Abstract
Bacillus subtilis J-15 is a plant growth-promoting rhizobacteria isolated from the soil rhizosphere of cotton and is resistant to cotton verticillium wilt. This study evaluated the effects of metabolites of J-15 (J-15-Ms), including mycosubtilin, on plant growth using Arabidopsis and cotton plants. The results showed that J-15-Ms promoted Arabidopsis seeding growth at lower concentrations of 0.2 μg/mL but inhibited the growth at higher concentrations, such as 20 μg/mL. Similar results were obtained in cotton. Thus, J-15-Ms-treated plants showed low-concentration-induced growth promotion and high-concentration-induced growth inhibition. The J-15-Ms components were analyzed by liquid chromatography–mass spectrometry. Correlation analysis using the J-15 genomic databases suggested that J-15 may synthesize indoleacetic acid via the indole-3-pymvate pathway and indole-3-acetamide pathway. Treatment with mycosubtilin, a purified peptide from J-15-Ms, showed that the peptide promoted Arabidopsis growth at a low concentration (0.1 μg/mL) and inhibited plant growth at high concentrations (higher than 1 μg/mL), which also significantly increased plant lateral root number. Transcriptomic analysis showed that mycosubtilin might promote lateral root development and inhibit plant primary root growth by regulating the expression of the plant hormone signaling pathway. This study reveals the mechanism of Bacillus subtilis J-15 in affecting plant growth.
1. Introduction
Plant rhizosphere soils contain various microorganisms such as bacteria, fungi, actinomycetes, and soil protozoa. These microorganisms include not only pathogenic bacteria that cause damage to plants, but also probiotics that promote host plant growth, nutrient absorption, resistance to stress, and pathogenic bacteria [1]. Among them, the beneficial bacteria that colonize plant rhizosphere and promote plant growth are known as PGPR [2]. PGPR colonizes the plant root system and promotes active plant growth through direct mechanisms, such as nutrient solubilization, nitrogen fixation, and growth production [3], or indirect mechanisms, such as root development stimulation, competitive exclusion of pathogens, or removal of phytotoxic substances [4]. Various PGPR strains, including Agrobacterium, Bacillus, Burkholderia, Pseudomonas, and Serratia, have been identified [5]. These PGPRs promote plant growth, inhibit pathogenic bacteria, and maintain the ecological balance of the root system by regulating the microenvironment of plant growth and development [6]. Root inoculation with Azotobacter chroococcum 76A enhanced the nutrient assimilation efficiency and promoted the growth of tomato plants under salt stress [7]. Bacillus subtilis strain GOT9 enhanced tolerance against drought and salt stresses and improved lateral root growth in Arabidopsis [8]. The rhizobacterial strains BKM20 and BKM04 also promoted Lolium perenne L. growth and improved soil fertility and microbial activity [9]. Pseudomonas sp. P8, Peribacillus sp. P10, and Streptomyces sp. X52 promoted growth and improved the adaptability of maize to salt stress [10]. Furthermore, Bacillus amyloliquefaciens B14 reduced the charcoal root rot incidences in common peas [11]. Enterobacter 64S1 and Pseudomonas 42P4 isolated from tomato roots showed strong phosphate solubilizing activity and promoted the growth of tomato seedlings with reduced fertilizer application [12].
Among the PGPRs, Bacillus is the most studied due to its ease of isolation from soil and plants, broad metabolite profile, rapid growth, and ability to colonize plant surfaces [13]. Bacillus can exert its growth-promoting activities on plants directly by producing plant growth regulators, such as growth hormones, cytokinins, and gibberellins [14], or through nitrogen fixation, phosphate solubilization, and ironophilins production [15]. Most Bacillus species can also synthesize and release indoleacetic acid (IAA) [16]; for example, Bacillus sp. UCMB5113 can produce cytokinin and indole-3-acetic, affecting plant root growth [17]. Bacillus strain J119 produces IAA and iron carriers to promote the growth of oilseed rape, maize, tomato, and other plants [18]. Bacillus megaterium BP17 promotes Arabidopsis growth by downregulating ethylene-responsive genes and upregulating nutrient absorption-related genes [19]. Inoculation of Bacillus subtilis strain L1 promoted nitrate utilization and plant growth [20]. Furthermore, Bacillus HNH7 and HNH9 reportedly promote the growth of land cotton by producing iron carriers to dissolve iron and upregulate cotton growth-related genes [21]. Bacillus aryabhattai LAD exhibits phosphate solubilization and nitrogen fixation activities, which facilitate root development and growth of maize seedlings [22]. Bacillus subtilis EA-CB0575 also promotes tomato plant growth by dissolving phosphorus, repairing nitrogen, and producing indole and iron cell-containing compounds [23]. The volatile compounds excreted by Bacillus subtilis also play an important role in regulating plant growth [24]. The volatiles emitted by Bacillus subtilis (GB03) have a lasting beneficial effect on the growth of Arabidopsis [25].
In addition to hormones, Bacillus also stimulates plant growth indirectly via indirect mechanisms by producing 1-aminocyclopropane-1-carboxylate (ACC) deaminases, antibiotics, cell wall-degrading enzymes, and hydrogen cyanide, as well as by activating host systemic resistance and ferrocarriers [6]. For example, Bacillus subtilis Rhizo SF 48 produces ACC deaminase, which promotes plant growth and induces drought resistance in tomatoes [26]. The beneficial soil bacterium Bacillus subtilis (GB03) treatment enhanced Arabidopsis Choline synthesis and improved plant tolerance to osmotic stress [27]. Bacillus subtilis 5YN8 and DSN012 have high antagonistic and hydrolytic enzyme activities, which significantly inhibit pepper grey mold and promote pepper growth [28]. Moreover, Bacillus subtilis CBR05 can directly inhibit the growth of pathogens and improve pathogen resistance by enhancing the systemic resistance in tomatoes [29]. Many Bacillus species have been reported to produce various antimicrobial compounds from the lipopeptide family [30], including surfactins, iturins, and fengycins, which have strong antagonistic activity against various plant pathogenic fungi [31,32,33]. Bacillus Fcl1 produces a mixture of iturin A and surfactants that act as antifungal agents [34]. Bacillus subtilis 6051 is able to form biofilm on roots and secrete surfactin to protect plants from pathogenic bacteria [35]. Bacillus subtilis SG_JW.03 triggers systemic resistance in plants by secreting antifungal lipopeptides to suppress pathogens and upregulate host plant pathogenesis-related genes [36]. The cyclic lipopeptides (CLPs) of Bacillus subtilis ABS-S14 can activate the plant defense pathways, improve resistance, and inhibit chloromycnosis caused by citrus penicillium [37]. Bacillus subtilis M4 also showed the potential to protect plants from fungal diseases, such as bean sprout wilt and apple gray mold caused by putrescence, via different pathogenic systems through its secreted fungicides [38]. Moreover, mycosubtilin and surfactin secreted by Bacillus subtilis can induce resistance against pathogenic fungal spores in grapes [39]. Despite these disease-resistance activities, the effects of lipopeptides on plant growth are not well studied.
Bacillus subtilis J-15, a probiotic strain isolated from the inter-rhizosphere soil of cotton, produces metabolites (J-15-Ms) that can effectively inhibit the infection by Verticillium dahliae, thus reducing the incidence of verticillium wilt in cotton [40]. J-15-Ms have broad-spectrum resistance to various plant pathogens [41] and have no deleterious effects on the abundance and diversity of soil microbial communities [42]. However, whether the J-15-Ms regulate plant growth and development is unknown. This study investigated the effects of J-15-Ms on the growth and development of Arabidopsis and cotton seedlings. The J-15-Ms metabolites were analyzed by LC-MS. The previous analysis of the J-15 genome found that the major antifungal gene clusters reported in Bacillus subtilis only contained the complete mycosubtilin and bacilibactin manipulators, suggesting that the two could be the major antifungal active substances [43]. Another study also isolated and purified the J-15-Ms and identified the antagonist as mycosubtilin [44]. Based on this, we investigate the mycosubtilin plant growth regulatory mechanism and provide a theoretical basis for applying the J-15 strain in plant growth improvement.
2. Results
2.1. Effect of J-15-Ms on Arabidopsis Growth
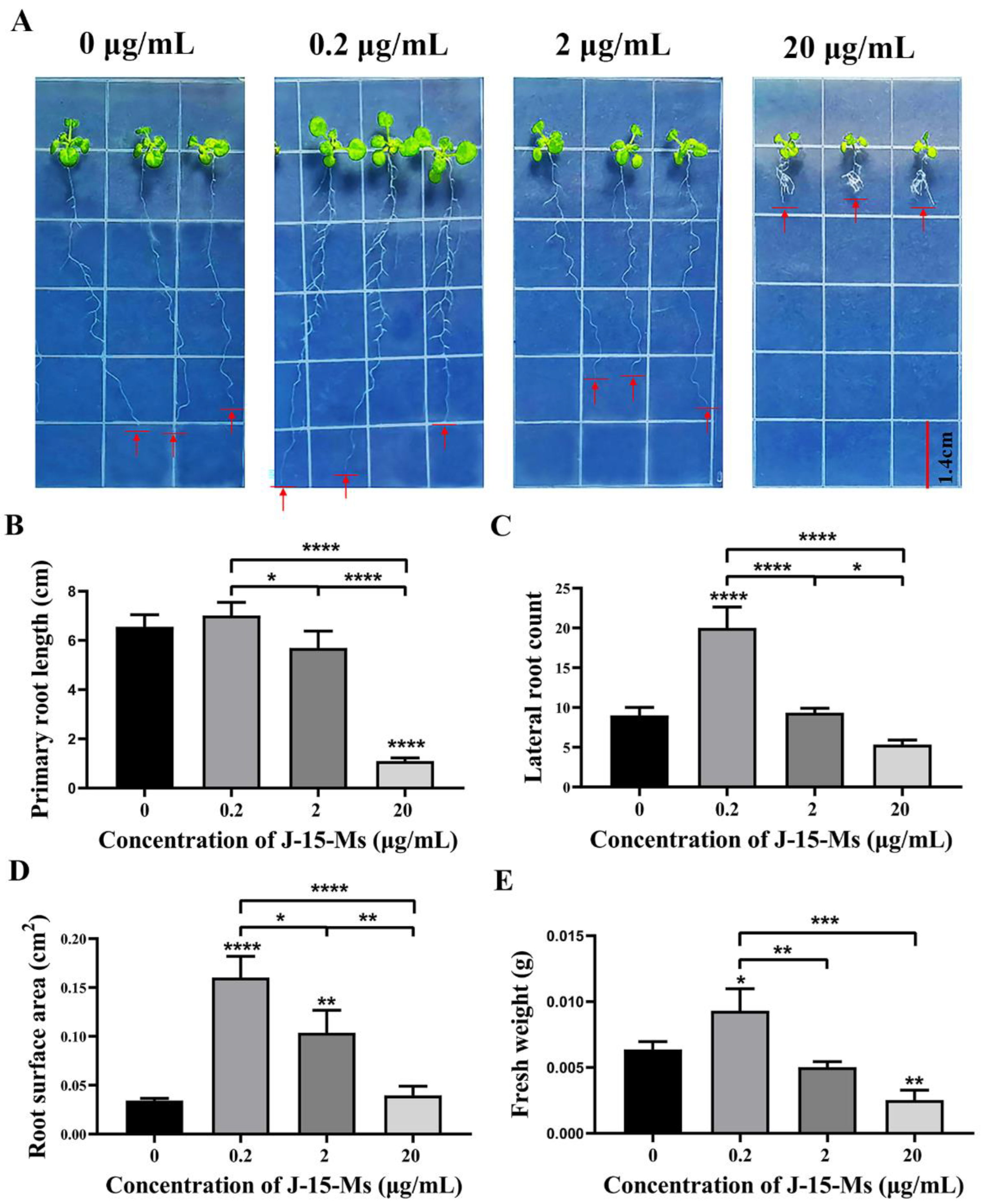
After treating Arabidopsis seedlings with J-15-Ms for 10 days, we found that Arabidopsis growth was promoted at lower J-15-Ms concentrations but inhibited at higher concentrations. At 0.2 μg/mL, the J-15-Ms promoted the growth of Arabidopsis primary roots, significantly increasing their root surface area and fresh weight and the number of lateral. However, when the concentration was increased to 2 μg/mL, J-15-Ms inhibited the growth of Arabidopsis primary roots and the development of Arabidopsis lateral root (Figure 1). These findings suggest that J-15-Ms acted in a concentration-dependent manner; the higher the concentration, the more the inhibition of Arabidopsis primary root, an effect that was more obvious at 20 μg/mL. This suggests the potential of J-15-Ms in controlling plant growth by regulating plant root development.
