Natural Populations of Astrocaryum aculeatum Meyer in Amazonia: Genetic Diversity and Conservation
Abstract
1. Introduction
2. Results
2.1. Genetic Diversity Indexes
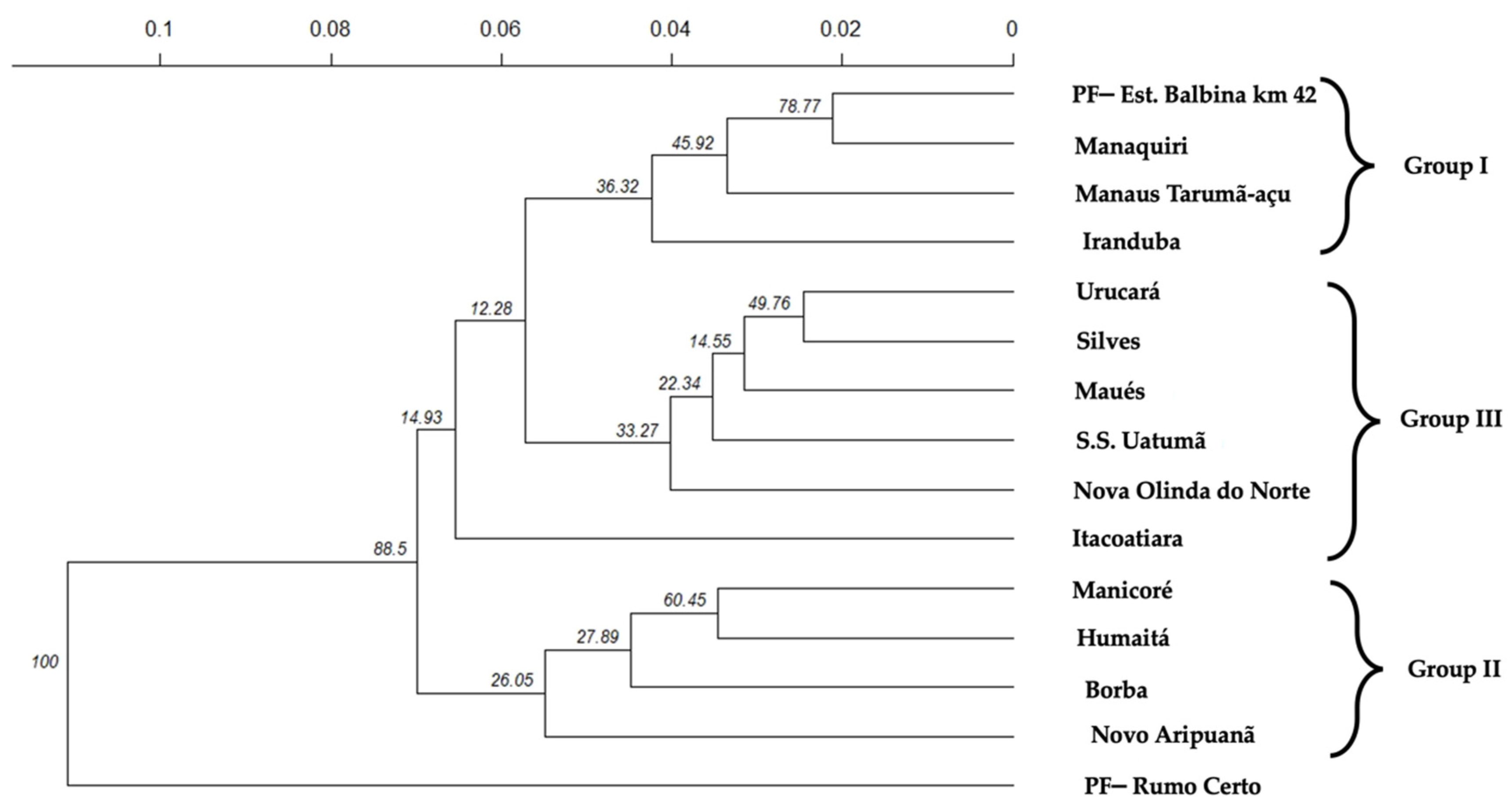
2.2. Genetic Structure
3. Discussion
4. Materials and Methods
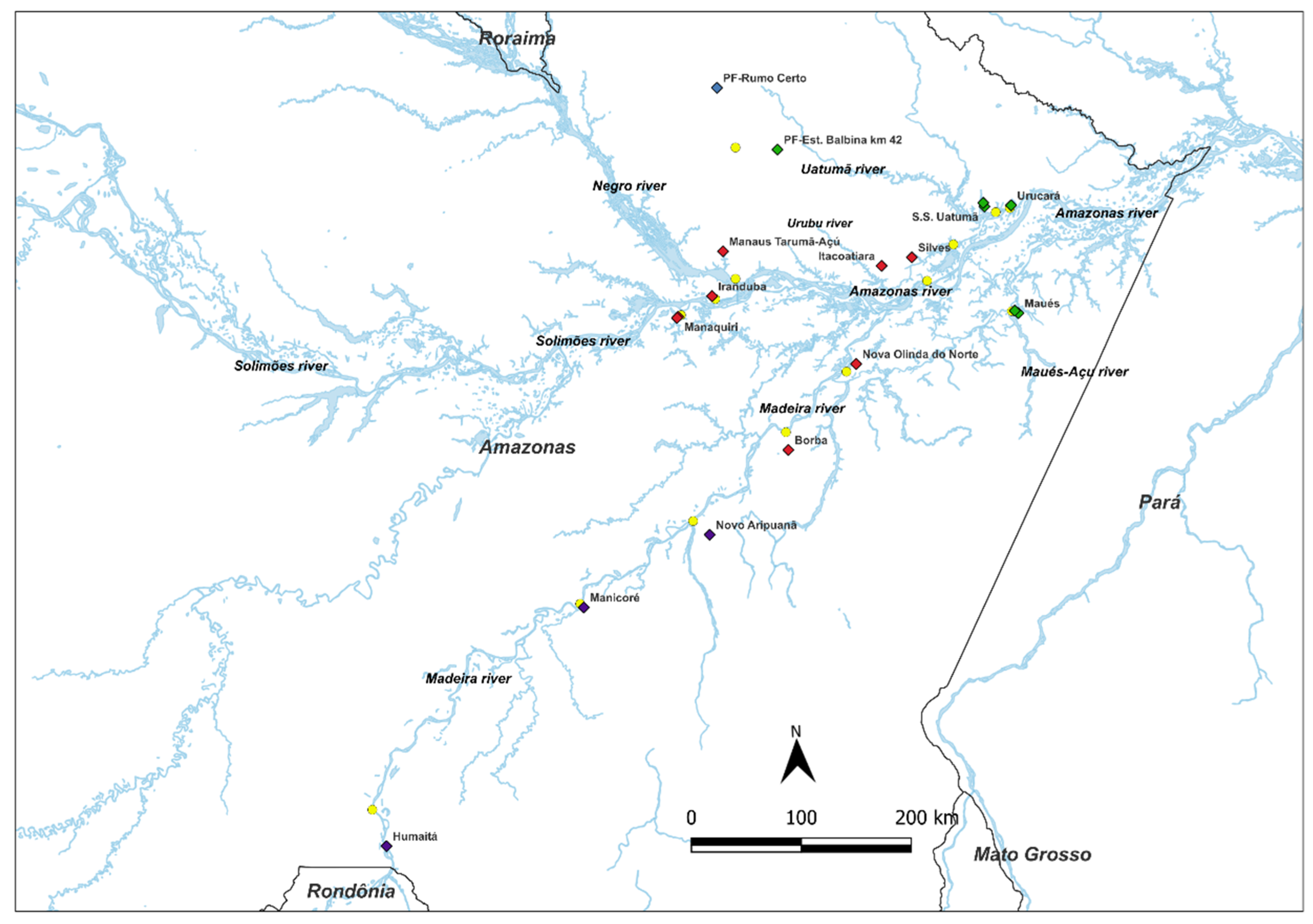
4.1. Study Area and Sampling
4.2. DNA Extraction and Genotyping
4.3. Analysis of Diversity and Genetic Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopes, M.T.G.; Macêdo, J.L.V.D.; Lopes, R.; Leeuwen, J.V.; Ramos, S.L.F.; Bernardes, L. Domestication, and breeding of the Tucum Palm. In Domestication and Breeding: Amazonian Species, 1st ed.; Borém, A., Lopes, M.T.G., Clement, C.R., Noda, H., Eds.; Editora UFV: Viçosa, Brazil, 2012; Volume 1, pp. 421–436. [Google Scholar]
- Casas, L.L.; de Jesus, R.P.; de Queiroz Costa Neto, P.; Corrêa, S.A.M. Nutritional, chemical and pharmacological aspects of Tucumã (Astrocaryum aculeatum Meyer and Astrocaryum vulgare Mart.). Braz. J. Dev. 2022, 8, 13667–13687. [Google Scholar] [CrossRef]
- Araújo, N.M.P.; Arruda, H.S.; Marques, D.R.P.; Oliveira, W.Q.; Pereira, G.A.; Pastore, G.M. Functional and nutritional properties of selected Amazon fruits: A review. Food Res. Int. 2021, 147, 110520. [Google Scholar] [CrossRef] [PubMed]
