Transcriptome and Regional Association Analyses Reveal the Effects of Oleosin Genes on the Accumulation of Oil Content in Brassica napus
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of Oleosin Family Genes in the Genome of B. napus
2.2. Analysis of Cis-Acting Elements in Oleosin Genes of B. napus
2.3. Prediction of TF Binding Sites in BnOLEO Genes
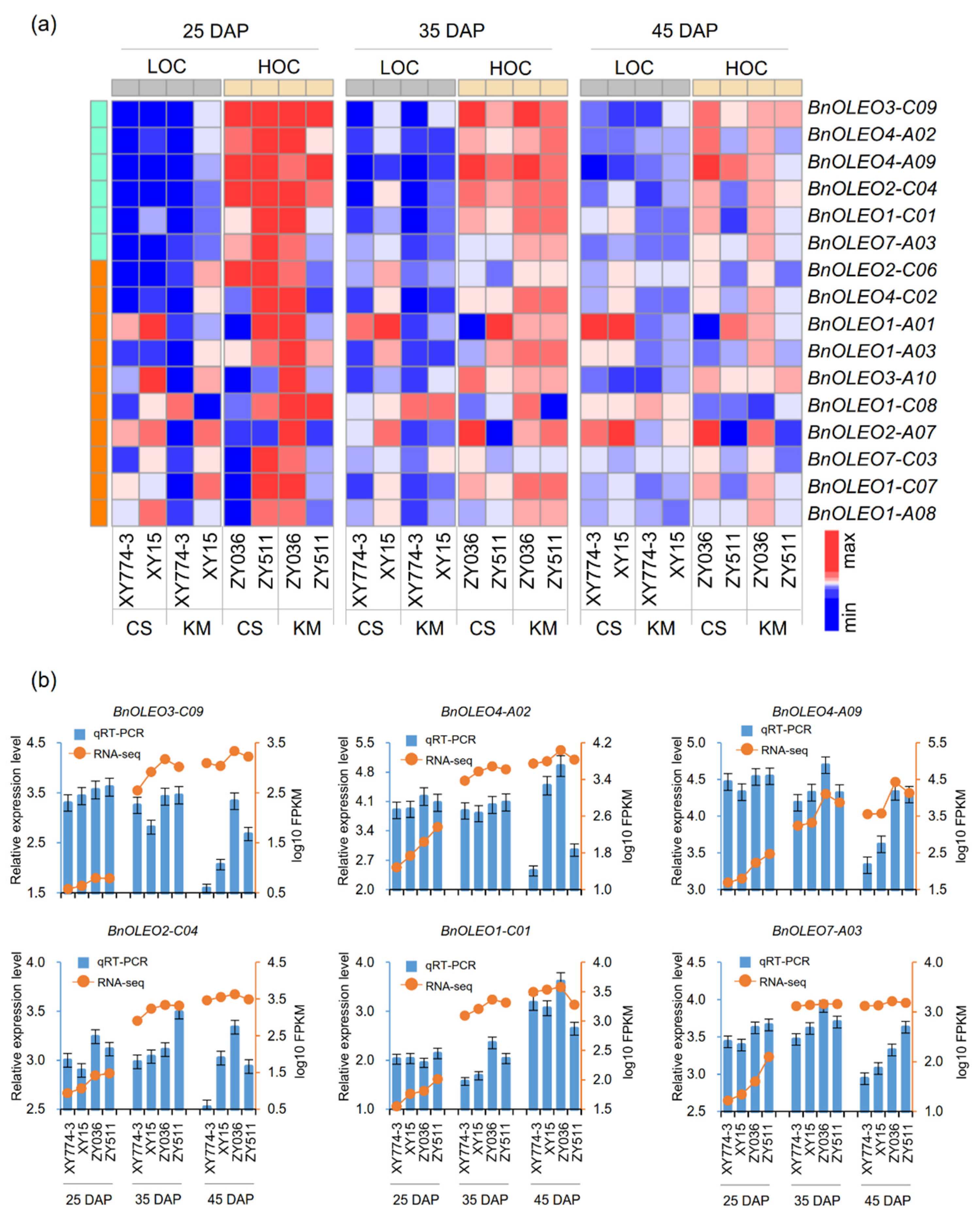
2.4. Expression Analysis of Oleosin Genes in B. napus with High and Low Oil Content
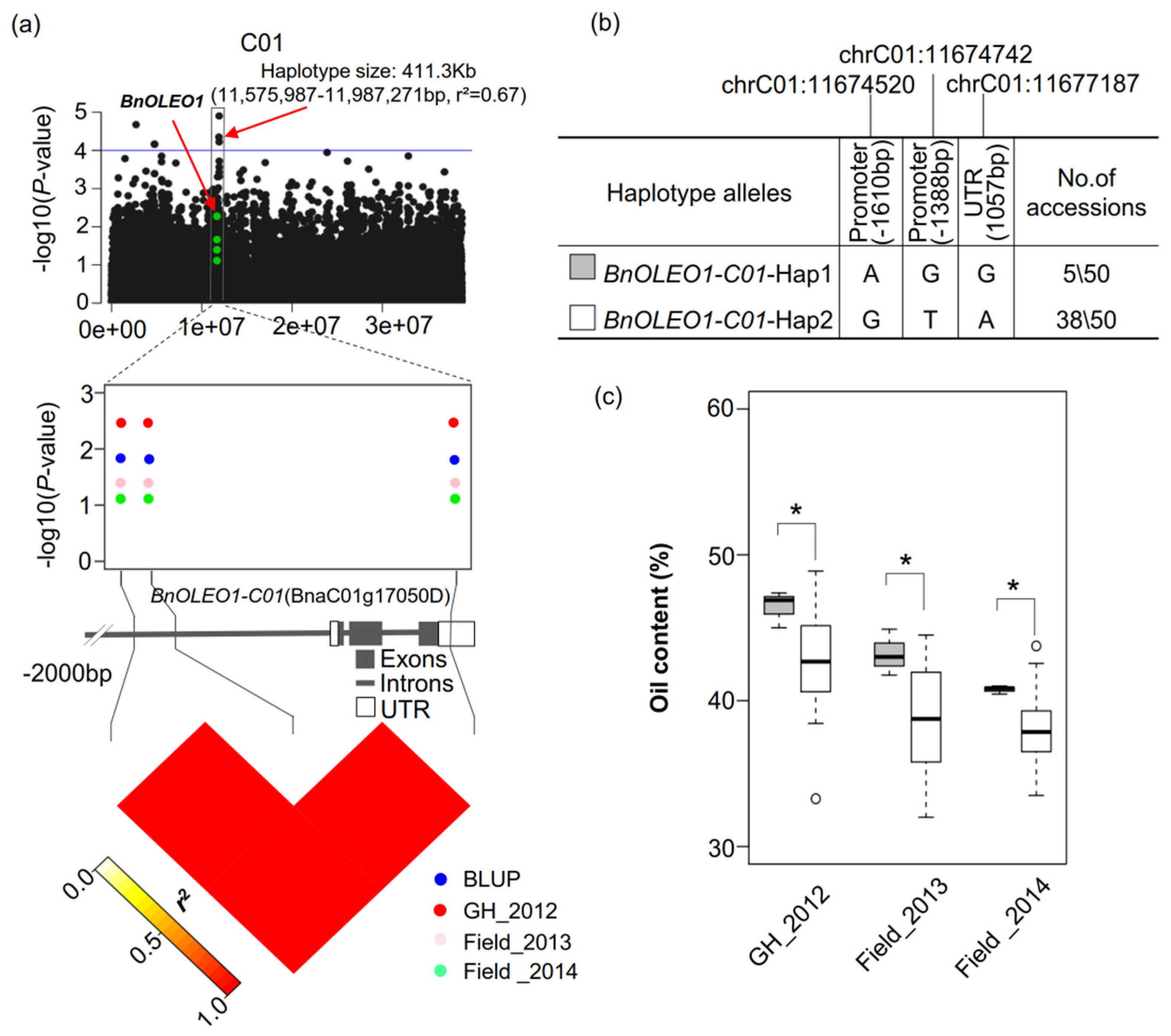
2.5. Regional Association Analysis of Oleosin Genes
2.6. Co-Expression Analysis of Oleosin Genes
3. Discussion
4. Materials and Methods
4.1. Identification of Oleosin Genes in Arabidopsis and B. napus
4.2. Phylogenetic Analysis and Regulatory GENE Prediction of oleosin Genes
4.3. Gene Structure and Conserved Motif Analysis
4.4. Transcriptome Sequencing
4.5. Expression Analysis of Oleosin Genes
4.6. Regional Association Analysis
4.7. Co-Expression Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Usda, E.R.S. Oil Crops Yearbook. 2014. Available online: http://www.ers.usda.gov/data-products/oil-crops-yearbook.aspx (accessed on 27 December 2021).