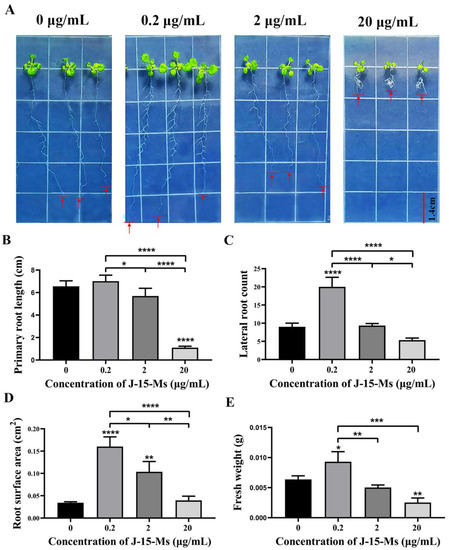
Figure 1.
Effect of J-15-Ms on the growth of Arabidopsis seedlings. (A) Arabidopsis seedling growth phenotype. (B) Effect on primary root length. (C) Effect on lateral root number. (D) Effect on root surface area. (E) Effect on fresh weight. Schemes follow the same formatting. Note: Means ± SEs, n = 50, different “*” indicate significant differences among treatments based on the least significant difference at p ≤ 0.05.
2.2. Effect of J-15-Ms on Cotton Growth
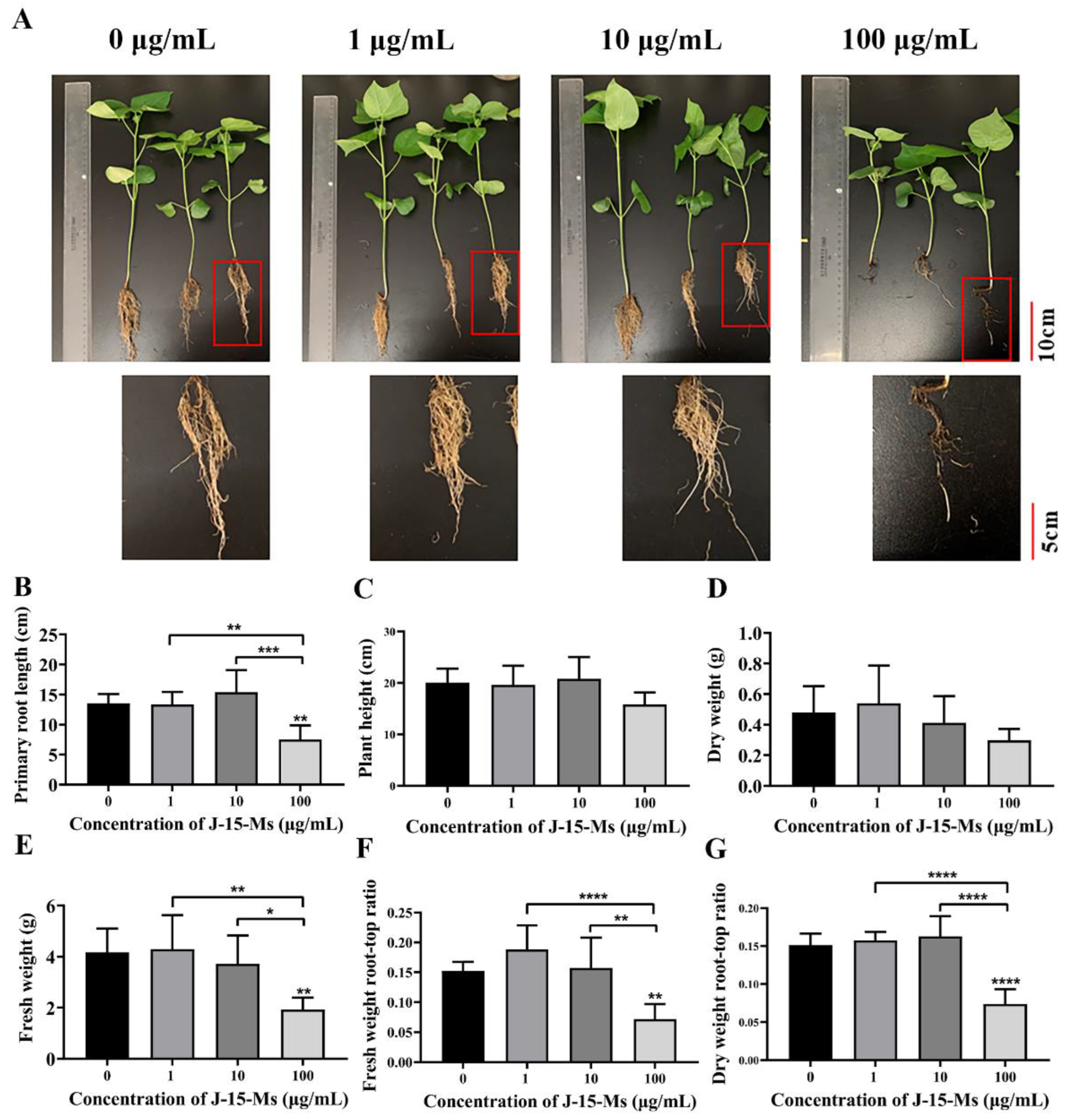
After 21 days of treating cotton seedlings with different J-15-Ms concentrations, the J-15-Ms at 1 μg/mL increased the fresh weight and dry weight as well as the fresh weight root-to-crown ratio in cotton. At 10 μg/mL, J-15-Ms increased the plant height, root length, and dry weight root-to-crown ratio in cotton; however, the growth of cotton was significantly inhibited at J-15-Ms concentration of 100 μg/mL (Figure 2). Thus, similar to Arabidopsis, J-15-Ms promoted cotton growth at lower concentrations but inhibited it at higher concentrations.
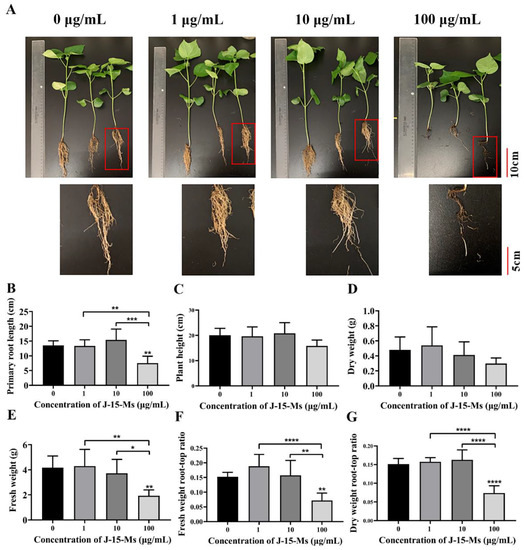
Figure 2.
Effects of J-15-Ms on the growth of cotton seedlings. (A) Cotton seedling growth phenotype. (B) Effect on primary root length. (C) Effect on plant height. (D) Effect on dry weight. (E) Effect on fresh weight. (F) Effect on fresh weight root-top ratio. (G) Effect on dry weight root-top ratio. Note: Means ± SEs, n = 50, different “*” indicate significant differences among treatments based on the least significant difference at p ≤ 0.05.