- Jantsch, M.H.; Bernardes, V.M.; Oliveira, J.S.; Passos, D.F.; Dornelles, G.L.; Manzoni, A.G.; Cabral, F.L.; Silva, J.L.G.; Schetinger, M.R.C.; Leal, D.B.R. Tucumã (Astrocaryum aculeatum) prevents memory loss and oxidative imbalance in the brain of rats with hyperlipidemia. J. Food Biochem. 2021, 45, e13636. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.P.F.; Nascimento, R.P.; Alves, M.R.; Reguengo, L.M.; Marostica Junior, M.R. Brazilian tucumã-do-Amazonas (Astrocaryum aculeatum) and tucuma-do-Pará (Astrocaryum vulgare) fruits: Bioactive composition, health benefits, and technological potential. Food Res. Int. 2022, 151, 110902. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.L.F.; Lopes, M.T.G.; Lopes, R.; Cunha, R.N.V.; Macêdo, J.L.V.; Contim, L.A.S.; Clement, C.R.; Rodrigues, D.P.; Bernardes, L.G. Determination of the mating system of Tucumã palm using microsatellite markers. Crop Breed. Appl. Biotechnol. 2011, 11, 181–185. [Google Scholar] [CrossRef]
- Barbosa, B.S.; Koolen, H.H.F.; Barreto, A.C.; Silva, J.D.; Figlluolo, R.; Nunomura, S.M. The use of tucumã of amazonas kernel oil in the biodiesel production. Acta Amaz. 2009, 39, 371–376. [Google Scholar] [CrossRef]