- Nagaharu, U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot 1935, 7, 389–452. [Google Scholar]
- Meyer, M. Rapeseed oil fuel-the crisis-proof home-made eco-fuel. Agrarforschung 2009, 16, 262–267. [Google Scholar]
- Weselake, R.J.; Taylor, D.C.; Rahman, M.H.; Shah, S.; Laroche, A.; McVetty, P.B.E.; Harwood, J.L. Increasing the flow of carbon into seed oil. Biotechnol. Adv. 2009, 27, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Mekhedov, S.; de Ilárduya, O.M.; Ohlrogge, J. Toward a functional catalog of the plant genome. A survey of genes for lipid biosynthesis. Plant Physiol. 2000, 122, 389–402. [Google Scholar] [CrossRef]
- Baud, S.; Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 2010, 49, 235–249. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Burns, M.J.; Barnes, S.R.; Bowman, J.G.; Clarke, M.H.E.; Werner, C.P.; Kearsey, M.J. QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) Seed oil content and fatty acid composition. Heredity 2003, 90, 39–48. [Google Scholar] [CrossRef]
- Delourme, R.; Falentin, C.; Huteau, V.; Clouet, V.; Horvais, R.; Gandon, B.; Specel, S.; Hanneton, L.; Dheu, J.E.; Deschamps, M. Genetic control of oil content in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2006, 113, 1331–1345. [Google Scholar] [CrossRef]
- Qiu, D.; Morgan, C.; Shi, J.; Long, Y.; Liu, J.; Li, R.; Zhuang, X.; Wang, Y.; Tan, X.; Dietrich, E. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor. Appl. Genet. 2006, 114, 67–80. [Google Scholar] [CrossRef]
- Chen, G.; Geng, J.; Rahman, M.; Liu, X.; Tu, J.; Fu, T.; Li, G.; McVetty, P.B.E.; Tahir, M. Identification of QTL for oil content, seed yield, and flowering time in oilseed rape (Brassica napus). Euphytica 2010, 175, 161–174. [Google Scholar] [CrossRef]
- Sun, M.; Hua, W.; Liu, J.; Huang, S.; Wang, X.; Liu, G.; Wang, H. Design of new genome-and gene-sourced primers and identification of QTL for seed oil content in a specially high-oil Brassica napus cultivar. PLoS ONE 2012, 7, e47037. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Huang, J.; Chen, F.; Xu, F.; Ni, X.; Xu, H.; Wang, Y.; Jiang, C.; Wang, H.; Xu, A. Molecular mapping of Arabidopsis thaliana lipid-related orthologous genes in Brassica napus. Theor. Appl. Genet. 2012, 124, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Shi, J.; Li, R.; Long, Y.; Wang, H.; Li, D.; Zhao, J.; Meng, J. Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.). Theor. Appl. Genet. 2014, 127, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.; Wang, H.; Wang, X.; Guo, L.; Gu, J.; Zhao, W.; Li, B.; Chen, D.; Raboanatahiry, N.; Li, M. Genetic dissection of seed oil and protein content and identification of networks associated with oil content in Brassica napus. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Liu, S.; Fan, C.; Li, J.; Cai, G.; Yang, Q.; Wu, J.; Yi, X.; Zhang, C.; Zhou, Y. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor. Appl. Genet. 2016, 129, 1203–1215. [Google Scholar] [CrossRef]
- Körber, N.; Bus, A.; Li, J.; Parkin, I.A.P.; Wittkop, B.; Snowdon, R.J.; Stich, B. Agronomic and seed quality traits dissected by genome-wide association mapping in Brassica napus. Front. Plant Sci. 2016, 7, 386. [Google Scholar] [CrossRef]