2.3. LC-MS Identification and Taxonomic Analysis of J-15-Ms
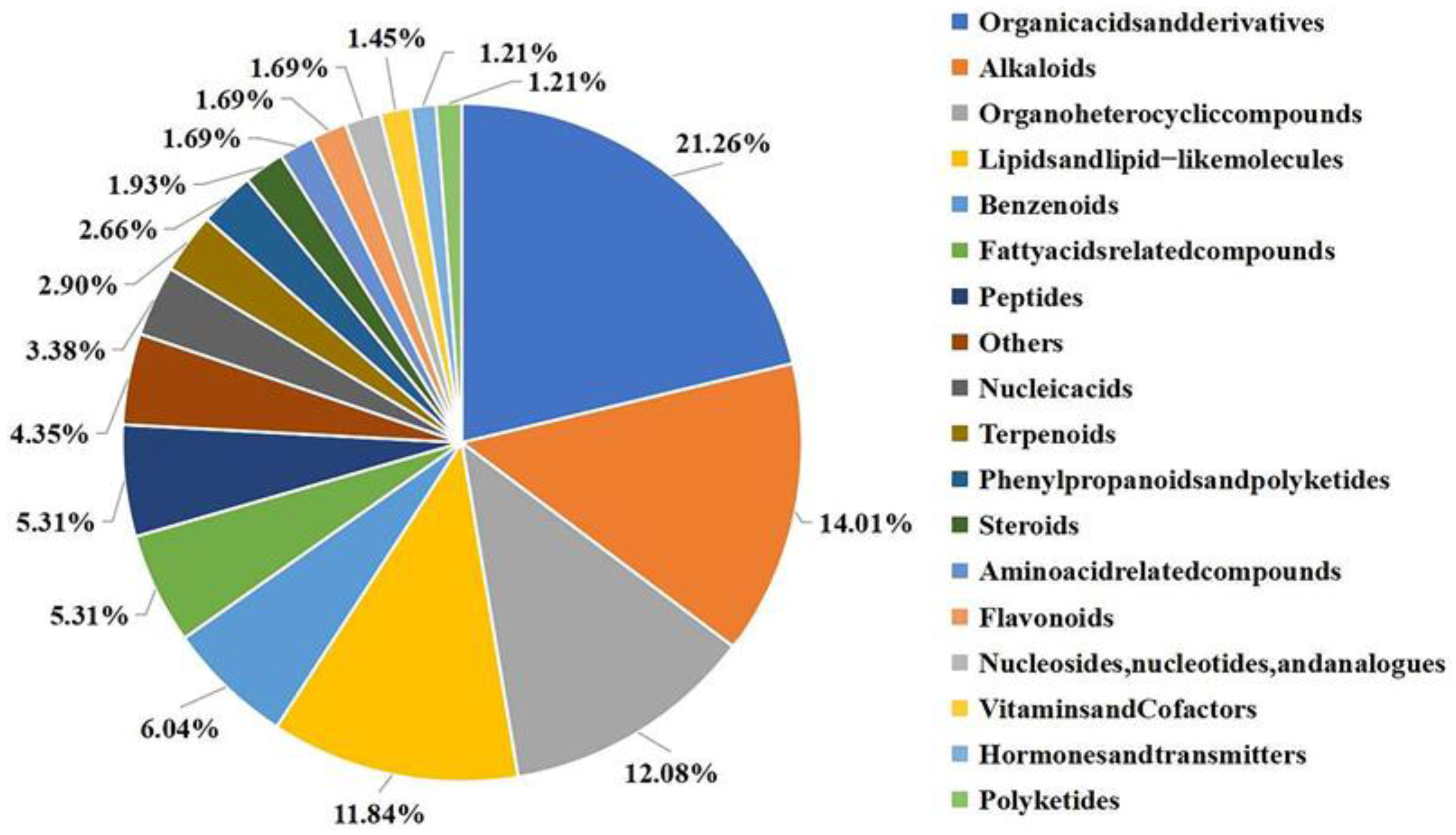
To investigate the components of J-15-Ms that regulate plant growth and determine their action mechanism, we examined the extracted J-15-Ms using LC-MS. We identified 414 metabolites, among which organic acids and their derivatives, organoheterocyclic compounds, lipids, lipid-like compounds, and alkaloids accounted for the highest proportion (more than 10% each). There were 27 benzenoids, 25 fatty acid-related compounds, 22 peptides, and 21 organic oxygen compounds (Figure 3). A few nucleic acids, terpenoids, organic nitrogen compounds, phenylpropanoids, flavonoids, amino acids, and nucleotides were also detected.
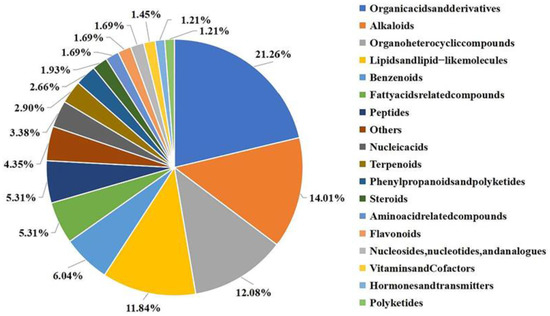
Figure 3.
Relative proportions of different types of metabolites in J-15. Different colors represent different classifications of compounds and their proportions. All compounds with their affiliation to the particular group can be found in Table S5.
The metabolites produced by J-15 included several primary metabolites, such as organic acids, amino acids, nucleic acids, lipids, and carbohydrates, and secondary metabolites, such as alkaloids, flavonoids, phenylpropanoids, terpenoids, and phytohormones. These primary metabolites are indispensable for plant growth and development, while the secondary ones are closely related to plant disease, stress resistance, and growth regulation. Thus, these compounds synergistically regulate plant growth.
Alkaloids are a class of metabolites with complex ring structures and are mainly involved in preventing stress effects in plants. Up to 42 alkaloids were obtained in J-15-Ms and were presented in the order of their abundance, as shown in Table 1.

Table 1.
Alkaloids in J-15-Ms.
Terpenoids are an important class of J-15-Ms consisting of isoprene as their basic component and can be used as phytoprobiotics to protect plants by reducing the infestation of pathogens and feeding animals and insects. The terpenoids identified in J-15-Ms included Gossypol, Glaucarubin, Withanolide, Agavoside A, Protodioscin, Musennin, and Soyasaponin A1 (Table 2).

Table 2.
Terpenoids in J-15-Ms.
Flavonoids are a broad class of J-15-Ms with bacteriostatic, antioxidant, antitumor, immune-enhancing, and other biological activities. Flavonoids protect plants against UV light and regulate the transportation of plant growth hormones. Mulberrofuran A, Genistein, Prenyl glucoside, Daidzein, (R)-Glabridin, Glycitein, and Dihydrokaempferol were the identified J-15-Ms flavonoids (Table 3).

Table 3.
Flavonoids in J-15-Ms.
Three plant hormones—indoleacetic acid (IAA), abscisic acid (ABA), and jasmine acid (JA)—and 14 indole plant growth regulators, such as indole-3-carboxaldehyde, indoleacetaldehyde, 3-Methylindole, indole-3-acetaldehyde, and 3-Methylindole, were detected in J-15-ms (Table 4).

Table 4.
Hormonal substance in J-15-Ms.
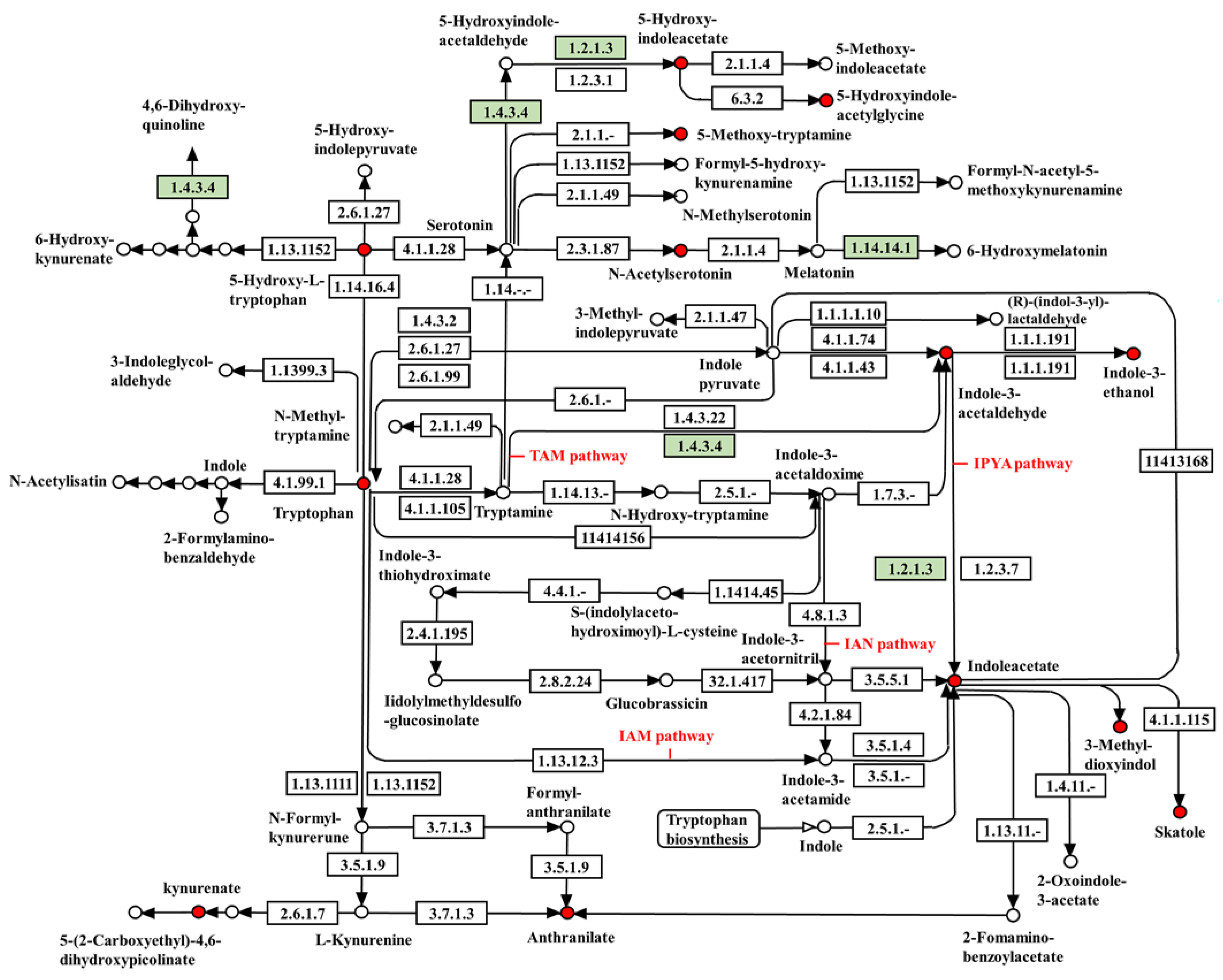
The KEGG maps of the target metabolites involved in the IAA synthesis pathway and the analysis of the tryptophan metabolic pathway in the J-15 genome identified the possible involvement of J-15 in the IAA biosynthetic pathway (Figure 4). The results showed that genes involved in the IPyA and TAM pathways were present, while the key genes involved in the IAM and IAN pathways were lacking in the J-15 genome. Since tryptophan, indole-3-acetaldehyde, 3-indoleacetic acid, 3-methylindole, and indoleacetaldehyde, which are all J-15-Ms, play key roles in the IPyA and TAM pathways, it can be concluded that J-15-Ms can biosynthesize IAA via the IPyA and TAM pathways.
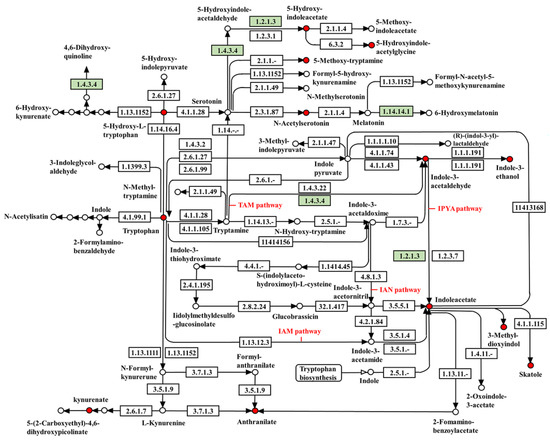
Figure 4.
