Genome-Wide Identification and Expression Analysis of TUA and TUB Genes in Wheat (Triticum aestivum L.) during Its Development
Abstract
:1. Introduction
2. Results
2.1. Identification and Physicochemical Characteristics of TUA and TUB Proteins in Wheat
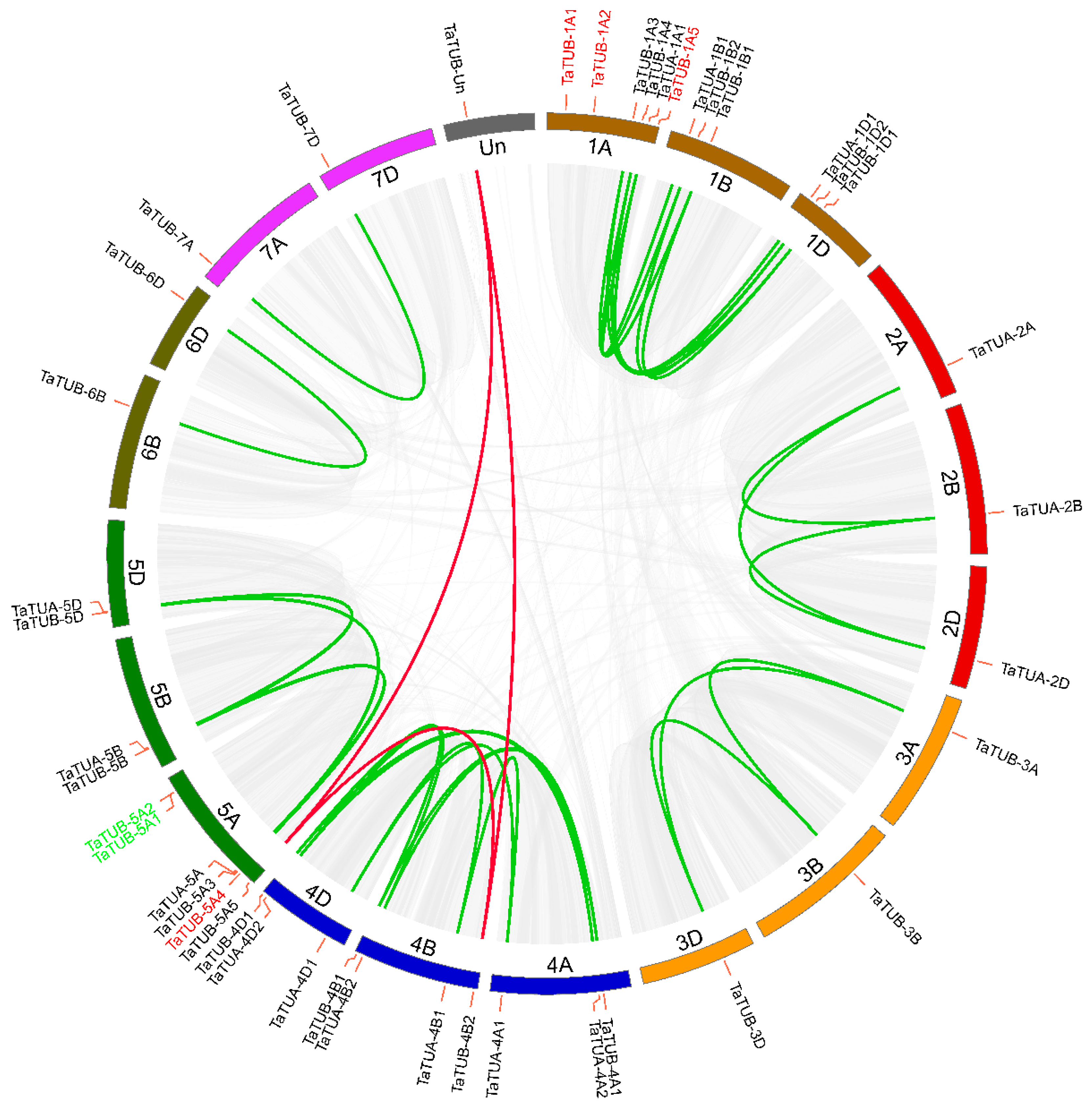
2.2. Gene Duplication and Synteny Analysis of TaTUA and TaTUB
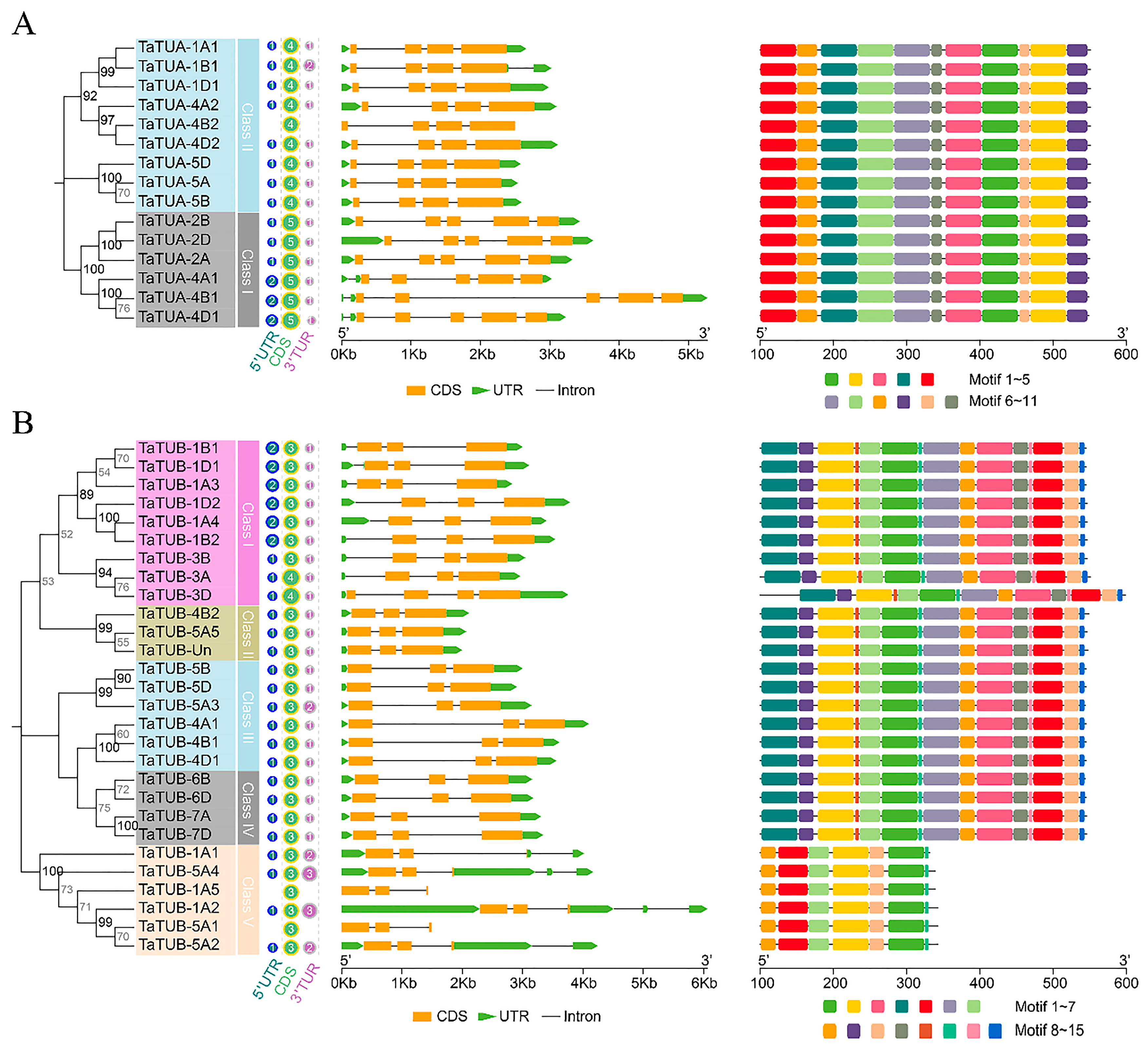
2.3. Gene Structure Analysis of TaTUA and TaTUB
2.4. Promoter Cis-Acting Element Analysis of TaTUA and TaTUB Genes
2.5. Expression Patterns of TaTUA and TaTUB Genes in Different Tissues and Developmental Stages of Wheat
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Identification of the Members of the TaTUA and TaTUB Gene Family
4.3. TaTUA and TaTUB Gene Structural, Evolution, and Protein Motifs Analysis
4.4. Promoter Cis-Acting Elements Analysis of TaTUA and TaTUB Genes
4.5. Gene Duplication and Synteny Analysis of TaTUA and TaTUB Genes
4.6. RNA Extraction, Reverse Transcription, and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fygenson, D.K.; Needleman, D.J.; Sneppen, K. Variability-based sequence alignment identifies residues responsible for functional differences in alpha and beta tubulin. Protein Sci. 2004, 13, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Tuszynski, J.A.; Carpenter, E.J.; Huzil, J.T.; Malinski, W.; Luchko, T.; Luduena, R.F. The evolution of the structure of tubulin and its potential consequences for the role and function of microtubules in cells and embryos. Int. J. Dev. Biol. 2006, 50, 341–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrotta, L.; Cresti, M.; Cai, G. Accumulation and post-translational modifications of plant tubulins. Plant Biol. 2014, 16, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Kopczak, S.D.; Haas, N.A.; Hussey, P.J.; Silflow, C.D.; Snustad, D.P. The small genome of Arabidopsis contains at least six expressed alpha-tubulin genes. Plant Cell 1992, 4, 539–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snustad, D.P.; Haas, N.A.; Kopczak, S.D.; Silflow, C.D. The small genome of Arabidopsis contains at least nine expressed beta-tubulin genes. Plant Cell 1992, 4, 549–556. [Google Scholar] [CrossRef] [Green Version]
