] [CrossRef]
- Cerón-Rojas, J.J.; Crossa, J. Linear Selection Indices in Modern Plant Breeding; Springer International Publishing: Cham, UK, 2018; pp. 1–256. [Google Scholar]
- Hazel, L.N.; Dickerson, G.E.; Freeman, A.E. The selection index—Then, now, and for the future. J. Dairy Sci. 1994, 77, 3236–3251. [Google Scholar] [CrossRef]
- Hazel, L.N.; Lush, J.L. The efficiency of three methods of selection. J. Hered. 1942, 33, 393–399. [Google Scholar] [CrossRef]
- Kempthorne, O.; Nordskog, A.W. Restricted selection indices. Biometrics 1959, 15, 10–19. [Google Scholar] [CrossRef]
- Cerón-Rojas, J.J.; Crossa, J. The statistical theory of linear selection indices from phenotypic to genomic selection. Crop Sci. 2022, 62, 537–563. [Google Scholar] [CrossRef] [PubMed]
- Cerón-Rojas, J.J.; Crossa, J. Expectation and variance of the estimator of the maximized selection response of linear selection indices with normal distribution. Theor. Appl. Genet. 2020, 133, 2743–2758. [Google Scholar] [CrossRef] [PubMed]
- Cerón-Rojas, J.J.; Crossa, J.; Sahagún-Castellanos, J. Statistical sampling properties of the coefficients of three phenotypic selection indices. Crop Sci. 2016, 56, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Cerón-Rojas, J.J.; Toledo, F.H.; Crossa, J. Optimum and decorrelated constrained multistage linear phenotypic selection indices theory. Crop Sci. 2019, 59, 2585–2600. [Google Scholar] [CrossRef] [Green Version]
- Cerón-Rojas, J.J.; Toledo, F.H.; Crossa, J. The relative efficiency of two multistage linear phenotypic selection indices to predict the net genetic merit. Crop Sci. 2019, 59, 1037–1051. [Google Scholar] [CrossRef] [Green Version]
- Smith, H.F. A discriminant function for plant selection. Ann. Eugen. 1936, 7, 240–250. [Google Scholar] [CrossRef]
- Iqbal, M.; Semagn, K.; Céron-Rojas, J.J.; Crossa, J.; Jarquin, D.; Howard, R.; Beres, B.L.; Strenzke, K.; Ciechanowska, I.; Spaner, D. Identification of spring wheat with superior performance for agronomic traits under contrasting nitrogen managements using linear phenotypic selection indices. Plants 2022, 11, 1887. [Google Scholar] [CrossRef]
- Brar, G.S.; Fetch, T.; McCallum, B.D.; Hucl, P.J.; Kutcher, H.R. Virulence dynamics and breeding for resistance to stripe, stem, and leaf rust in Canada since 2000. Plant Dis. 2019, 103, 2981–2995. [Google Scholar] [CrossRef] [Green Version]
- Laroche, A.; Frick, M.; Graf, R.J.; Larsen, J.; Laurie, J.D. Pyramiding disease resistance genes in elite winter wheat germplasm for Western Canada. Crop J. 2019, 7, 739–749. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Dubcovsky, W.J.; Rogers, W.J.; Raupp, W.J. Catalogue of gene symbols for wheat: 2020 supplement. Annu. Wheat Newsl. 2020, 66, 109–128. Available online: https://wheat.pw.usda.gov/GG3/wgc (accessed on 8 August 2022).