Diagram of the KEGG pathway of tryptophan metabolism from the J-15 genome combined with the metabolome. Red dots indicate that the material is the material tested in this LC-MS. Green markers are genes related to the IAA synthesis pathway identified in the J-15 genome. Not all target genes and metabolites are included in the figure.
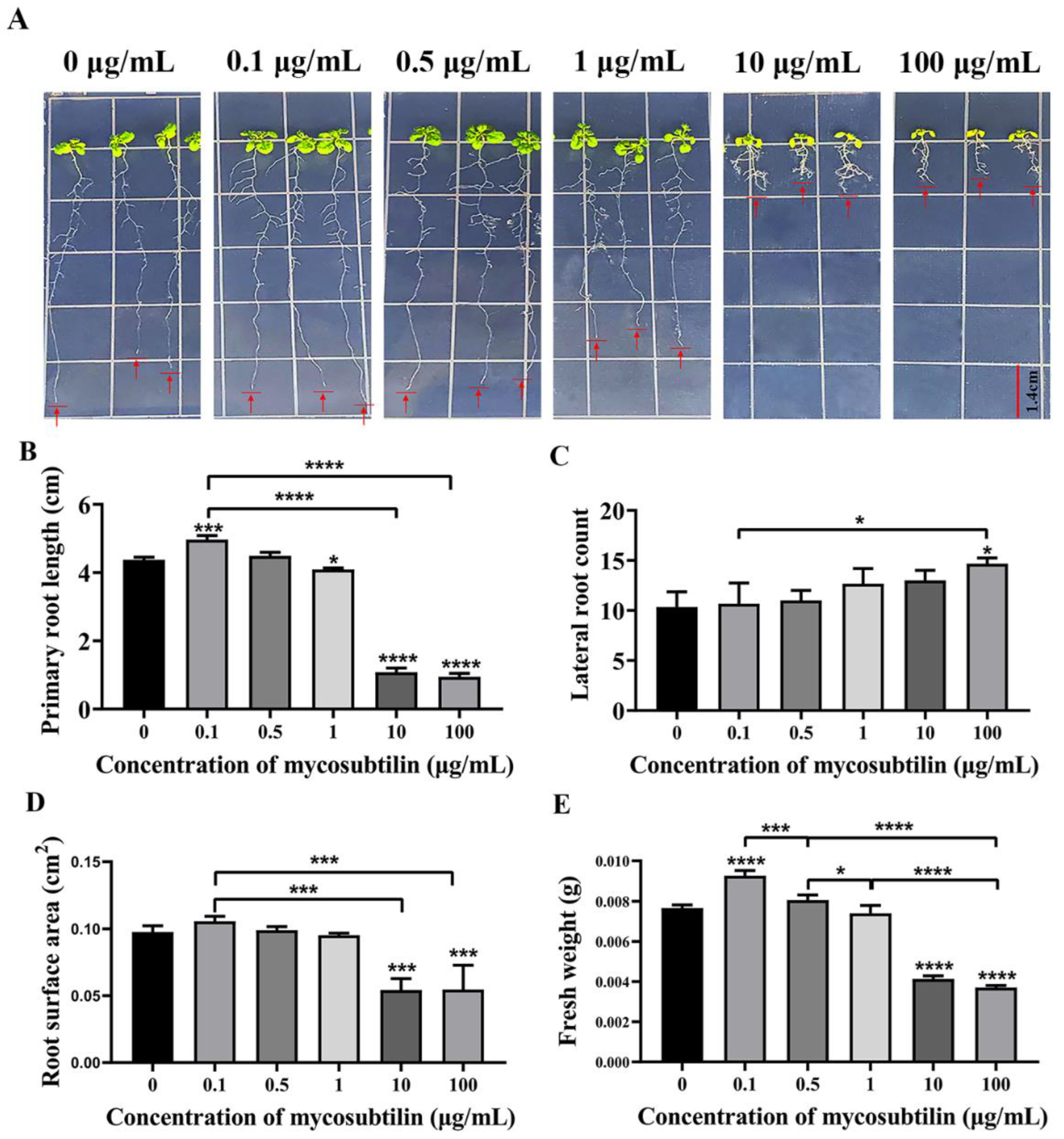
2.4. Effect of Mycosubtilin on Arabidopsis Growth
Arabidopsis seedlings were treated with different concentrations of mycosubtilin for 10 days to investigate its effect on plant growth. The results showed that at a very low concentration of 0.1 μg/mL, mycosubtilin improved the growth of Arabidopsis seedlings, increasing their primary roots, root surface area, and fresh weight, compared to the control group. As the concentration increased (1 μg/mL), mycosubtilin inhibited the growth of the primary roots, the root surface area, and the fresh weight of Arabidopsis seedlings but enhanced the development of lateral roots (Figure 5). Thus, mycosubtilin also had the same low-concentration-promoting and high-concentration-inhibiting, but not lethal, effects on the growth and development of Arabidopsis.
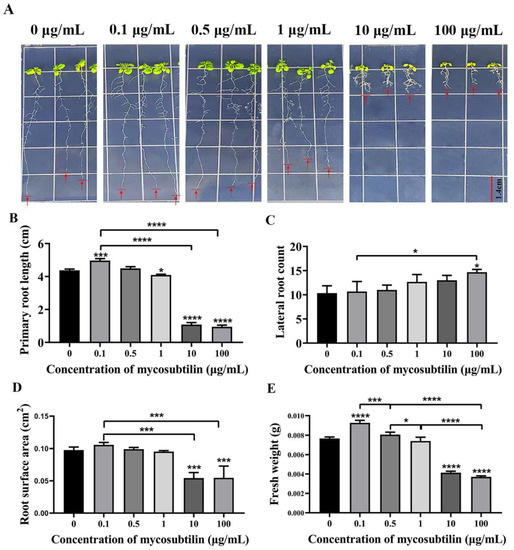
Figure 5.
Effect of mycosubtilin on the growth of Arabidopsis seedlings. (A) Growth phenotype of Arabidopsis seedlings. (B) Effect on primary root length. (C) Effect on the number of lateral roots. (D) Effect on root surface area. (E) Effect on fresh weight. Note: Means ± SEs, n = 50, different “*” indicate significant differences among treatments based on the least significant difference at p ≤ 0.05.
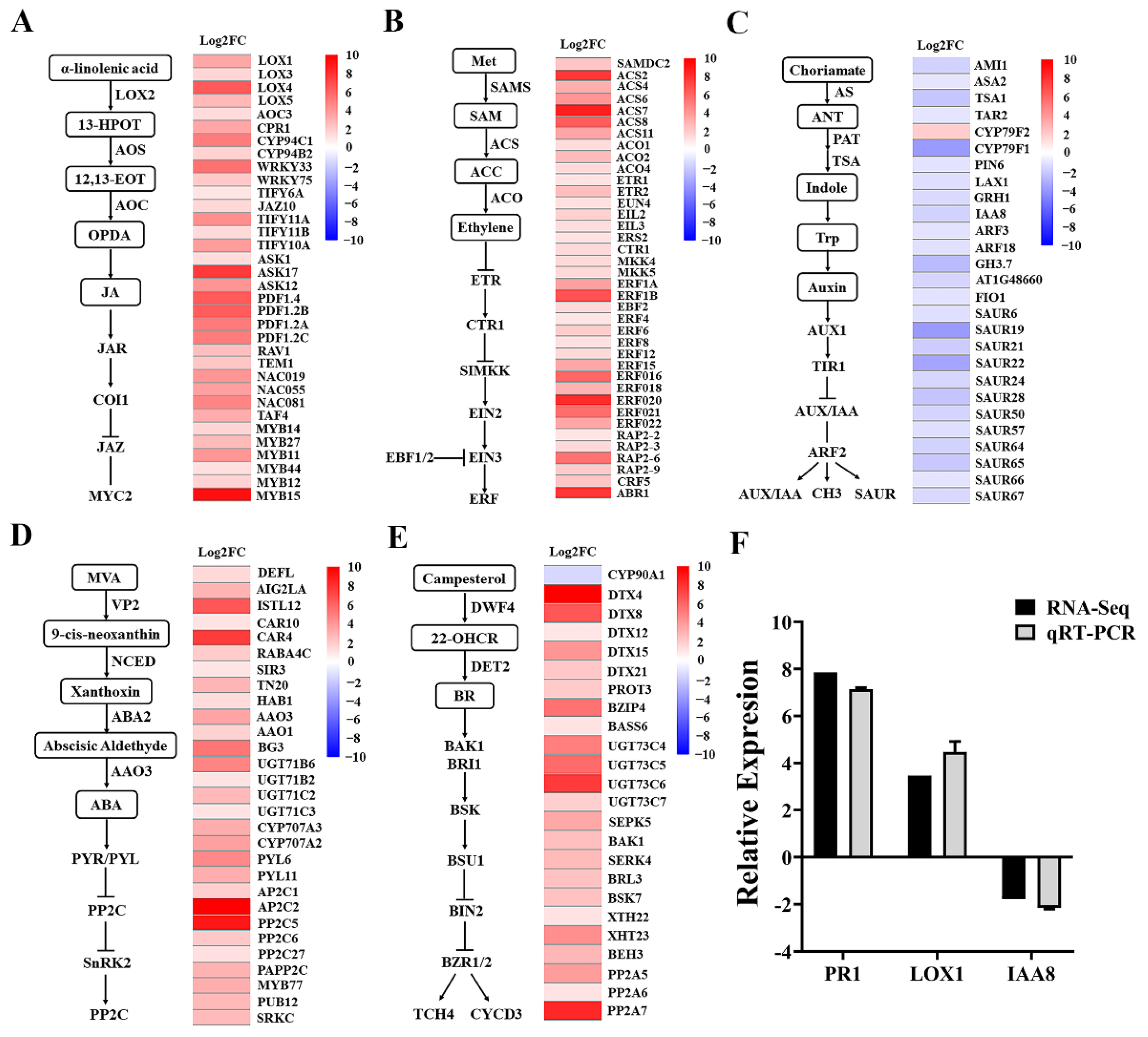
2.5. Expression Pattern of the Genes Associated with Growth Regulatory Mechanisms of Mycosubtilin in Arabidopsis
Mycosubtilin exhibited significant inhibition of primary root growth and promotion of lateral root development in Arabidopsis. To investigate the plant growth regulatory mechanism by mycosubtilin, we treated Arabidopsis seedlings with 50 μg/mL of mycosubtilin for 10 days and analyzed the changes in gene expression of the phytohormone pathway (Figure 6). The results showed that the mycosubtilin treatment upregulated LOX4, AOC3, CYP94C1, JAZ10, ASK17, and MYB15 genes of the JA signaling pathway in Arabidopsis. Similarly, ACS2, ACS7, ETR2, ERS2, ERF18, and ERF20 of the ET signaling pathway and DT4, DT8, UGT73, and PP2A of the BR signaling pathway were highly upregulated. For the ABA signaling pathway, genes such as CTR4, AAO3, BG3, PYL6, and AP2A were upregulated. To ensure the reliability of the transcriptome data, PR1, LOX1, and IAA8 of the phytohormone signaling pathway were verified by qRT-PCR, and the results showed consistent expression trends with the transcriptome data. Thus, mycosubtilin inhibited the primary root growth and promoted lateral root development in Arabidopsis by upregulating various genes involved in the JA, ET, BR, and ABA signaling pathways and downregulating those involved in the IAA signaling pathway. This suggests that mycosubtilin may affect plant growth by regulating phytohormone levels.