- Oakley, R.V.; Wang, Y.S.; Ramakrishna, W.; Harding, S.A.; Tsai, C.J. Differential expansion and expression of alpha- and beta-tubulin gene families in Populus. Plant Physiol. 2007, 145, 961–973. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, M.; Yang, G.X.; Kawaguchi, K.; Komatsu, S. Expression analyses of beta-tubulin isotype genes in rice. Plant Cell Physiol. 2003, 44, 1202–1207. [Google Scholar] [CrossRef] [Green Version]
- He, X.C.; Qin, Y.M.; Xu, Y.; Hu, C.Y.; Zhu, Y.X. Molecular cloning, expression profiling, and yeast complementation of 19 beta-tubulin cDNAs from developing cotton ovules. J. Exp. Bot. 2008, 59, 2687–2695. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.P.; Srivastava, R.; Kumar, J. Male sterility systems in wheat and opportunities for hybrid wheat development. Acta Physiol. Plant 2015, 37, 108006. [Google Scholar] [CrossRef]
- Li, H.X.; Guo, J.L.; Zhang, C.Y.; Zheng, W.J.; Song, Y.L.; Wang, Y. Identification of Differentially Expressed miRNAs between a Wheat K-type Cytoplasmic Male Sterility Line and Its Near-Isogenic Restorer Line. Plant Cell Physiol. 2019, 60, 1604–1618. [Google Scholar] [CrossRef]
- Liu, Z.; Li, S.; Li, W.; Liu, Q.; Zhang, L.; Song, X. Comparative transcriptome analysis indicates that a core transcriptional network mediates isonuclear alloplasmic male sterility in wheat (Triticum aestivum L.). BMC Plant Biol. 2020, 20, 10. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhang, G.; Song, Q.; Zhang, Y.; Li, Z.; Guo, J.; Niu, N.; Ma, S.; Wang, J. Abnormal development of tapetum and microspores induced by chemical hybridization agent SQ-1 in wheat. PLoS ONE 2015, 10, e0119557. [Google Scholar] [CrossRef]
- Galindo-Trigo, S.; Grand, T.M.; Voigt, C.A.; Smith, L.M. A malectin domain kinesin functions in pollen and seed development in Arabidopsis. J. Exp. Bot. 2020, 71, 1828–1841. [Google Scholar] [CrossRef] [Green Version]