- Tang, S.; Zhao, H.; Lu, S.; Yu, L.; Zhang, G.; Zhang, Y.; Yang, Q.Y.; Zhou, Y.; Wang, X.; Ma, W. Genome-and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol. Plant 2021, 14, 470–487. [Google Scholar] [CrossRef]
- Xiao, Z.; Zhang, C.; Tang, F.; Yang, B.; Zhang, L.; Liu, J.; Huo, Q.; Wang, S.; Li, S.; Wei, L. Identification of candidate genes controlling oil content by combination of genome-wide association and transcriptome analysis in the oilseed crop Brassica napus. Biotechnol. Biofuels 2019, 12, 1–16. [Google Scholar] [CrossRef]
- Yao, M.; Guan, M.; Yang, Q.; Huang, L.; Xiong, X.; Jan, H.U.; Voss-Fels, K.P.; Werner, C.R.; He, X.; Qian, W. Regional association analysis coupled with transcriptome analyses reveal candidate genes affecting seed oil accumulation in Brassica napus. Theor. Appl. Genet. 2021, 134, 1545–1555. [Google Scholar] [CrossRef]
- Yip, A.M.; Horvath, S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinform. 2007, 8, 22. [Google Scholar] [CrossRef]
- Liu, W.; Lin, L.; Zhang, Z.; Liu, S.; Gao, K.; Lv, Y.; Tao, H.; He, H. Gene co-expression network analysis identifies trait-related modules in Arabidopsis thaliana. Planta 2019, 249, 1487–1501. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zeng, X.; Xiong, Q.; Wei, D.; Liao, J.; Xu, Y.; Chen, G.; Zhou, Y.; Dong, H.; Wan, H. Combining quantitative trait locus and co-expression analysis allowed identification of new candidates for oil accumulation in rapeseed. J. Exp. Bot. 2021, 72, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Liu, A.; Zhang, Z.; Ge, Q.; Fan, S.; Gong, W.; Li, J.; Gong, J.; Shi, Y.; Tian, B. Co-expression network analysis and hub gene selection for high-quality fiber in upland cotton (Gossypium hirsutum) using RNA sequencing analysis. Genes 2019, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Zhang, Z.; Wang, Z.; Yu, J.; Qin, H.; Mao, X.; Jiang, H.; Xin, D.; Yin, Z.; Zhu, R. Meta-analysis and transcriptome profiling reveal hub genes for soybean seed storage composition during seed development. Plant Cell Environ. 2018, 41, 2109–2127. [Google Scholar] [CrossRef]
- Capuano, F.; Beaudoin, F.; Napier, J.A.; Shewry, P.R. Properties and exploitation of oleosins. Biotechnol. Adv. 2007, 25, 203–206. [Google Scholar] [CrossRef]
- Huang, M.D.; Huang, A.H.C. Bioinformatics reveal five lineages of oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants. Plant Physiol. 2015, 169, 453–470. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.H.C. Plant lipid droplets and their associated proteins: Potential for rapid advances. Plant Physiol. 2018, 176, 1894–1918. [Google Scholar] [CrossRef]
- Chen, K.; Yin, Y.; Liu, S.; Guo, Z.; Zhang, K.; Liang, Y.; Zhang, L.; Zhao, W.; Chao, H.; Li, M. Genome-wide identification and functional analysis of oleosin genes in Brassica napus L. BMC Plant Biol. 2019, 19, 294. [Google Scholar] [CrossRef]
- Huang, A.H.C. Oleosins and Oil Bodies in Seeds and Other Organs. Plant Physiol. 1996, 110, 1055–1061. [Google Scholar] [CrossRef]