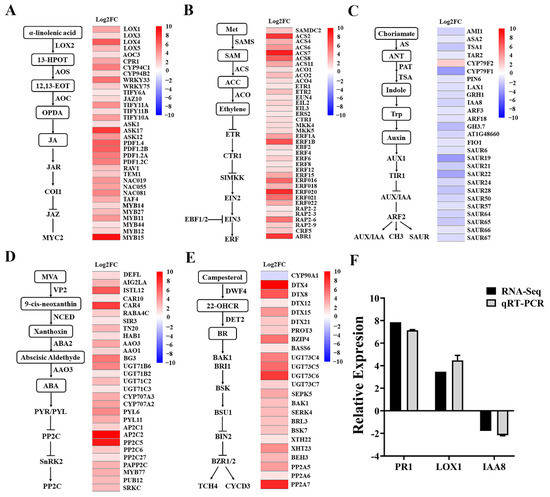
Figure 6.
Fold changes in gene expression of major plant hormone signallingsignaling pathways by mycosubtilin treatment. After 12 h treatment with mycosubtilin in Arabidopsis: (A) JA signaling pathway fold change in gene expression; (B) ET signaling pathway fold change in gene expression; (C) IAA signaling pathway fold change in gene expression; (D) ABA signaling pathway fold change in gene expression; (E) BR signaling pathway fold change in gene expression; and (F) transcriptome data qRT-PCR verification.
3. Discussion
PGPR is essential for improved plant growth, either through the direct secretion of plant growth and development-related substances or indirectly by regulating the living plant environment [45]. ACC deaminase-producing Burkholderia cepacia P10 promotes plant growth by producing compounds such as IAA and iron carriers [46]. Two PGPR strains from the pokeweed root zone, WM13-24 and M30-35, promote ryegrass growth and root development by regulating phytohormone distribution [47]. Moreover, Bacillus pallidus PP7S promotes plant growth and increases anthocyanin biosynthesis in Arabidopsis by triggering specific induced systemic resistance (ISR) in the plant [48]. Previous studies have shown that J-15-Ms can induce V. dahliae resistance in cotton [40] and inhibit various pathogenic bacteria [41]. In this study, Arabidopsis seedlings were treated with different concentrations of J-15-Ms for 10 days. The results showed that, at 0.2 μg/mL, J-15-Ms had a significant pro-growth effect on Arabidopsis, increasing the primary root length, lateral root number, root surface area, and fresh weight of the Arabidopsis seedlings compared to the control. However, as the concentration increased, the root surface area, root length, and lateral root number decreased, inhibiting the growth of Arabidopsis seedlings (Figure 1). This indicates that lower J-15-Ms concentrations promote the growth of Arabidopsis roots, while higher concentrations inhibit it. Similar results were also obtained on cotton growth analysis. The treatment with 1 μg/mL of J-15-Ms increased the biomass of the treated cotton compared to the control, and 10 μg/mL resulted in taller cotton plants with longer roots and an increased root-to-crown ratio. However, at the J-15-Ms concentration of 100 μg/mL, the cotton growth was inhibited, resulting in reduced root length, plant height, root-to-crown ratio, and biomass of the treated cotton compared to the control, and it even appeared that the roots turned black and the primary roots broke off (Figure 2). Although low concentrations of J-15-Ms are beneficial to the growth of cotton, too high concentrations can cause damage to the cotton, but this damage does not lead to death. This suggests that the J-15-Ms have regulatory effects on plant growth and development. Thus, we speculate that a certain component of J-15-Ms could be directly involved in plant growth and development promotion by regulating the dynamic balance of plant hormone levels and nutrients in the plant root system.
To investigate how the J-15-Ms regulate plant growth, we analyzed and identified the J-15-Ms components by LC-MS. The results showed that various organic acids and their derivatives, organic heterocyclic compounds, lipids, and lipid-like compounds, which are all primary metabolites related to plant growth and development, were contained among the J-15-Ms. There were also secondary metabolites, such as alkaloids, flavonoids, peptides, a few nucleic acids, terpenoids, phenylpropanoids, flavonoids, amino acids, and phytohormones (Figure 3). We found that the J-15-Ms exhibited broad-spectrum resistance to various pathogens [41]. The alkaloids xanthine, angularine, vincristine, caffeine, and 6-acetylmorphine found in the J-15-Ms (Table 1) also have excellent inhibitory effects against various plant pathogens [49,50,51]. The terpenoid (Table 2) gossypol can effectively inhibit the germination of V. dahliae spores and the growth of V. striatum [52]. Furthermore, saponins, such as agave saponin, proto-diosgenin, and soy saponin, have antibacterial, antiviral, and insecticidal effects and strong inhibitory activity against phytopathogenic bacteria causing wheat root rot [53,54]. The flavonoid (Table 3) photoglycyrrhizin also has inhibitory effects against various plant pathogens [55,56]. Genistein regulates plant growth and mutual recognition between plants and microorganisms, (R)-Glabridin inhibits various plant pathogens, and Glycitein exhibits concentration-dependent inhibition of plant radicle length. These substances indirectly promote plant growth by inhibiting the growth of pathogenic bacteria and inducing resistance in plants [57].
It has been reported that PGPR directly promotes plant growth by secreting growth-regulating substances such as IAA. Pseudomonas aeruginosa HMR1 produces IAA and, thus, has a high plant growth-promoting potential [58]. It was found that Bacillus cereus CJCL2 and RJGP41 could positively regulate the expression of plant hormones such as ABA, thereby significantly improving plant growth under cold stress [59]. Moreover, inoculation with Pseudomonas spp. significantly increased the levels of hormones such as ABA but reduced ET levels in soybean plants to enhance the drought stress tolerance of soybeans [60]. In this study, LC-MS analysis showed that J-15-SMs contain phytohormones, such as IAA, ABA, and JA, and that indole-3-acetaldehyde, 3-indoleacetic acid, 3-methylindole, and other plant growth regulators regulate plant growth (Table 4). IAA has a crucial regulatory role in almost all plant growth and development stages, especially root development and structure. At lower concentrations, IAA stimulates primary root elongation, but reduces primary root length, increases root hair formation, and stimulates the formation of lateral roots at higher concentrations. Overall, IAA regulates the surface area and length of roots, allowing plants greater access to soil nutrients [61]. Conversely, ABA inhibits seed germination and primary root growth by suppressing lateral root germination and the activity of lateral root meristems. ABA also inhibits lateral root growth and development by cross-interacting with various plant hormones and environmental signals [62]. Although it is an important hormone that enhances plant defense, higher levels of JA can inhibit plant growth. Indoles, such as indole-3-butyric acid, are also important broad-spectrum plant growth regulators. Since most indoles are intermediates in the synthesis of indoleacetic acid from tryptophan, J-15 could likely use tryptophan to synthesize IAA to regulate plant growth. When the IAA concentration is in the right range, it can stimulate the production of root hairs and increase the length and number of lateral roots in plants; however, the development of primary roots is inhibited to some extent when the concentration of produced IAA is high [63]. This is consistent with our previous results that low concentrations of J-15-Ms promote plant growth and high concentrations of J-15-Ms inhibit growth. Therefore, we considered that the key substance in J-15-Ms that regulates plant growth is IAA. The tryptophan-dependent IAA production is widely considered the most dominant mode of IAA secretion by plant bacteria [64]. Therefore, we performed a combined KEGG analysis of the J-15 genome and J-15-MS. The results showed that J-15 contained genes related to the IPyA and TAM pathway of tryptophan metabolism, and that tryptophan, indole-3-acetaldehyde, 3-indoleacetic acid, 3-methylindole, and indole acetaldehyde in the J-15-Ms were all present in the IAA synthesis pathway (Figure 4). This suggests that J-15 can use these substances to synthesize IAA. Thus, J-15 may regulate plant growth by promoting IAA biosynthesis through the IPyA and TAM pathways.
Mycosubtilin, a lipopeptide purified from J-15-Ms, has strong antifungal activity [44]; however, its effect on plant growth has not been reported. The treatment of Arabidopsis seedlings and cotton with different mycosubtilin concentrations showed the same pattern of pro-growth at lower concentrations and inhibition at higher concentrations. At a lower concentration of 0.1 μg/mL, the growth of Arabidopsis seedlings was promoted, increasing the primary root length, root surface area, and fresh weight compared to the control. However, the plant growth decreased as the concentration increased; for example, mycosubtilin treatment at 1 μg/mL inhibited the primary root length, root surface area, and fresh weight but promoted the development of lateral roots of the Arabidopsis seedlings. Thus, the higher the concentration, the more pronounced the growth inhibition was. Although higher mycosubtilin concentrations strongly inhibited Arabidopsis growth, it did not cause lethal effects at 100 μg/mL (Figure 5).