- Goates, B.J. Identification of new pathogenic races of common bunt and dwarf bunt fungi, and evaluation of known races using an expanded set of differential wheat lines. Plant Dis. 2012, 96, 361–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamari, L.; Strelkov, S.E.; Yahyaoui, A.; Orabi, J.; Smith, R.B. The identification of two new races of Pyrenophora tritici-repentis from the host center of diversity confirms a one-to-one relationship in tan spot of wheat. Phytopathology 2003, 93, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Batra, R.; Sharma, S.; Saripalli, G.; Gautam, T.; Singh, R.; Pal, S.; Malik, P.; Kumar, M.; Jan, I.; et al. WheatQTLdb: A QTL database for wheat. Mol. Genet. Genom. 2021, 296, 1051–1056. [Google Scholar] [CrossRef]
- Perez-Lara, E.; Semagn, K.; Tran, A.N.; Ciechanowska, I.; Chen, H.; Iqbal, M.; N’Diaye, A.; Pozniak, C.; Strelkov, S.E.; Hucl, P.J.; et al. Population structure and genomewide association analysis of resistance to disease and insensitivity to Ptr toxins in Canadian spring wheat using 90K SNP array. Crop Sci. 2017, 57, 1522–1539. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J.; et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef]
- Alaux, M.; Rogers, J.; Letellier, T.; Flores, R.; Alfama, F.; Pommier, C.; Mohellibi, N.; Durand, S.; Kimmel, E.; Michotey, C.; et al. Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 2018, 19, 111. [Google Scholar] [CrossRef] [PubMed]
- Semagn, K.; Iqbal, M.; Alachiotis, N.; N’Diaye, A.; Pozniak, C.; Spaner, D. Genetic diversity and selective sweeps in historical and modern Canadian spring wheat cultivars using the 90K SNP array. Sci. Rep. 2021, 11, 23773. [Google Scholar] [CrossRef] [PubMed]
- Rutkoski, J.E. A practical guide to genetic gain. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 157, pp. 217–249. [Google Scholar]
- Sharma, R.C.; Pandey-Chhetri, B.; Duveiller, E. Heritability estimates of spot blotch resistance and its association with other traits in spring wheat crosses. Euphytica 2006, 147, 317–327. [Google Scholar] [CrossRef]
- Zou, J.; Semagn, K.; Chen, H.; Iqbal, M.; Asif, M.; N’Diaye, A.; Navabi, A.; Perez-Lara, E.; Pozniak, C.; Yang, R.C.; et al. Mapping of QTLs associated with resistance to common bunt, tan spot, leaf rust, and stripe rust in a spring wheat population. Mol. Breed. 2017, 37, 144. [Google Scholar] [CrossRef]
- Ramos Guimarães, P.H.; Guimarães Santos Melo, P.; Centeno Cordeiro, A.C.; Pereira Torga, P.; Nakano Rangel, P.H.; Pereira de Castro, A. Index selection can improve the selection efficiency in a rice recurrent selection population. Euphytica 2021, 217, 95. [Google Scholar] [CrossRef]
- Pesek, J.; Baker, R.J. An application of index selection to the improvement of self-pollinated species. Can. J. Plant Sci. 1970, 50, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Aoun, M.; Chen, X.; Somo, M.; Xu, S.S.; Li, X.; Elias, E.M. Novel stripe rust all-stage resistance loci identified in a worldwide collection of durum wheat using genome-wide association mapping. Plant Genome 2021, 14, e20136. [Google Scholar] [CrossRef]
- Luo, P.; Hu, X.; Zhang, H.; Ren, Z. Genes for resistance to stripe rust on chromosome 2B and their application in wheat breeding. Prog. Nat. Sci. 2009, 19, 9–15. [Google Scholar] [CrossRef]