- Niu, B.X.; Wang, L.D.; Zhang, L.S.; Ren, D.; Ren, R.; Copenhaver, G.P.; Ma, H.; Wang, Y.X. Arabidopsis Cell Division Cycle 20.1 Is Required for Normal Meiotic Spindle Assembly and Chromosome Segregation. Plant Cell 2015, 27, 3367–3382. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.A.; Allen, T.; Kim, G.J.; Sidorova, A.; Borg, M.; Park, S.K.; Twell, D. Arabidopsis Fused kinase and the Kinesin-12 subfamily constitute a signalling module required for phragmoplast expansion. Plant J. 2012, 72, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.A.; Allen, T.; Twell, D. A ticket for the live show: Microtubules in male gametophyte development. Plant Signal. Behav. 2010, 5, 614–617. [Google Scholar] [CrossRef] [Green Version]
- Tchorzewska, D.; Derylo, K.; Blaszczyk, L.; Winiarczyk, K. Tubulin cytoskeleton during microsporogenesis in the male-sterile genotype of Allium sativum and fertile Allium ampeloprasum L. Plant Reprod. 2015, 28, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.F.; Han, J.P.; Wang, E.F.; Xiao, J.; Hu, R.; Yang, G.X.; He, G.Y. Genome-Wide Identification and Homoeologous Expression Analysis of PP2C Genes in Wheat (Triticum aestivum L.). Front. Genet. 2019, 10, 561. [Google Scholar] [CrossRef] [Green Version]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A Customizable RNA-seq Data Analysis and Visualization Platform. Plant Physiol. 2016, 170, 2172. [Google Scholar] [CrossRef]
- Ramírez-González, R.; Borrill, P.; Lang, D.; Harrington, S.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef] [Green Version]
- Radchuk, V.V. The transcriptome of the tubulin gene family in plants. In NATO Science for Peace and Security Series C; Springer: Berlin/Heidelberg, Germany, 2008; pp. 219–241. [Google Scholar] [CrossRef]
- Breviario, D.; Giani, S.; Ponzoni, E.; Mastromauro, F.; Morello, L. Plant tubulin intronics. Cell Biol. Int. 2008, 32, 571–573. [Google Scholar] [CrossRef] [PubMed]