- Krahmer, N.; Guo, Y.; Wilfling, F.; Hilger, M.; Lingrell, S.; Heger, K.; Newman, H.W.; Schmidt-Supprian, M.; Vance, D.E.; Mann, M. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP: Phosphocholine cytidylyltransferase. Cell Metab. 2011, 14, 504–515. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Chou, Y.R.; Wang, C.-S.; Tseng, T.H.; Chen, L.J.; Tzen, J.T.C. Different effects on triacylglycerol packaging to oil bodies in transgenic rice seeds by specifically eliminating one of their two oleosin isoforms. Plant Physiol. Biochem. 2010, 48, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Siloto, R.M.P.; Findlay, K.; Lopez-Villalobos, A.; Yeung, E.C.; Nykiforuk, C.L.; Moloney, M.M. The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 2006, 18, 1961–1974. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, H.; Hu, Z.; Chu, S.; Yu, K.; Lv, L.; Yang, Y.; Zhang, X.; Chen, X.; Kan, G. Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet. 2019, 15, e1008267. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.L.; Shimada, T.; Takahashi, H.; Fukao, Y.; Hara-Nishimura, I. A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 2008, 55, 798–809. [Google Scholar] [CrossRef]
- Li, Y.; Beisson, F.; Pollard, M.; Ohlrogge, J. Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 2006, 67, 904–915. [Google Scholar] [CrossRef]
- Ashfaq, M.; Khan, S. Role of phytohormones in improving the yield of oilseed crops. In Oils Seed Crops: Yield and Adaptation under Environmental Stress, 1st ed.; Ahmad, P., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 165–183. [Google Scholar]
- Menkens, A.E.; Schindler, U.; Cashmore, A.R. The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 1995, 20, 506–510. [Google Scholar] [CrossRef]
- Wu, G.; Gao, Z.; Du, H.; Lin, B.; Yan, Y.; Li, G.; Guo, Y.; Fu, S.; Wei, G.; Wang, M. The effects of abscisic acid, salicylic acid and jasmonic acid on lipid accumulation in two freshwater Chlorella strains. J. Gen. Appl. Microbiol. 2018, 64, 42–49. [Google Scholar] [CrossRef]
- Raboanatahiry, N.; Chao, H.; Guo, L.; Gan, J.; Xiang, J.; Yan, M.; Zhang, L.; Yu, L.; Li, M. Synteny analysis of genes and distribution of loci controlling oil content and fatty acid profile based on QTL alignment map in Brassica napus. BMC Genom. 2017, 18, 776. [Google Scholar] [CrossRef]
- Kumar, N.; Chaudhary, A.; Singh, D.; Teotia, S. Transcriptional regulation of seed oil accumulation in Arabidopsis thaliana: Role of transcription factors and chromatin remodelers. J. Plant Biochem. Biotechnol. 2020, 29, 754–768. [Google Scholar] [CrossRef]
- Elahi, N.; Duncan, R.W.; Stasolla, C. Modification of oil and glucosinolate content in canola seeds with altered expression of Brassica napus LEAFY COTYLEDON1. Plant Physiol. Biochem. 2016, 100, 52–63. [Google Scholar] [CrossRef]
- Cernac, A.; Benning, C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004, 40, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Li, Y.; Gilday, A.D.; Graham, S.; Graham, I.A. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 2006, 18, 1887–1899. [Google Scholar] [CrossRef] [PubMed]
- Alonso, R.; Oñate-Sánchez, L.; Weltmeier, F.; Ehlert, A.; Diaz, I.; Dietrich, K.; Vicente-Carbajosa, J.; Droge-Laser, W. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 2009, 21, 1747–1761. [Google Scholar] [CrossRef]