- Naruoka, Y.; Ando, K.; Bulli, P.; Muleta, K.T.; Rynearson, S.; Pumphrey, M.O. Identification and Validation of SNP Markers Linked to the Stripe Rust Resistance Gene Yr5 in Wheat. Crop Sci. 2016, 56, 3055–3065. [Google Scholar] [CrossRef]
- Gardiner, L.-J.; Bansept-Basler, P.; El-Soda, M.; Hall, A.; O’Sullivan, D.M. A framework for gene mapping in wheat demonstrated using the Yr7 yellow rust resistance gene. PLoS ONE 2020, 15, e0231157. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chang, Z.; Zhang, X.; Yang, Z.; Li, X.; Jia, J.; Zhan, H.; Guo, H.; Wang, J. Putative Thinopyrum intermedium-derived stripe rust resistance gene Yr50 maps on wheat chromosome arm 4BL. Theor. Appl. Genet. 2013, 126, 265–274. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, M.N.; Chen, X.M.; See, D.; Chao, S.; Jing, J. Mapping of Yr62 and a small-effect QTL for high-temperature adult-plant resistance to stripe rust in spring wheat PI 192252. Theor. Appl. Genet. 2014, 127, 1449–1459. [Google Scholar] [CrossRef]
- Suenaga, K.; Singh, R.P.; Huerta-Espino, J.; William, H.M. Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology 2003, 93, 881–890. [Google Scholar] [CrossRef] [Green Version]
- Lagudah, E.S.; Krattinger, S.G.; Herrera-Foessel, S.; Singh, R.P.; Huerta-Espino, J.; Spielmeyer, W.; Brown-Guedira, G.; Selter, L.L.; Keller, B. Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor. Appl. Genet. 2009, 119, 889–898. [Google Scholar] [CrossRef] [Green Version]
- McCallum, B.D.; Fetch, T.; Chong, J. Cereal rust control in Canada. Aust. J. Agric. Res. 2007, 58, 639–647. [Google Scholar] [CrossRef]
- McCallum, B.D.; Hiebert, C.W. Interactions between Lr67 or Lr34 and other leaf rust resistance genes in wheat (Triticum aestivum). Front. Plant Sci. 2022, 13, 871970. [Google Scholar] [CrossRef] [PubMed]
- German, S.E.; Kolmer, J.A. Effect of gene Lr34 in the enhancement of resistance to leaf rust of wheat. Theor. Appl. Genet. 1992, 84, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Huerta-Espino, J. Effect of leaf rust resistance gene Lr34 on components of slow rusting at seven growth stages in wheat. Euphytica 2003, 129, 371–376. [Google Scholar] [CrossRef]
- Bokore, F.E.; Knox, R.E.; Hiebert, C.W.; Cuthbert, R.D.; DePauw, R.M.; Meyer, B.; N’Diaye, A.; Pozniak, C.J.; McCallum, B.D. A combination of leaf rust resistance genes, including Lr34 and Lr46, is the key to the durable resistance of the Canadian wheat cultivar, Carberry. Front. Plant Sci. 2022, 12, 775383. [Google Scholar] [CrossRef] [PubMed]
- McCallum, B.D.; Humphreys, D.G.; Somers, D.J.; Dakouri, A.; Cloutier, S. Allelic variation for the rust resistance gene Lr34/Yr18 in Canadian wheat cultivars. Euphytica 2012, 183, 261–274. [Google Scholar] [CrossRef]
- McCartney, C.A.; Somers, D.J.; McCallum, B.D.; Thomas, J.; Humphreys, D.G.; Menzies, J.G.; Brown, P.D. Microsatellite tagging of the leaf rust resistance gene Lr16 on wheat chromosome 2BSc. Mol. Breed. 2005, 15, 329–337. [Google Scholar] [CrossRef]
- Hiebert, C.W.; Thomas, J.B.; McCallum, B.D. Stacking pairs of disease resistance genes in wheat populations using telocentric chromosomes. Mol. Breed. 2010, 26, 681–692. [Google Scholar] [CrossRef]