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, A.B. Intron-mediated regulation of gene expression. Curr. Top. Microbiol. Immunol. 2008, 326, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.S.; Lee, S.; Jung, K.H.; Jun, S.H.; Kim, C.; An, G. Tissue-preferential expression of a rice alpha-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol. 2000, 123, 1005–1014. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, E.; Zeng, L.; Baird, W.V. Alpha-tubulin missense mutations correlate with antimicrotubule drug resistance in Eleusine indica. Plant Cell 1998, 10, 297–308. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Wang, X.L.; Huang, G.Q.; Li, X.B. Molecular characterization of cotton GhTUA9 gene specifically expressed in fibre and involved in cell elongation. J. Exp. Bot. 2007, 58, 3227–3238. [Google Scholar] [CrossRef]
- Ridha Farajalla, M.; Gulick, P.J. The alpha-tubulin gene family in wheat (Triticum aestivum L.) and differential gene expression during cold acclimation. Genome 2007, 50, 502–510. [Google Scholar] [CrossRef]
- Innan, H.; Kondrashov, F. The evolution of gene duplications: Classifying and distinguishing between models. Nat. Rev. Genet. 2010, 11, 97–108. [Google Scholar] [CrossRef]
- Murat, F.; Van de Peer, Y.; Salse, J. Decoding Plant and Animal Genome Plasticity from Differential Paleo-Evolutionary Patterns and Processes. Genome Biol. Evol. 2012, 4, 917–928. [Google Scholar] [CrossRef] [Green Version]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [Green Version]
- Buy, D.D.; Demkovych, A.E.; Pirko, Y.V.; Blume, Y.B. Analysis of α-tubulin gene expression during cold acclimation of winter and spring soft wheat. Cytol. Genet. 2019, 53, 23–33. [Google Scholar] [CrossRef]
- Christov, N.K.; Imai, R.; Blume, Y. Differential expression of two winter wheat alpha-tubulin genes during cold acclimation. Cell Biol. Int. 2008, 32, 574–578. [Google Scholar] [CrossRef]