To elucidate the molecular mechanism of the growth-regulating effect of mycosubtilin, we analyzed the genes regulating plant growth using the Arabidopsis transcriptome data obtained after 12 h of mycosubtilin treatment. We found that mycosubtilin treatment upregulated the expression of genes in the JA, ET, ABA, and BR signaling pathways and downregulated the expression of those in the IAA signaling pathway. In the JA signaling pathway, the expression of LOX1, LOX4, AOC3, CYP94, and other genes regulating jasmonic acid synthesis were upregulated. When synthesized in large quantities, JA significantly inhibits the elongation of Arabidopsis primary roots in a concentration-dependent manner, with higher concentrations causing severe inhibition of root growth [65]. JA regulates the growth of lateral roots via the JA-COI1-JAZ pathway, and the JAZs proteins downstream of JA form a complex with transcription factors, such as RHD6 and RSL1, which regulate root hair development [66]. Mycosubtilin treatment significantly upregulated JAZ10, ASK17, PDF1.2, and MYB15 genes of the JA-COI1-JAZ pathway, which inhibited primary root growth and promoted lateral root development. ET promotes the biosynthesis and transport of growth hormone, leading to local accumulation of growth hormone in the plant, thus inhibiting cell elongation and root growth [67]. Several ET receptor genes, such as ETR1, ETR2, and ERS2, were upregulated by mycosubtilin treatment, activating the ET response [68]. Similarly, several genes of the ERF family of ethylene response factors were upregulated by mycosubtilin, promoting the regulatory effect of ET on the plant root system. IAA plays a crucial role in plant development, such as embryo development, root development, apical dominance, and various directional responses, which can be regulated through the IAA signaling pathway [69]. PIN6, IAA8, ARF3, and ARF18 genes are the key regulatory components exerting multiple IAA effects on plant growth and development [70]. The IAAs and ARFs interacting with AUX/IAAs play an important role in lateral root development [71,72]. Furthermore, downregulating GH3 and SUAR family genes downstream of IAA inhibits the growth of primary roots and promotes the development of lateral and adventitious roots by regulating the transportation of growth hormones [73,74]. ABA is involved in many plant growth and development processes, such as the inhibition of plant seed germination and primary root growth. Mycosubtilin treatment upregulated the expression of CAR4 and AAO3 genes of the ABA pathway, which promoted ABA synthesis [75]. The ABA-regulated lateral root growth in Arabidopsis requires the activation of ABA receptor PYL family genes [76], and MYB77 can be activated by the growth hormone to promote lateral root growth. Furthermore, interactions between PYL8 (of the PYL family) and MYB77 regulate lateral root growth recurrence after inhibition [77]. BR is essential for normal plant growth and development [78]. Studies have shown that BR plays an important regulatory role in maintaining the size of Arabidopsis root meristems, root cell elongation, lateral root primordia initiation, root hair formation, and root weight orientation [79]. In the presented study, mycosubtilin treatment upregulated the expression of DTX4, DTX8, PROT3, and other genes that promote BR production. Upregulation of BAK1, BRL3, and BSK7 genes of the BR signaling pathway, such as BRI1 and BSKs, has been shown to promote endogenous BR levels and root elongation [80]. As part of the BR signaling pathway, the BRI1-related receptor kinase BAK1 is involved in root development, and its mutants show a root-inhibiting phenotype [81]. Moreover, BR, IAA, ET, and other hormonal signals can synergistically or antagonistically regulate the growth and development of Arabidopsis roots [82]. In summary, the mycosubtilin treatment activated gene expression changes in the IAA, JA, ET, ABA, and BR signaling pathways, and these hormonal pathways acted synergistically to inhibit primary root and promote lateral root development in Arabidopsis.
4. Materials and Methods
4.1. Strain Activation and Extraction of Metabolites
Bacillus subtilis J-15 was isolated from the rhizosphere soil of a healthy continuous cotton field in Xinjiang and stored in the key laboratory of special species conservation and regulatory biology at Xinjiang Normal University, China. Bacillus subtilis J-15 was activated by scribing, and single colonies were picked and cultured in beef paste peptone medium at 37 °C with shaking at 180 revolutions/min for 18 h. The J-15-Ms were extracted with acetone, chloroform (to remove the heteroproteins), and n-butanol [83]. The J-15-Ms was freeze-dried and solubilized using dimethyl sulfoxide, then purified by high-performance liquid chromatography to obtain the lipopeptide mycosubtilin [44].
4.2. Cultivation and Treatment of Arabidopsis Seedlings
This study used the wild-type Arabidopsis, Columbia-0 (Col-0), obtained from the key laboratory of special species conservation and regulatory biology at Xinjiang Normal University, China. The 7-day Arabidopsis Col-0 seedlings grown on MS (Murashige and Skoog) medium were transferred to a new Petri dish with 1/2 MS medium and grown in a controlled light incubator (GXM-508, produced in Ningbo Jiangnan Instrument Factory, China) at 23 ± 1 °C under 50 μmol/m2·s photon flux density of photosynthetically active radiation with a relative humidity of 70% under 18 h light/6 h dark photoperiod.
4.3. Cultivation and Treatment of Cotton Seedlings
The cotton strain Xinluzao 72 used in this study was kindly donated by the Xinjiang Academy of Agricultural Sciences, China. The full-grained lint-free cotton seeds were surface disinfected with 2% sodium hypochlorite for 20 min and rinsed 4–5 times in sterile water. The seeds were soaked in sterile water for 6–12 h and wrapped in moist triple gauze for germination. After the germination, the seeds were transferred in a sterile soil bowl (containing vermiculite: nutrient soil: flower soil = 1:1:2) and were incubated in a plant culture room at 25 °C/21 °C day/night temperature under 75 μmol/m2·s photon flux density of photosynthetically active radiation, relative humidity of 60–75%, and 16 h day/8 h dark photoperiod, with regular watering.
4.4. Growth Indicators in Arabidopsis Treated with J-15-Ms
Three-day-old Arabidopsis Col-0 seedlings, grown on MS medium, were transferred to Petri dishes containing 1/2 MS medium with different concentrations of J-15-Ms (0 μg/mL, 0. 2 μg/mL, 2 μg/mL, and 20 μg/mL), with three replicates for each concentration, containing at least 50 Arabidopsis seedlings in each concentration. The plates were kept vertically so that the roots of the plants grew downwards at 23 ± 1 °C under 50 μmol/m2·s photon flux density of photosynthetically active radiation with a relative humidity of 70% under 18 h light/6 h dark photoperiod. After 7 days, images of the Arabidopsis seedlings were taken, and their growth indicators, such as primary root length, lateral roots, and root surface area, were measured at different concentrations using ImageJ software.
4.5. Growth Indicators in Arabidopsis Treated with J-15-Ms
The leaf surface of the cotton seedlings at the two-leaf and one-center stages were sprayed with different concentrations of J-15-Ms (0 μg/mL, 1 μg/mL, 10 μg/mL, and 100 μg/mL) once a day for three days under 75 μmol/m2·s photon flux density of photosynthetically active radiation, relative humidity of 60–75%, and 16 h day/8 h dark photoperiod, with regular watering. At least 20 cotton plants were in each experiment, and three replicates were performed. After 21 days of treatment, the growth indicators, such as root length, plant height, fresh weight, and dry weight, were measured according to the method used in Section 4.4.
4.6. Full-Spectrum LC-MS Detection of J-15-Ms
A mass of J-15-Ms was extracted at low temperature, accurately measured, and then centrifuged to extract the supernatant metabolite solvent for liquid–liquid mass spectrometry. The sample processing or chromatographic conditions (http://www.majorbio.com/, accessed on 21 September 2020). included: ACQUITY UPLC HSS T3 (100 mm × 2.1 mm i.d 1.8 µm; Waters, Milford, USA), mobile phase A, and mobile phase B (Table S1). The mobile phase A was 95% water + 5% acetonitrile containing 0.1% formic acid, while the mobile phase B was 47 5% acetonitrile + 47.5% isopropanol + 5% water containing 0.1% formic acid. The injection volume and the column temperature were 2 µL and 40 °C, respectively, and the mobile phase elution gradient is shown in Table S2. Mass spectrometric conditions: the samples were subjected to electrospray ionization, and the mass spectrometric signals were acquired in positive and negative ion scanning mode, respectively, with the parameters shown in Table S3.
4.7. Growth Indicators in Arabidopsis Treated with Mycosubtilin
Mycosubtilin concentrations were set at 0 μg/mL, 0.1 μg/mL, 0.5 μg/mL, 1 μg/mL, 5 μg/mL, 10 μg/mL, and 100 μg/mL, with three replicates for each concentration, containing at least 50 Arabidopsis seedlings in each concentration. Three-day-old Arabidopsis Col-0 seedlings grown on MS medium were transferred to different concentrations of mycosubtilin in Petri dishes, and the plates were kept vertically to allow for downwards growth of the roots. After 7 days, images of the seedlings were taken, and their growth indicators, such as primary root length, lateral root length, and root surface area, were measured as described in Section 4.4.
4.8. Gene Expression of the Hormone Signaling Pathways of Arabidopsis Treated with Mycosubtilin
Arabidopsis Col-0 seedlings grown on MS medium for 10 days were planted in six-well plates containing 50 μg/mL mycosubtilin, and those planted in plates with dd H2O served as controls. The whole plant samples were collected at 12 h after treatment and sent to Shanghai Meiji Biologicals (http://www.majorbio.com/, accessed on 31 July 2020), China, for transcriptome sequencing. Three biological replicates were performed per sample.
Arabidopsis Col-0 seedlings were treated with 50 μg/mL mycosubtilin for 12 h. Total RNA was extracted from each group of samples by TRIzol regent (Tiangen, Beijing, China), and the nucleic acid concentration was measured using an ultra-micro spectrophotometer (Thermo Fisher™ NanoDrop One, Waltham, MA, USA). The RNA was reverse transcribed into cDNA using a reverse transcription kit (Tiangen, Beijing, China ), The qRT-PCR reaction was performed on the Real-Time PCR instrument (ABI StepOne™, FosterCity, CA; USA). Data were standardized using AtActin and three replicates were made. Specific primers were designed according to the gene sequence, and primer sequences are shown in Table S4.
4.9. Statistics
Data were subjected to one-way analysis of variance (ANOVA) using SPSS 20 (IBM, Armonk, NY, USA). The significant differences between treatments were tested according to the least significant difference (LSD) at p ≤ 0.05. The asterisk “*” indicates a significant difference between the groups.
5. Conclusions
This study found that lower J-15-Ms concentrations promoted the growth of Arabidopsis and cotton seedlings, while higher concentrations inhibited the growth. LC-MS analysis found that J-15 regulates plant growth by synthesizing IAA via the IPyA and TAM pathways. The mycosubtilin purified from J-15-Ms also promoted plant growth at lower concentrations and inhibited growth but promoted lateral root development at higher concentrations. The transcriptome data showed that mycosubtilin regulates plant growth by upregulating the genes associated with JA, ET, ABA, and BR pathways and downregulating IAA pathway-related genes. Therefore, these findings provide preliminary insights into the growth-promoting mechanism of J-15 and a scientific basis for developing efficient.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11233205/s1, Table S1: Main reagents; Table S2. Mobile phase elution gradients; Table S3. Mass spectrometry parameters; Table S4. qRT-PCR primer sequences (Sangon Biotech); Table S5 Classification of compounds identified by LC-MS.