- Kassa, M.T.; You, F.M.; Hiebert, C.W.; Pozniak, C.J.; Fobert, P.R.; Sharpe, A.G.; Menzies, J.G.; Humphreys, D.G.; Rezac Harrison, N.; Fellers, J.P.; et al. Highly predictive SNP markers for efficient selection of the wheat leaf rust resistance gene Lr16. BMC Plant Biol. 2017, 17, 45. [Google Scholar] [CrossRef] [Green Version]
- Lewarne, M.K. Characterization and Genetic Mapping of Leaf Rust (Puccinia triticina) Resistance Genes Lr2a and Lr46 in Canadian Spring Wheat (Triticum aestivum) Germplasm. Master’s Thesis, Department of Plant Science, University of Manitoba, Winnipeg, CA, USA, 2021. [Google Scholar]
- Menzies, J.G.; Knox, R.E.; Popovic, Z.; Procunier, J.D. Common bunt resistance gene Bt10 located on wheat chromosome 6D. Can. J. Plant Sci. 2006, 86, 1409–1412. [Google Scholar] [CrossRef]
- Steffan, P.M.; Torp, A.M.; Borgen, A.; Backes, G.; Rasmussen, S.K. Mapping of common bunt resistance gene Bt9 in wheat. Theor. Appl. Genet. 2017, 130, 1031–1040. [Google Scholar] [CrossRef] [Green Version]
- Gaudet, D.A.; Puchalski, B.L. Races of common bunt (Tilletia caries and T. foetida) of wheat in western Canada. Can. J. Plant Pathol. 1989, 11, 415–418. [Google Scholar] [CrossRef]
- Laroche, A.; Demeke, T.; Gaudet, D.A.; Puchalski, B.; Frick, M.; McKenzie, R. Development of a PCR marker for rapid identification of the Bt-10 gene for common bunt resistance in wheat. Genome 2000, 43, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-X.; Barbier, H.; Rouse, M.N.; Singh, S.; Singh, R.P.; Bhavani, S.; Huerta-Espino, J.; Sorrells, M.E. A consensus map for Ug99 stem rust resistance loci in wheat. Theor. Appl. Genet. 2014, 127, 1561–1581. [Google Scholar] [CrossRef] [Green Version]
- Kosgey, Z.C.; Edae, E.A.; Dill-Macky, R.; Jin, Y.; Bulbula, W.D.; Gemechu, A.; Macharia, G.; Bhavani, S.; Randhawa, M.S.; Rouse, M.N. Mapping and validation of stem rust resistance loci in spring wheat line CI 14275. Front. Plant Sci. 2021, 11, 2147. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, C.D.G.; Garvin, D.F.; Kolmer, J.A. Genetics of stem rust resistance in the spring wheat cultivar Thatcher and the enhancement of stem rust resistance by Lr34. Euphytica 2008, 159, 391–401. [Google Scholar] [CrossRef]
- Zurn, J.D.; Rouse, M.N.; Chao, S.; Aoun, M.; Macharia, G.; Hiebert, C.W.; Pretorius, Z.A.; Bonman, J.M.; Acevedo, M. Dissection of the multigenic wheat stem rust resistance present in the Montenegrin spring wheat accession PI 362698. BMC Genom. 2018, 19, 67. [Google Scholar] [CrossRef] [Green Version]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [Green Version]
- Beavis, W.D. QTL analyses: Power, precision and accuracy. In Molecular Dissection of Complex Traits; Paterson, A.H., Ed.; CRC Press: Boca Raton, FL, USA, 1998; pp. 145–161. [Google Scholar]
- Josephs, E.B.; Stinchcombe, J.R.; Wright, S.I. What can genome-wide association studies tell us about the evolutionary forces maintaining genetic variation for quantitative traits? New Phytol. 2017, 214, 21–33. [Google Scholar] [CrossRef] [Green Version]
- Xu, S. Theoretical basis of the Beavis effect. Genetics 2003, 165, 2259–2268. [Google Scholar] [CrossRef]