- Lira, C.S.; Berruti, F.M.; Palmisano, P.; Berruti, F.; Briens, C.; Pécora, A.A.B. Fast pyrolysis of Amazon tucumã (Astrocaryum aculeatum) seeds in a bubbling fluidized bed reactor. JAAP 2013, 99, 23–31. [Google Scholar] [CrossRef]
- Freitas, F.A.D.; Mendonça, I.R.S.; Barros, S.D.S.; Pessoa, W.G.A.; Sá, I.S.C.; Gato, L.B.; Silva, E.P.; Farias, M.A.S.; Nobre, F.X.; Maia, P.J.S.; et al. Biodiesel production from tucumã (Astrocaryum aculeatum Meyer) almond oil applying the electrolytic paste of spent batteries as a catalyst. Renew. Energy 2022, 191, 919–931. [Google Scholar] [CrossRef]
- Ramos, S.L.F.; de Macêdo, J.L.V.; Martins, C.C.; Lopes, R.; Lopes, M.T.G. Tratamentos pré-germinativos e procedência de sementes do tucumã-do-Amazonas para a produção de mudas. Rev. Bras. Frut. 2011, 33, 962–969. [Google Scholar] [CrossRef][Green Version]
- Ramos, S.L.F.; Dequigiovanni, G.; Sebbenn, A.M.; Lopes, M.T.G.; Kageyama, P.Y.; Macêdo, J.L.V.D.; Veasey, E.A. Spatial genetic structure, genetic diversity and pollen dispersal in a harvested population of Astrocaryum aculeatum in the Brazilian Amazon. BMC Genet. 2016, 17, 63. [Google Scholar] [CrossRef]
- Brazilian Institute of Geography and Statistics. Panorama of Cities and States. Available online: https://cidades.ibge.gov.br/brasil/am/manaus/panorama (accessed on 15 March 2022).