- Liu, Y.F.; Li, Q.T.; Lu, X.; Song, Q.X.; Lam, S.M.; Zhang, W.K.; Ma, B.; Lin, Q.; Man, W.Q.; Du, W.G. Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol. 2014, 14, 1–16. [Google Scholar] [CrossRef]
- Barthole, G.; To, A.; Marchive, C.; Brunaud, V.; Soubigou-Taconnat, L.; Berger, N.; Dubreucq, B.; Lepiniec, L.; Baud, S. MYB118 represses endosperm maturation in seeds of Arabidopsis. Plant Cell 2014, 26, 3519–3537. [Google Scholar] [CrossRef]
- Bhatla, S.C.; Kaushik, V.; Yadav, M.K. Use of oil bodies and oleosins in recombinant protein production and other biotechnological applications. Biotechnol. Adv. 2010, 28, 293–300. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K. The Pfam protein families database. Nucleic Acids Res. 2010, 38, D211–D222. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Nesi, N.; Lucas, M.O.; Auger, B.; Baron, C.; Lécureuil, A.; Guerche, P.; Kronenberger, J.; Lepiniec, L.; Debeaujon, I.; Renard, M. The promoter of the Arabidopsis thaliana BAN gene is active in proanthocyanidin-accumulating cells of the Brassica napus seed coat. Plant Cell Rep. 2009, 28, 601–617. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Y.; Fang, H.; Shi, H.; Chen, K.; Zhang, Z.; Tan, X. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Mol. Genet. Genom. 2014, 289, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Tan, C.; Li, Y.; He, Y.; Wei, S.; Cui, Y.; Chen, Y.; Wei, D.; Fu, Y.; He, Y. Genome-wide association study reveals both overlapping and independent genetic loci to control seed weight and silique length in Brassica napus. Front. Plant Sci. 2018, 9, 921. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.D. qqman: An R package for visualizing GWAS results using QQ and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Strimmer, K. fdrtool: A versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 2008, 24, 1461–1462. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis (2nd ed.). Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Y.; Yao, M.; He, X.; Xiong, X.; Guan, M.; Liu, Z.; Guan, C.; Qian, L. Transcriptome and Regional Association Analyses Reveal the Effects of Oleosin Genes on the Accumulation of Oil Content in Brassica napus. Plants 2022, 11, 3140. https://doi.org/10.3390/plants11223140
Jia Y, Yao M, He X, Xiong X, Guan M, Liu Z, Guan C, Qian L. Transcriptome and Regional Association Analyses Reveal the Effects of Oleosin Genes on the Accumulation of Oil Content in Brassica napus. Plants. 2022; 11(22):3140. https://doi.org/10.3390/plants11223140
Chicago/Turabian StyleJia, Yuan, Min Yao, Xin He, Xinghua Xiong, Mei Guan, Zhongsong Liu, Chunyun Guan, and Lunwen Qian. 2022. "Transcriptome and Regional Association Analyses Reveal the Effects of Oleosin Genes on the Accumulation of Oil Content in Brassica napus" Plants 11, no. 22: 3140. https://doi.org/10.3390/plants11223140
APA StyleJia, Y., Yao, M., He, X., Xiong, X., Guan, M., Liu, Z., Guan, C., & Qian, L. (2022). Transcriptome and Regional Association Analyses Reveal the Effects of Oleosin Genes on the Accumulation of Oil Content in Brassica napus. Plants, 11(22), 3140. https://doi.org/10.3390/plants11223140