- Brachi, B.; Morris, G.P.; Borevitz, J.O. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011, 12, 232. [Google Scholar] [CrossRef] [Green Version]
- Schaid, D.J.; Chen, W.; Larson, N.B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018, 19, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Money, D.; Gardner, K.; Migicovsky, Z.; Schwaninger, H.; Zhong, G.-Y.; Myles, S. LinkImpute: Fast and accurate genotype imputation for nonmodel organisms. G3 Genes|Genomes|Genet. 2015, 5, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Semagn, K.; Iqbal, M.; Chen, H.; Perez-Lara, E.; Bemister, D.H.; Xiang, R.; Zou, J.; Asif, M.; Kamran, A.; N’Diaye, A.; et al. Physical mapping of QTL in four spring wheat populations under conventional and organic management systems. I. Earliness. Plants 2021, 10, 853. [Google Scholar] [CrossRef]
- Semagn, K.; Iqbal, M.; Jarquin, D.; Crossa, J.; Howard, R.; Ciechanowska, I.; Henriquez, M.H.; Randhawa, H.; Aboukhaddour, R.; McCallum, B.D.; et al. Genomic predictions for common bunt, FHB, stripe rust, leaf rust, and leaf spotting resistance in spring wheat. Genes 2022, 13, 565. [Google Scholar] [CrossRef]
- Bokore, F.E.; Knox, R.E.; Cuthbert, R.D.; Pozniak, C.J.; McCallum, B.D.; N’Diaye, A.; DePauw, R.M.; Campbell, H.L.; Munro, C.; Singh, A.; et al. Mapping quantitative trait loci associated with leaf rust resistance in five spring wheat populations using single nucleotide polymorphism markers. PLoS ONE 2020, 15, e0230855. [Google Scholar] [CrossRef] [Green Version]
- Friesen, T.L.; Rasmussen, J.B.; Kwon, C.Y.; Ali, S.; Francl, L.J.; Meinhardt, S.W. Reaction of Ptr ToxA-insensitive wheat mutants to Pyrenophora tritici-repentis race 1. Phytopathology 2002, 92, 38–42. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, M.R.; Fox, S.L.; Hucl, P.; Singh, A.K. Leaf spotting reaction of spring common, durum and spelt wheat, and Kamut under organic management in western Canada. Can. J. Plant Sci. 2014, 94, 929–935. [Google Scholar] [CrossRef]
- Singh, P.K.; Mergoum, M.; Ali, S.; Adhikari, T.B.; Elias, E.M.; Anderson, J.A.; Glover, K.D.; Berzonsky, W.A. Evaluation of elite wheat germ plasm for resistance to tan spot. Plant Dis. 2006, 90, 1320–1325. [Google Scholar] [CrossRef] [Green Version]
- Gaudet, D.A.; Puchalski, B.J.; Kozub, G.C.; Schaalje, G.B. Susceptibility and resistance in Canadian spring wheat cultivars to common bunt (Tilletia tritici and T.laevis). Can. J. Plant Sci. 1993, 73, 1217–1224. [Google Scholar] [CrossRef] [Green Version]
- Alvarado, G.; Rodríguez, F.M.; Pacheco, A.; Burgueño, J.; Crossa, J.; Vargas, M.; Pérez-Rodríguez, P.; Lopez-Cruz, M.A. META-R: A software to analyze data from multi-environment plant breeding trials. Crop J. 2020, 8, 745–756. [Google Scholar] [CrossRef]
- Jones, B.; Sall, J. JMP statistical discovery software. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 188–194. [Google Scholar] [CrossRef]
- Semagn, K.; Iqbal, M.; N’Diaye, A.; Pozniak, C.; Ciechanowska, I.; Barbu, S.-P.; Spaner, D. Genome-wide association mapping of agronomic traits and grain characteristics in spring wheat under conventional and organic management systems. Crop Sci. 2022, 62, 1069–1087. [Google Scholar] [CrossRef]