- Ramos, S.L.F.; Dequigiovanni, G.; Sebbenn, A.M.; Lopes, M.T.G.; Macêdo, J.L.V.; Veasey, E.A.; Alves-Pereira, A.; Silva, P.P.; Garcia, J.N.; Kageyama, P.Y. Paternity analysis, pollen flow, and spatial genetic structure of a natural population of Euterpe precatoria in the Brazilian Amazon. Ecol. Evol. 2018, 8, 11143–11157. [Google Scholar] [CrossRef] [PubMed]
- Novello, M.; Viana, J.P.G.; Alves-Pereira, A.; Silvestre, E.A.; Nunes, H.F.; Pinheiro, J.B.; Zucchi, M.I. Genetic conservation of a threatened Neotropical palm through community-management of fruits in agroforests and second-growth Forests. For. Ecol. Manag. 2018, 407, 200–209. [Google Scholar] [CrossRef]
- Schroth, G.; Mota, M.S.S.; Lopes, R.; Freitas, A.F. Extractive use, management and in situ domestication of a weedy palm, Astrocaryum tucuma, in the Central Amazon. For. Ecol. Manag. 2004, 202, 161–179. [Google Scholar] [CrossRef]
- Ouborg, N.J.; Pertoldi, C.; Loeschcke, V.; Bijlsma, R.; Hedrick, P.W. Conservation genetics in transition to conservation genomics. Trends Genet. 2010, 26, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.L.F.; Macedo, J.L.V.; Lopes, M.T.G.; Batista, J.S.; Formiga, K.M.; Da Silva, P.P.; Saulo-Machado, A.C.; Veasey, E.A. Microsatellite loci for tucumã of Amazonas (Astrocaryum aculeatum) and amplification in other Arecaceae. Am. J. Bot. 2012, 99, e508–e510. [Google Scholar] [CrossRef][Green Version]
- Oliveira, N.P.; Oliveira, M.S.P.; Davide, L.C.; Kalisz, S. Population genetic structure of three species in the genus Astrocaryum G. Mey. (Arecaceae). Genet. Mol. Res. 2017, 16, gmr16039676. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Nazareno, A.G.; dos Reis, M.S. At Risk of Population Decline? An Ecological and Genetic Approach to the Threatened Palm Species Butia eriospatha (Arecaceae) of Southern Brazil. J. Hered. 2014, 105, 120–129. [Google Scholar] [CrossRef]
- Eguiarte, L.E.; Perez-Nasser, N.; Piñero, D. Genetic—Structure, outcrossing rate and heterosis in Astrocaryum mexicanum (Tropical Palm): Implications for evolution and conservation. Heredity 1992, 69, 217–228. [Google Scholar] [CrossRef]
- Silva, M.S.; Vieira, F.A.; Carvalho, D. Diversity and genetic structure in natural population of Geonoma schottian Mart (ARECACEAE): Implications for conservation. Cerne 2011, 17, 195–201. [Google Scholar] [CrossRef][Green Version]
- Arabnezhad, H.; Bahar, M.; Mohammadi, H.R.; Latifian, M. Development, characterization and use of microsatellite markers for germplasm analysis in date palm (Phoenix dactylifera L.). Sci. Hortic. 2012, 134, 150–156. [Google Scholar] [CrossRef]