Author Contributions
H.Z. (Hui Zhang) and Q.Y. contributed equally to this work. Conceptualization, H.Z. (Huixin Zhao); methodology, H.Z. (Hui Zhang); validation, H.Z. (Hui Zhang) and Q.Y.; formal analysis, H.Z. (Hui Zhang), J.Z. and Q.Y.; data curation, H.Z. (Hui Zhang), J.C., S.W., M.M. and H.L.; writing—original draft preparation, H.Z. (Hui Zhang); writing—review and editing, H.Z. (Hui Zhang), Q.Z., H.Z. (Heping Zhao), D.Z. and X.W.; visualization, J.G.; supervision, J.G.; project administration, H.Z. (Huixin Zhao); funding acquisition, H.Z. (Huixin Zhao). All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Scientific Research Program of Colleges and Universities in Xinjiang (No. XJEDU2021I023), the Natural Science Foundation of China (No. 32160074), and the Open Project of Key Laboratory in Xinjiang (No. 2020D4010).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets supporting the results of this article are available in the CGNB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with accession number: CNP0003287. (https://db.cngb.org/search/project/CNP0003287/ (accessed on 27 August 2022)).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Enhanced plant growth by siderophores produced by plant growth promoting rhizobacteria. Nature 1980, 268, 885–886. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, R.J.; You, M.P.; Barbetti, M.J. Pathogen Biocontrol Using Plant Growth-Promoting Bacteria (PGPR): Role of Bacterial Diversity. Microorganisms 2021, 9, 1988. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.J.; Smith, D.L. Intracellular and extracellular PGPR: Commonalities and distinctions in the plant-bacterium signalling processes. Soil Biol. Biochem. 2005, 37, 395–412. [Google Scholar] [CrossRef]
- Olenska, E.; Malek, W.; Wojcik, I.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 2020, 743, 140682. [Google Scholar] [CrossRef]
- Van Oosten, M.J.; Di Stasio, E.; Cirillo, V.; Silletti, S.; Ventorino, V. Root inoculation with Azotobacter chroococcum 76A enhances tomato plants adaptation to salt stress under low N conditions. BMC Plant Biol. 2018, 18, 205. [Google Scholar] [CrossRef]
- Woo, O.G.; Kim, H.; Kim, J.S.; Keum, H.L.; Lee, K.C. Bacillus subtilis strain GOT9 confers enhanced tolerance to drought and salt stresses in Arabidopsis thaliana and Brassica campestris. Plant Physiol. Biochem. 2020, 148, 359–367. [Google Scholar] [CrossRef]
- Benidire, L.; Madline, A.; Pereira, S.I.A.; Castro, P.M.L.; Boularbah, A. Synergistic effect of organo-mineral amendments and plant growth-promoting rhizobacteria (PGPR) on the establishment of vegetation cover and amelioration of mine tailings. Chemosphere 2020, 262, 127803. [Google Scholar] [CrossRef]
- Peng, J.I.; Ma, J.; Wei, X.Y.; Zhang, C.M.; Jia, N.; Wang, X.; Wang, E.T.; Hu, D.; Wang, Z.W. Accumulation of beneficial bacteria in the rhizosphere of maize (Zea mays L.) grown in a saline soil in responding to a consortium of plant growth promoting rhizobacteria. Ann. Microbiol. 2021, 71, 40. [Google Scholar] [CrossRef]
- Sabaté, D.C.; Brandan, C.P.; Petroselli, G.; Balsells, R.E.; Audisio, M.C. Decrease in the incidence of charcoal root rot in common bean (Phaseolus vulgaris L.) by Bacillus amyloliquefaciens B14, a strain with PGPR properties. Biol. Control 2017, 113, 1–8. [Google Scholar] [CrossRef]
- Pérez-Rodriguez, M.M.; Piccoli, P.; Anzuay, M.S.; Baraldi, R.; Neri, L. Native bacteria isolated from roots and rhizosphere of Solanum lycopersicum L. increase tomato seedling growth under a reduced fertilization regime. Sci. Rep. 2020, 10, 15642. [Google Scholar] [CrossRef]
- Kumar, A.; Prakash, A.; Johri, B.N. Bacillus as PGPR in Crop Ecosystem. In Bacteria in Agrobiology: Crop Ecosystems; Springer: Berlin/Heidelberg, Germany, 2011; pp. 37–59. [Google Scholar]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Ambawade, M.S.; Pathade, G.R. Production of gibberellic acid by Bacillus siamensis BE 76 isolated from banana plant (Musa spp.). Int. J. Sci. Res. 2015, 4, 394–398. [Google Scholar]
- Patten, C.L.; Glick, B.R. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 1996, 42, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Asari, S.; Tarkowská, D.; Rolčík, J.; Novák, O.; Palmero, D.V. Analysis of plant growth-promoting properties of Bacillus amyloliquefaciens UCMB5113 using Arabidopsis thaliana as host plant. Planta 2016, 245, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.F.; He, L.Y.; Wang, Q.Y.; Ye, H.S.; Jiang, C.Y. Effects of inoculation of biosurfactant-producing Bacillus sp. J119 on plant growth and cadmium uptake in a cadmium-amended soil. J. Hazard. Mater. 2008, 155, 17–22. [Google Scholar] [CrossRef]
- Vibhuti, M.; Kumar, A.; Sheoran, N.; Nadakkakath, A.V.; Eapen, S.J. Molecular Basis of Endophytic Bacillus megaterium-induced Growth Promotion in Arabidopsis thaliana: Revelation by Microarray-based Gene Expression Analysis. J. Plant Growth Reg. 2017, 36, 118–130. [Google Scholar] [CrossRef]
- Lee, S.; Trịnh, C.S.; Lee, W.J.; Jeong, C.Y.; Truong, H.A. Bacillus subtilis strain L1 promotes nitrate reductase activity in Arabidopsis and elicits enhanced growth performance in Arabidopsis, lettuce, and wheat. J. Plant Res. 2020, 133, 231–244. [Google Scholar] [CrossRef]
- Hasan, N.; Khan, I.U.; Farzand, A.; Heng, Z.; Moosa, A. Bacillus altitudinis HNH7 and Bacillus velezensis HNH9 promote plant growth through upregulation of growth-promoting genes in upland cotton. J. Appl. Microbiol. 2022, 132, 3812–3824. [Google Scholar] [CrossRef]
- Deng, C.; Zhang, N.; Liang, X.L.; Huang, T.; Li, B.X. Bacillus aryabhattai LAD impacts rhizosphere bacterial community structure and promotes maize plant growth. J. Sci. Food Agric. 2022, 102, 6650–6657. [Google Scholar] [CrossRef] [PubMed]
- Franco-Sierra, N.D.; Posada, L.F.; Santa-María, G.; Romero-Tabarez, M. Bacillus subtilis EA-CB0575 genome reveals clues for plant growth promotion and potential for sustainable agriculture. Funct. Integr. Genom. 2020, 20, 575–589. [Google Scholar] [CrossRef]
- Li, F.; Tang, M.; Tang, X.X.; Sun, W.; Gong, J.Y.; Yi, Y. Bacillus subtilis—Arabidopsis thaliana: A model interaction system for studying the role of volatile organic compounds in the interchange between plants and bacteria. Botany 2019, 97, 661–669. [Google Scholar] [CrossRef]
- Xie, X.T.; Zhang, H.M.; Paré, P.W. Sustained growth promotion in Arabidopsis with long-term exposure to the beneficial soil bacterium Bacillus subtilis (GB03). Plant Signal. Behav. 2009, 4, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Gowtham, H.G.; Brijesh, S.S.; Murali, M.; Shilpa, N.; Melvin, P.; Mohammed, A.; Amruthesh, K.N.; Niranjana, S.R. Induction of drought tolerance in tomato upon the application of ACC deaminase producing plant growth promoting rhizobacterium Bacillus subtilis Rhizo SF 48. Microbiol. Res. 2020, 234, 126422. [Google Scholar]
- Zhang, H.M.; Murzello, C.; Sun, Y.; Kim, M.S.; Xie, X.T. Choline and osmotic-stress tolerance induced in Arabidopsis by the soil microbe Bacillus subtilis (GB03). Mol. Plant Microbe Interact. 2010, 23, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.H.; Liao, M.J.; Wang, H.K.; Zheng, M.Z.; Xu, J.J.; Guo, J.H. Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol. Control 2018, 126, 147–157. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Paramasivan, M.; Chun, S.C. Bacillus subtilis CBR05 induces Vitamin B6 biosynthesis in tomato through the de novo pathway in contributing disease resistance against Xanthomonas campestris pv. vesicatoria. Sci. Rep. 2019, 9, 6495. [Google Scholar] [CrossRef]
- Deleu, M.; Razafindralambo, H.L.; Popineau, Y.; Jacques, P.; Thonart, P.; Paquot, M. Interfacial and emulsifying properties of lipopeptides from Bacillus subtilis. Colloids Surf. A Physicochem. Eng. Asp. 1999, 152, 3–10. [Google Scholar] [CrossRef]
- Arrebola, E.; Jacobs, R.; Korsten, L. Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J. Appl. Microbiol. 2010, 108, 386–395. [Google Scholar] [CrossRef]
- Deleu, M.; Paquot, M.; Nylander, T. Effect of fengycin, a lipopeptide produced by Bacillus subtilis, on model biomembranes. Biophys. J. 2008, 94, 2667–2679. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jourdan, E.; Adam, A.; Paquot, M.; Brans, A. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 2007, 9, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Jayakumar, A.; Krishna, A.; Mohan, M.; Nair, I.C.; Radhakrishnan, E.K. Plant Growth Enhancement, Disease Resistance, and Elemental Modulatory Effects of Plant Probiotic Endophytic Bacillus sp. Fcl1. Probiotics Antimicrob. Proteins 2019, 11, 526–534. [Google Scholar] [CrossRef]
- Bais, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Gond, S.K.; Bergen, M.S.; Torres, M.S.; White, J.F. Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 2014, 172, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Tunsagool, P.; Jutidamrongphan, W.; Phaonakrop, N.; Jaresitthikunchai, J.; Roytrakul, S. Insights into stress responses in mandarins triggered by Bacillus subtilis cyclic lipopeptides and exogenous plant hormones upon Penicillium digitatum infection. Plant Cell Rep. 2019, 38, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jacques, P.; Touré, Y.; Destain, J.; Jabrane, A. Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Appl. Microbiol. Biotechnol. 2005, 69, 29–38. [Google Scholar] [CrossRef]
- Farace, G.; Fernandez, O.; Jacquens, L.; Coutte, F.; Krier, F. Cyclic lipopeptides from Bacillus subtilis activate distinct patterns of defence responses in grapevine. Mol. Plant Pathol. 2015, 16, 177–187. [Google Scholar] [CrossRef]