- Wang, S.; Dvorkin, D.; Da, Y. SNPEVG: A graphical tool for GWAS graphing with mouse clicks. BMC Bioinform. 2012, 13, 319. [Google Scholar] [CrossRef]





| Genotype | Pedigree | Wheat Class * | Year ** | Stripe Rust (Yr) | Leaf Rust (Lr) | Common Bunt (Cb) | Leaf Spot (Ls) | Yield (t ha−1) | Selected Based on Each Disease |
|---|---|---|---|---|---|---|---|---|---|
| 5605HR CL | 99S2232-10/99S3228-4 | CNHR | 2013 | 2.2 | 1.0 | 1.5 | 2.3 | 4.9 | Both Lr and Ls |
| 5701PR | N89-3004/N87-0446//Oslo | CPSR | 2002 | 1.8 | 1.4 | 2.5 | 3.2 | 5.0 | Both Yr and Lr |
| AAC Bailey | 9505-LP03A/Journey//Lillian | CWRS | 2012 | 3.1 | 1.0 | 1.3 | 3.8 | 4.5 | Only Lr |
| AAC Brandon | Superb/CDC Osler//ND744 | CWRS | 2013 | 1.5 | 1.1 | 2.5 | 3.7 | 5.2 | Both Yr and Lr |
| AAC Connery | Somerset/BW865 | CWRS | 2015 | 1.6 | 3.1 | 1.0 | 3.5 | 4.5 | Both Yr and Cb |
| AAC Elie | Superb/CDC Osler//ND744 | CWRS | 2013 | 1.6 | 1.9 | 1.8 | 3.5 | 4.9 | Both Yr and Lr |
| AAC Penhold | 5700PR/HY644-BE//HY469 | CPSR | 2014 | 2.2 | 1.0 | 1.3 | 3.0 | 5.2 | Only Lr |
| AAC Concord | Lillian/Journey//9505-LP03A | CNHR | 2016 | 1.7 | 1.6 | 1.8 | 3.3 | 4.7 | Both Yr and Lr |
| AAC Redberry | Stettler/Glenn | CWRS | 2016 | 1.5 | 1.2 | 1.5 | 3.7 | 5.0 | Both Yr and Lr |
| BYT14-11 | Peace/Carberry | CWRS | Unregistered | 1.2 | 1.6 | 1.0 | 3.2 | 4.3 | Yr, Lr, and Cb |
| Carberry | Alsen/Superb | CWRS | 2009 | 1.8 | 1.6 | 1.3 | 3.7 | 4.5 | Both Yr and Lr |
| CDC Alsask | AC Elsa/AC Cora | CWRS | 2005 | 2.4 | 1.8 | 1.0 | 2.7 | 4.6 | Lr, Cb, and Ls |
| CDC Bradwell | 5602HR/W02330 | CWRS | 2015 | 2.3 | 1.8 | 1.5 | 3.5 | 4.3 | Only Lr |
| GP112 | 99S3148-1/00S3075-3-13 *** | CWSP | Unregistered | 1.3 | 1.1 | 1.0 | 2.7 | 5.1 | Yr, Lr, Cb, and Ls |
| AAC Castle | Conquer/CDN Bison//5701PR | CPSR | 2018 | 1.7 | 1.9 | 1.3 | 3.2 | 5.0 | Both Yr and Lr |
| Kane | AC Domain/McKenzie | CNHR | 2006 | 2.8 | 1.8 | 1.0 | 3.8 | 4.5 | Both Lr and Cb |
| Muchmore | Alsen/Superb | CNHR | 2009 | 1.9 | 1.8 | 1.0 | 4.0 | 4.9 | Yr, Lr, and Cb |
| Pasteur | Cadenza/(Palermo/KS91WGRC11) | CWSP | 2011 | 2.1 | 1.0 | 1.8 | 2.8 | 5.9 | Both Lr and Ls |
| Jake | McKenzie/Alsen//BW297 | CWRS | 2018 | 1.8 | 1.7 | 2.3 | 2.2 | 4.5 | Yr, Lr, and Ls |
| Tracker | Peace/CDC Stanley | CWRS | 2018 | 1.6 | 1.5 | 1.5 | 2.5 | 4.9 | Yr, Lr, and Ls |
| SY637 | BW337/AC ELSA | CWRS | 2016 | 2.3 | 1.0 | 1.0 | 3.0 | 4.3 | Both Lr and Cb |
| SY995 | 99S3144-7/5701PR **** | CPSR | 2014 | 2.3 | 1.5 | 1.0 | 3.2 | 5.1 | Both Lr and Cb |
| Region | No. of Significant SNPs | Chr | Min Position (bp) | Max Position (bp) | Phenotypic Variance (%) | |||
|---|---|---|---|---|---|---|---|---|
| Cb | Lr | Ls | Yr | |||||
| QCbt.dms-1A.1 | 1 | 1A | 4,383,342 | 4,383,342 | 6.9 | |||
| QCbt.dms-1A.2 | 2 | 1A | 13,371,025 | 14,027,876 | 7.2 | |||
| QCbt.dms-1A.3 | 1 | 1A | 556,873,095 | 556,873,095 | 7.4 | |||
| QLs.dms-1B | 1 | 1B | 18,203,933 | 18,203,933 | 6.6 | |||
| QYr.dms-1B | 1 | 1B | 540,583,488 | 540,583,488 | 7.0 | |||
| QLr.dms-1B | 4 | 1B | 547,332,148 | 549,647,134 | 7.5 | |||
| QCbt.dms-1B | 1 | 1B | 645,251,547 | 645,251,547 | 6.6 | |||
| QCbt.dms-1D | 1 | 1D | 10,669,243 | 10,669,243 | 6.6 | |||
| QLs.dms-2A.1 | 1 | 2A | 19,019,241 | 19,019,241 | 7.9 | |||