- Ottewell, K.; Grey, E.; Castillo, F.; Karubian, J. The pollen dispersal kernel and mating system of an insect-pollinated tropical palm, Oenocarpus bataua. Heredity 2012, 109, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.S.P.D.; Santos, J.B.D.; Amorim, E.P.; Ferreira, D.F. Genetic variability among accessions of assai palm based on microsatellite markers. Cienc. Agrotecnol. 2010, 34, 1253–1260. [Google Scholar] [CrossRef][Green Version]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Vallejo, M.I.; Galeano, G.; Bernal, R.; Zuidema, P.A. The fate of populations of Euterpe oleracea harvested for palm heart in Colombia. For. Ecol. Manag. 2014, 318, 274–284. [Google Scholar] [CrossRef]
- Kottek, M.; Grieser, J.; Beck, C.; Rudolf, B.; Rubel, F. World Map of the Köppen-Geiger climate classification updated. Meteorol. Z. 2006, 15, 259–263. [Google Scholar] [CrossRef]
- Anderson, J.T.; Willis, J.H.; Mitchell-Olds, T. Evolutionary genetics of plant adaptation. Trends Genet. 2011, 27, 258–266. [Google Scholar] [CrossRef]
- Hartl, D.L.; Clark, A.G. Princípios de Genética de Populações, 4th ed.; Artmed Editora: Porto Alegre, Brazil, 2010; 660p. [Google Scholar]
- Ramos, S.L.F.; Dequigiovanni, G.; Lopes, M.T.G.; Aguiar, A.V.D.; Lopes, R.; Veasey, E.A.; Macêdo, J.L.V.D.; Alves-Pereira, A.; Fraxe, T.D.J.P.; Wrege, M.S.; et al. Genetic Structure in Populations of Euterpe precatoria Mart. in the Brazilian Amazon. Front. Ecol. Evol. 2021, 8, 603448. [Google Scholar] [CrossRef]
- Pickersgill, B. Domestication of plants in the Americas: Insights from Mendelian and molecular genetics. Ann. Bot. 2007, 100, 925–940. [Google Scholar] [CrossRef]
- Leducq, J.B.; Llaurens, V.; Castric, V.; Saumitou-Laprade, P.; Hardy, O.J.; Vekemans, X. Effect of balancing selection on spatial genetic structure within populations: Theoretical investigations on the self-incompatibility locus and empirical studies in Arabidopsis halleri. Heredity 2011, 106, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Freeland, J.R.; Kirk, H.; Petersen, S. Molecular Ecology, 2nd ed.; Wiley & Sons: Chichester, UK, 2011; 449p. [Google Scholar]
- Pannell, J.R.; Fields, P.D. Evolution in subdivided plant populations: Concepts, recent advances and future directions. New Phytol. 2014, 201, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Caviglia-Harris, J.L.; Sills, E.O. Land use and income diversification: Comparing traditional and colonist populations in the Brazilian Amazon. Agric. Econ. 2005, 32, 221–237. [Google Scholar] [CrossRef]
- Egler, M.; Egler, C.A.G.; Franz, B.; de Araújo, M.S.M.; de Freitas, M.A.V. Indicators of deforestation in the Southern Brazilian Pre-Amazon. Reg. Environ. Change 2013, 13, 263–271. [Google Scholar] [CrossRef]
- Lieberman, M.; Lieberman, D. Nearest-neighbor tree species combinations in tropical forest: The role of chance, and some consequences of high diversity. Oikos 2007, 116, 377–386. [Google Scholar] [CrossRef]
- Altieri, M.A.; Nicholls, C.I. The adaptation and mitigation potential of traditional agriculture in a changing climate. Clim. Change 2017, 140, 33–45. [Google Scholar] [CrossRef]