- Wu, M.J.; Yang, Q.L.; Li, Y.H.; Ge, F.W.; Zhao, Q.Q.; Yuan, L.L.; Ning, H.C.; Chen, Z.Y.; Li, J.Y.; Zhao, H.X. Effect of BS-Z15 Metabolites on Cotton Verticillium wilt Prevention and Cotton Growth. Mol. Plant Breed. 2019, 17, 8237–8244. [Google Scholar]
- Zhao, Q.Q.; Wu, M.J.; Dong, Y.Y.; Wang, Q.Y.; Lin, R.R.; Zhao, H.P.; Zhao, H.X. Antifungal Activity of BS-Z15 Metabolites and Its Safety in Mice. Nat. Prod. Res. Dev. 2018, 30, 1608–1613, 1620. [Google Scholar]
- Abuduaini, X.; Nuer, H.; Zhao, Q.Q.; Li, S.T.; Liu, J.Q.; Zhao, H.P.; Zhao, H.X. Effects of Bacillus subtilis J-15 secondary metabolites against Verticillium dahliae on diversity of soil fungi. Microbiol. China 2021, 48, 1997–2007. [Google Scholar]
- Chen, Z.Y.; Abuduaini, X.; Mamat, N.; Yang, Q.L. Genome sequencing and functional annotation of Bacillus sp. strain BS-Z15 isolated from cotton rhizosphere soil having antagonistic activity against Verticillium dahliae. Arch. Microbiol. 2021, 203, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.R.; Zhang, Q.; Yin, L.; Zhang, Y.W.; Yang, Q.L. Isolation and characterization of a mycosubtilin homologue antagonizing Verticillium dahliae produced by Bacillus subtilis strain Z15. PLoS ONE 2022, 17, e0269861. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.D.; Zu, M.T.; Sun, L.P.; Zuo, J.J.; Tao, J. Isolation and Screening of 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase producing PGPR from Paeonia lactiflora rhizosphere and enhancement of plant growth. Sci. Hortic. 2022, 297, 110956. [Google Scholar] [CrossRef]
- Han, L.Z.; Zhang, H.; Xu, Y.; Li, Y.; Zhou, J. Biological characteristics and salt-tolerant plant growth-promoting effects of an ACC deaminase-producing Burkholderia pyrrocinia strain isolated from the tea rhizosphere. Archives of microbiology. Arch. Microbiol. 2021, 203, 2279–2290. [Google Scholar] [CrossRef]
- He, A.; Niu, S.Q.; Yang, D.; Ren, W.; Zhao, L.Y. Two PGPR strains from the rhizosphere of Haloxylon ammodendron promoted growth and enhanced drought tolerance of ryegrass. Plant Physiol. Biochem. 2021, 161, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Trinh, C.S.; Jeong, C.Y.; Lee, W.J.; Truong, H.A.; Chung, N. Paenibacillus pabuli strain P7S promotes plant growth and induces anthocyanin accumulation in Arabidopsis thaliana. Plant Physiol. Biochem. 2018, 129, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.; Jozwiak, A.; Sonawane, P.D.; Szymanski, J.; Kazachkova, A. Steroidal alkaloids defence metabolism and plant growth are modulated by the joint action of gibberellin and jasmonate signalling. New Phytol. 2022, 233, 1220–1237. [Google Scholar] [CrossRef]
- Nakagawa, A.; Sakamoto, S.; Takahashi, M.; Morikawa, H.; Sakamoto, A. The RNAi-mediated silencing of xanthine dehydrogenase impairs growth and fertility and accelerates leaf senescence in transgenic Arabidopsis plants. Plant Cell Physiol. 2007, 48, 1484–1495. [Google Scholar] [CrossRef]
- Wang, Y.C.; Qian, W.J.; Li, N.N.; Hao, X.Y.; Wang, L.; Xiao, B.; Wang, X.C.; Yang, Y.G. Metabolic Changes of Caffeine in Tea Plant (Camellia sinensis (L.) O. Kuntze) as Defense Response to Colletotrichum fructicola. J. Agric. Food Chem. 2016, 64, 6685–6693. [Google Scholar] [CrossRef]
- Puckhaber, L.S.; Dowd, M.K.; Stipanovic, R.D.; Howell, C.R. Toxicity of (+)- and (-)-gossypol to the plant pathogen, Rhizoctonia solani. J. Agric. Food Chem. 2002, 50, 7017–7021. [Google Scholar] [CrossRef] [PubMed]
- Zaynab, M.; Sharif, Y.; Abbas, S.; Afzal, M.; Qasim, M. Saponin toxicity as key player in plant defense against pathogens. Toxicon 2021, 193, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Ta, C.A.K.; Guerrero-Analco, J.A.; Roberts, E.; Liu, R.; Mogg, C.D.; Saleem, A. Antifungal Saponins from the Maya Medicinal Plant Cestrum schlechtendahlii G. Don (Solanaceae). Phytother. Res. 2016, 30, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Simmler, C.; Pauli, G.F.; Chen, S.N. Phytochemistry and biological properties of glabridin. Fitoterapia 2013, 90, 160–184. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; He, Q.; Wang, B.; Yang, Y.h.; Yang, C.P.; Chen, H.B.; Yue, G.Z. Synthesis and Antifungal Activities of Isoflavenes Derivatives. Chin. J. Synth. Chem. 2022, 1–10. [Google Scholar]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Bhojiya, A.A.; Joshi, H.; Upadhyay, S.K.; Srivastava, A.K.; Pathak, V.V.; Pandey, V.C.; Jain, D. Screening and Optimization of Zinc Removal Potential in Pseudomonas aeruginosa-HMR1 and its Plant Growth-Promoting Attributes. Bull. Environ. Contam. Toxicol. 2021, 108, 468–477. [Google Scholar] [CrossRef]
- Zubair, M.; Hanif, A.; Farzand, A.; Sheikh, T.M.M.; Khan, A.R.; Suleman, M.; Ayaz, M.; Gao, X.W. Genetic screening and expression analysis of psychrophilic Bacillus spp. reveal their potential to alleviate cold stress and modulate phytohormones in wheat. Microorganisms 2019, 7, 337. [Google Scholar] [CrossRef]
- Vaishnav, A.; Choudhary, D.K. Regulation of drought-responsive gene expression in Glycine max L. Merrill is mediated through Pseudomonas simiae strain AU. J. Plant Growth Regul. 2019, 38, 333–342. [Google Scholar] [CrossRef]
- Vejan, P.; Abdullah, R.; Abdullah, R.; Khadiran, T.; Ismail, S. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef]
- De Smet, I.; Zhang, H.M.; Inzé, D.; Beeckman, T. A novel role for abscisic acid emerges from underground. Trends Plant Sci. 2006, 11, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.J. Plant Hormones: Physiology, Biochemistry and Molecular Biology; Kluwer Academic: Boston, MA, USA, 1995. [Google Scholar]
- Malfanova, N.; Kamilova, F.; Validov, S.; Shcherbakov, A.; Chebotar, V.; Tikhonovich, I.; Lugtenberg, B. Characterization of Bacillus subtilis HC8, a novel plant-beneficial endophytic strain from giant hogweed. Microb. Biotechnol. 2011, 4, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, Y.; Ye, S.; Jiang, H.; Chen, Q.; Liu, F.; Zhou, W.; Chen, R.; Li, X.; Tietz, O.; et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 2009, 21, 1495–1511. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, M.; Yang, M.; Hu, Y. Arabidopsis JAZ Proteins interact with and suppress RHD6 transcription factor to regulate jasmonate-stimulated root hair development. Plant Cell 2020, 32, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Růžička, K.; Ljung, K.; Vanneste, S.; Podhorská, R.; Beeckman, T.; Friml, J.; Benková, E. Ethylene regulates root growth through effects on auxin biosynthesi and transport-dependent auxin distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef]
- Rzewuski, G.; Sauter, M. Ethylene biosynthesis and signaling in rice. Plant Sci. 2008, 175, 32–42. [Google Scholar] [CrossRef]
- Marta, D.B.; Stefan, K. Context, specificity, and self-organization in auxin response. Cold Spring Harb. Perspect. Biol. 2011, 3, a001578. [Google Scholar]
- Woodward, A.W.; Bonnie, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef]
- Khan, S.; Stone, J.M. Arabidopsis thaliana GH3.9 influences primary root growth. Planta 2007, 226, 21–34. [Google Scholar] [CrossRef]
- Mishra, B.S.; Singh, M.; Aggrawal, P.; Laxmi, A. Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS ONE 2009, 4, e4502. [Google Scholar] [CrossRef]
- Yamauchi, T.; Tanaka, A.; Inahashi, H.; Nishizawa, N.K.; Tsutsumi, N.; Inukai, Y.; Nakazono, M. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 20770–20775. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Liu, H.B.; Ruan, S.L.; Ali, B.; Gill, R.A.; Ma, H.S.; Zheng, Z.F.; Zhou, W.J. A zinc finger protein, interacted with cyclophilin, affects root development via IAA pathway in rice. J. Integr. Plant Biol. 2017, 59, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Spence, C.; Bais, H. Role of plant growth regulators as chemical signals in plant-microbe interactions: A double edged sword. Curr. Opin. Plant Biol. 2015, 27, 52–58. [Google Scholar] [CrossRef]
- Xing, L.; Zhao, Y.; Gao, J.H.; Xiang, C.B.; Zhu, J.K. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci. Rep. 2016, 6, 27177. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.G.; Hou, Y.J.; Gao, G.H.; Wang, P.C.; Duan, C.G.; Zhu, X.H.; Zhu, J.K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef]
- Belkhadir, Y.; Jaillais, Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015, 206, 522–540. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.Y.; Li, J. Brassinosteroids Regulate Root Growth, Development, and Symbiosis. Mol. Plant. 2015, 9, 86–100. [Google Scholar] [CrossRef]
- Zhang, C.; He, M.L.; Wang, S.L.; Chu, L.Y.; Wang, C.; Yang, N.M.; Ding, G.D.; Cai, H.M.; Shi, L.; Xu, F.S. Boron deficiency-induced root growth inhibition is mediated by brassinosteroid signalling regulation in Arabidopsis. Plant J. 2021, 107, 564–578. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.Q.; Lease, K.A.; Doke, J.T.; Tax, F.E. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- Li, M.X.; Zhu, Y.C.; Li, S.S.; Zhang, W.; Yin, C.X.; Lin, Y.J. Regulation of Phytohormones on the Growth and Development of Plant Root Hair. Front. Plant Sci. 2022, 13, 865302. [Google Scholar] [CrossRef]
- Zhao, J.J.; Zeng, W.J.; Li, Y.H.; Ge, F.W.; Du, Y.; Yuan, L.L.; Zhao, Q.Q. Isolation and identification of antagonistic Bacillus spp. Against Verticillium dahliae: The antibacterial properties of two strains. J. Beijing Norm. Univ. 2017, 53, 294–300. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).