| QLs.dms-2A.2 | 6 | 2A | 45,928,136 | 45,932,241 | 6.7 | |||
| QLr.dms-2A | 1 | 2A | 758,316,082 | 758,316,082 | 7.9 | |||
| QLs.dms-2B | 1 | 2B | 19,530,481 | 19,530,481 | 7.4 | |||
| QLr.dms-2B.1 | 1 | 2B | 690,898,021 | 690,898,021 | 6.7 | |||
| QLr.dms-2B.2 | 10 | 2B | 771,850,360 | 778,230,186 | 8.0 | |||
| QCbt.dms-2B | 1 | 2B | 811,019,075 | 811,019,075 | 7.1 | |||
| QYr.dms-2B | 1 | 2B | 812,244,914 | 812,244,914 | 7.4 | |||
| QLr.dms-2D | 5 | 2D | 624,625,220 | 624,952,858 | 8.3 | |||
| QCbt.dms-3A.1 | 1 | 3A | 10,279,544 | 10,279,544 | 10.4 | |||
| QCbt.dms-3A.2 | 1 | 3A | 671,290,927 | 671,290,927 | 8.5 | |||
| QLr.dms-3A | 1 | 3A | 51,644,908 | 51,644,908 | 7.1 | |||
| QLs.dms-3B | 1 | 3B | 557,835,101 | 557,835,101 | 6.6 | |||
| QLr.dms-3B.1 | 1 | 3B | 616,148,037 | 616,148,037 | 6.6 | |||
| QLr.dms-3B.2 | 4 | 3B | 743,632,624 | 743,922,021 | 7.0 | |||
| QLr.dms-3D | 1 | 3D | 550,280,985 | 550,280,985 | 7.0 | |||
| QLs.dms-4A | 1 | 4A | 580,845,356 | 580,845,356 | 7.3 | |||
| QYr.dms-4B | 3 | 4B | 638,809,518 | 638,813,440 | 8.9 | |||
| QLr.dms-5A.1 | 2 | 5A | 331,454,313 | 331,884,318 | 7.2 | |||
| QLr.dms-5A.2 | 1 | 5A | 338,666,459 | 338,666,459 | 6.8 | |||
| QYr.dms-5A | 1 | 5A | 547,615,657 | 547,615,657 | 7.4 | |||
| QLr.dms-5B | 2 | 5B | 281,176,164 | 284,717,655 | 7.2 | |||
| QCbt.dms-5D.1 | 1 | 5D | 244,100,829 | 244,100,829 | 7.0 | |||
| QCbt.dms-5D.2 | 1 | 5D | 565,867,455 | 565,867,455 | 6.6 | |||
| QCbt.dms-6D | 1 | 6D | 7,431,984 | 7,431,984 | 16.9 | |||
| QCbt.dms-7A | 1 | 7A | 15,774,259 | 15,774,259 | 6.7 | |||
| QLr.dms-7B | 1 | 7B | 36,162,027 | 36,162,027 | 6.8 | |||
| QYr.dms-7D | 1 | 7D | 48,955,909 | 48,955,909 | 8.6 | |||
| QLs.dms-7D | 1 | 7D | 266,720,824 | 266,720,824 | 6.6 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iqbal, M.; Semagn, K.; Jarquin, D.; Randhawa, H.; McCallum, B.D.; Howard, R.; Aboukhaddour, R.; Ciechanowska, I.; Strenzke, K.; Crossa, J.; et al. Identification of Disease Resistance Parents and Genome-Wide Association Mapping of Resistance in Spring Wheat. Plants 2022, 11, 2905. https://doi.org/10.3390/plants11212905
Iqbal M, Semagn K, Jarquin D, Randhawa H, McCallum BD, Howard R, Aboukhaddour R, Ciechanowska I, Strenzke K, Crossa J, et al. Identification of Disease Resistance Parents and Genome-Wide Association Mapping of Resistance in Spring Wheat. Plants. 2022; 11(21):2905. https://doi.org/10.3390/plants11212905
Chicago/Turabian StyleIqbal, Muhammad, Kassa Semagn, Diego Jarquin, Harpinder Randhawa, Brent D. McCallum, Reka Howard, Reem Aboukhaddour, Izabela Ciechanowska, Klaus Strenzke, José Crossa, and et al. 2022. "Identification of Disease Resistance Parents and Genome-Wide Association Mapping of Resistance in Spring Wheat" Plants 11, no. 21: 2905. https://doi.org/10.3390/plants11212905
APA StyleIqbal, M., Semagn, K., Jarquin, D., Randhawa, H., McCallum, B. D., Howard, R., Aboukhaddour, R., Ciechanowska, I., Strenzke, K., Crossa, J., Céron-Rojas, J. J., N’Diaye, A., Pozniak, C., & Spaner, D. (2022). Identification of Disease Resistance Parents and Genome-Wide Association Mapping of Resistance in Spring Wheat. Plants, 11(21), 2905. https://doi.org/10.3390/plants11212905