- Jorge, M.L.S.P.; Howe, H.F. Can forest fragmentation disrupt a conditional mutualism? A case from central Amazon. Oecologia 2009, 161, 709–718. [Google Scholar] [CrossRef]
- Barluenga, M.; Austerlitz, F.; Elzinga, J.A.; Teixeira, S.; Goudet, J.; Bernasconi, G. Fine-scale spatial genetic structure and gene dispersal in Silene latifolia. Heredity 2011, 106, 13–24. [Google Scholar] [CrossRef]
- Corrêa, C.E.; Fischer, E.; dos Santos, F.A.M. Seed banks on Attalea phalerata (Arecaceae) stems in the Pantanal wetland, Brazil. Ann. Bot. 2012, 109, 729–734. [Google Scholar] [CrossRef]
- Uhl, C. Restoration of degraded lands in the Amazon Basin. In Biodiversity, 1st ed.; Wilson, E.O., Peter, F.M., Eds.; National Academy Press: Washington, DC, USA, 1988; Volume 1, pp. 326–332. [Google Scholar]
- Scariot, A. Forest fragmentation effects on palm diversity in Central Amazonia. J. Ecol. 1999, 87, 66–76. [Google Scholar] [CrossRef]
- Vieira, D.L.M.; Scariot, A. Principles of natural regeneration of tropical dry forests for restoration. Restor. Ecol. 2006, 14, 11–20. [Google Scholar] [CrossRef]
- Uhl, C.; Clark, K.; Clark, H.; Murphy, P. Early plant succession after cutting and burning in the upper Rio Negro region of the Amazon Basin. J. Ecol. 1981, 69, 631–649. [Google Scholar] [CrossRef]
- Jaganathan, G.K. Ecological insights into the coexistence of dormancy and desiccation-sensitivity in Arecaceae species. Ann. For. Sci. 2021, 78, 10. [Google Scholar] [CrossRef]
- Liu, B.; Su, J.; Chen, J.; Cui, G.; Ma, J. Anthropogenic halo disturbances alter landscape and plant richness: A ripple effect. PLoS ONE 2013, 8, e56109. [Google Scholar] [CrossRef]
- Audigeos, D.; Brousseau, L.; Traissac, S.; Scotti-Saintagne, C.; Scotti, I. Molecular divergence in tropical tree populations occupying environmental mosaics. J. Evol. Biol. 2013, 26, 529–544. [Google Scholar] [CrossRef]
- Fearnside, P.M. Brazil’s Balbina Dam: Environment versus the legacy of the Pharaohs in Amazonia. Environ. Manag. 1989, 13, 401–423. [Google Scholar] [CrossRef]
- Futuyma, D.J. Evolution; Sinauer & Associates, Inc.: Sunderland, MA, USA, 2005; 603p. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodohl, P.A. DiveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing tables of statistical test. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistic for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [PubMed]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Smouse, P.E.; Long, J.C.; Sokal, R.R. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 1986, 35, 627–632. [Google Scholar] [CrossRef]
- Chessel, D.; Dufour, A.; Thioulouse, J. The ade4 package—I: One-table methods. R News 2004, 4, 5–10. [Google Scholar]
- Dray, S.; Dufour, A. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Soft. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.; Chessel, D. The ade4 package—II: Two-table and K-table methods. R News 2007, 7, 47–52. [Google Scholar]
- Bougeard, S.; Dray, S. Supervised Multiblock Analysis in R with the ade4 Package. J. Stat. Soft. 2018, 86, 1–17. [Google Scholar] [CrossRef]
- Villordon, A. GIS on the cheap: DIVA-GIS and other free data visualization tools for research. HortScience 2006, 41, 497–520. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P.J. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinform. 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef]
| Municipality—Population | n | AT | A | Ap | Ho | HE | f |
|---|---|---|---|---|---|---|---|
| Humaitá | 15 | 51 | 5.1 | 2 | 0.64 | 0.56 | –0.131 |
| Manicoré | 15 | 48 | 4.8 | 1 | 0.65 | 0.55 | –0.182 |
| Novo Aripuanã | 15 | 48 | 4.8 | 2 | 0.63 | 0.52 | –0.203 |
| Borba | 15 | 45 | 4.5 | 1 | 0.67 | 0.56 | –0.194 |
| Nova Olinda do Norte | 15 | 47 | 4.7 | 2 | 0.68 | 0.57 | –0.204 |
| Manaquiri | 14 | 44 | 4.4 | 1 | 0.69 | 0.52 | –0.317 |
| Iranduba | 13 | 45 | 4.5 | 1 | 0.67 | 0.59 | –0.143 |
| Itacoatiara | 15 | 49 | 4.9 | 2 | 0.55 | 0.55 | –0.015 |
| Silves | 15 | 43 | 4.3 | - | 0.63 | 0.53 | –0.195 |
| Maués | 15 | 44 | 4.4 | 1 | 0.62 | 0.58 | –0.077 |
| Urucará | 15 | 42 | 4.2 | 1 | 0.62 | 0.53 | –0.162 |
| São Sebastião do Uatumã (S.S. Uatumã) | 14 | 42 | 4.2 | 1 | 0.65 | 0.53 | –0.231 |
| PF–Rumo Certo | 15 | 35 | 3.5 | 1 | 0.55 | 0.42 | –0.296 |
| PF–Est. Balbina km 42 | 14 | 47 | 4.7 | - | 0.74 | 0.56 | –0.322 |
| Manaus Tarumã-Açú | 13 | 39 | 3.9 | 1 | 0.59 | 0.50 | –0.197 |
| Average | - | - | 4.5 | - | 0.64 | 0.54 | - |
| FIS | FST | FIT | |
|---|---|---|---|
| Below all loci | –0.1521 | 0.0700 | –0.0714 |
| Upper (CI95%) | –0.1277 | 0.0825 | –0.0477 |
| Lower (CI95%) | –0.1772 | 0.0587 | –0.0950 |
| Sampled Locations | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | P12 | P13 | P14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P2 = Manicoré | 0.0214 | |||||||||||||
| P3 = Novo Aripuanã | 0.0557 * | 0.0478 * | ||||||||||||
| P4 = Borba | 0.0276 | 0.0427 * | 0.0610 * | |||||||||||
| P5 = Nova Olinda do Norte | 0.0557 * | 0.0769 * | 0.0951 * | 0.0442 * | ||||||||||
| P6 = Manaquiri | 0.0460 * | 0.0439 * | 0.0503 * | 0.0317 * | 0.0464 * | |||||||||
| P7 = Iranduba | 0.0705 * | 0.0767 * | 0.0831 * | 0.0586 * | 0.0586 * | 0.0355 * | ||||||||
| P8 = Itacoatiara | 0.0718 * | 0.0834 * | 0.1278 * | 0.0512 * | 0.0407 * | 0.0557 * | 0.0629 * | |||||||
| P9 = Silves | 0.0711 * | 0.0756 * | 0.1140 * | 0.0374 * | 0.0219 | 0.0285 * | 0.0393 * | 0.0421 * | ||||||
| P10 = Maués | 0.0520 * | 0.0735 * | 0.1110 * | 0.0332 * | 0.0342 * | 0.0574 * | 0.0245 | 0.0554 * | 0.0185 | |||||
| P11 = Urucará | 0.0682 * | 0.0698 * | 0.1041 * | 0.0618 * | 0.0249 | 0.0597 * | 0.0545 * | 0.0633 * | 0.0124 | 0.0177 | ||||
| P12 = S.S. Uatumã | 0.0832 * | 0.0864 * | 0.0883 * | 0.0603 * | 0.0377 * | 0.0656 * | 0.0476 * | 0.0727 * | 0.0335 | 0.0296 | 0.0142 | |||
| P13 = PF–Rumo Certo | 0.1535 * | 0.1661 * | 0.1668 * | 0.1807 * | 0.1411 * | 0.1519 * | 0.1594 * | 0.1835 * | 0.1612 * | 0.1556 * | 0.1505 * | 0.1180 * | ||
| P14 = PF–Est. Balbina km 42 | 0.0517 * | 0.0671 * | 0.0530 * | 0.0561 * | 0.0595 * | 0.0094 | 0.0335 | 0.0845 * | 0.0547 * | 0.0542 * | 0.0734 * | 0.0609 * | 0.1177 * | |
| P15 = Manaus Tarumã-Açú | 0.0729 * | 0.0677 * | 0.0990 * | 0.0585 * | 0.0649 * | 0.0224 * | 0.0248 | 0.0814 * | 0.0520 * | 0.0518 * | 0.0757 * | 0.0683 * | 0.1527 * | 0.0328 |
| Watershed | RMa | RSo | RAm | RUr | Rmu | RUa |
|---|---|---|---|---|---|---|
| RSo = Solimões River | 0.0264 | |||||
| Ram = Amazon River | 0.0337 | 0.0331 | ||||
| RUr = Urubu River | 0.0402 | 0.0238 | 0.0087 * | |||
| Rmu = Maues-Açu River | 0.0412 | 0.0339 | 0.0208 * | 0.0185 * | ||
| RUa = Uatumã River | 0.0439 | 0.0378 | 0.0499 | 0.0508 | 0.0526 | |
| RNe = Negro River | 0.0465 | 0.0127 * | 0.0588 | 0.0520 | 0.0518 | 0.0491 |
| Source of Variation | df | MS | Est. Var. | PV (%) |
|---|---|---|---|---|
| Among populations (Among Pops) | 14 | 8.4043 | 0.192 | 6.38 |
| Within populations (Within Pops) | 421 | 2.8200 | 2.820 | 93.62 |
| Total | 435 | 3.012 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, S.L.F.; Lopes, M.T.G.; Meneses, C.; Dequigiovanni, G.; de Macêdo, J.L.V.; Lopes, R.; Sebbenn, A.M.; da Silva, R.F.; de Jesus Pinto Fraxe, T.; Veasey, E.A. Natural Populations of Astrocaryum aculeatum Meyer in Amazonia: Genetic Diversity and Conservation. Plants 2022, 11, 2957. https://doi.org/10.3390/plants11212957
Ramos SLF, Lopes MTG, Meneses C, Dequigiovanni G, de Macêdo JLV, Lopes R, Sebbenn AM, da Silva RF, de Jesus Pinto Fraxe T, Veasey EA. Natural Populations of Astrocaryum aculeatum Meyer in Amazonia: Genetic Diversity and Conservation. Plants. 2022; 11(21):2957. https://doi.org/10.3390/plants11212957
Chicago/Turabian StyleRamos, Santiago Linorio Ferreyra, Maria Teresa Gomes Lopes, Carlos Meneses, Gabriel Dequigiovanni, Jeferson Luis Vasconcelos de Macêdo, Ricardo Lopes, Alexandre Magno Sebbenn, Rogério Freire da Silva, Therezinha de Jesus Pinto Fraxe, and Elizabeth Ann Veasey. 2022. "Natural Populations of Astrocaryum aculeatum Meyer in Amazonia: Genetic Diversity and Conservation" Plants 11, no. 21: 2957. https://doi.org/10.3390/plants11212957
APA StyleRamos, S. L. F., Lopes, M. T. G., Meneses, C., Dequigiovanni, G., de Macêdo, J. L. V., Lopes, R., Sebbenn, A. M., da Silva, R. F., de Jesus Pinto Fraxe, T., & Veasey, E. A. (2022). Natural Populations of Astrocaryum aculeatum Meyer in Amazonia: Genetic Diversity and Conservation. Plants, 11(21), 2957. https://doi.org/10.3390/plants11212957