- Song, Q.; Wang, S.; Zhang, G.; Li, Y.; Li, Z.; Guo, J.; Niu, N.; Wang, J.; Ma, S. Comparative proteomic analysis of a membrane-enriched fraction from flag leaves reveals responses to chemical hybridization agent SQ-1 in wheat. Front. Plant Sci. 2015, 6, 669. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Rychlik, W. OLIGO 7 primer analysis software. Methods Mol. Biol. 2007, 402, 35–60. [Google Scholar] [CrossRef] [PubMed]
- Rauf, M.; Awais, M.; Ud-Din, A.; Ali, K.; Gul, H.; Rahman, M.; Hamayun, M.; Arif, M. Molecular Mechanisms of the 1-Aminocyclopropane-1-Carboxylic Acid (ACC) Deaminase Producing Trichoderma asperellum MAP1 in Enhancing Wheat Tolerance to Waterlogging Stress. Front. Plant Sci. 2021, 11, 614971. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Gene ID | Gene Name | Chr | Length | MW (kDa) | pI | Gravy | Location | Gene Duplicate Patterns |
|---|---|---|---|---|---|---|---|---|
| TraesCS1A02G350200 | TaTUA-1A1 | 1A | 451 | 49.71 | 4.65 | −0.20 | GA | WGD/segmental |
| TraesCS1B02G364500 | TaTUA-1B1 | 1B | 451 | 49.71 | 4.65 | −0.20 | cytosol | WGD/segmental |
| TraesCS1D02G353100 | TaTUA-1D1 | 1D | 451 | 49.71 | 4.65 | −0.20 | cytosol | WGD/segmental |
| TraesCS2A02G185500 | TaTUA-2A | 2A | 450 | 49.61 | 4.72 | −0.15 | cytosol | WGD/segmental |
| TraesCS2B02G221800 | TaTUA-2B | 2B | 450 | 49.61 | 4.72 | −0.15 | cytosol | WGD/segmental |
| TraesCS2D02G202200 | TaTUA-2D | 2D | 450 | 49.61 | 4.72 | −0.15 | cytosol | WGD/segmental |
| TraesCS4A02G065700 | TaTUA-4A1 | 4A | 449 | 49.87 | 4.84 | −0.19 | cytosol | WGD/segmental |
| TraesCS4A02G257800 | TaTUA-4A2 | 4A | 451 | 49.73 | 4.65 | −0.20 | cytosol | WGD/segmental |
| TraesCS4B02G056700 | TaTUA-4B2 | 4B | 451 | 49.73 | 4.65 | −0.20 | cytosol | WGD/segmental |
| TraesCS4B02G243100 | TaTUA-4B1 | 4B | 449 | 49.89 | 4.84 | −0.19 | cytosol | WGD/segmental |
| TraesCS4D02G057000 | TaTUA-4D2 | 4D | 451 | 49.73 | 4.65 | −0.20 | cytosol | WGD/segmental |
| TraesCS4D02G242700 | TaTUA-4D1 | 4D | 449 | 49.89 | 4.84 | −0.19 | cytosol | WGD/segmental |
| TraesCS5A02G407800 | TaTUA-5A | 5A | 451 | 49.79 | 4.57 | −0.22 | GA | WGD/segmental |
| TraesCS5B02G412700 | TaTUA-5B | 5B | 451 | 49.79 | 4.57 | −0.22 | cytosol | WGD/segmental |
| TraesCS5D02G417700 | TaTUA-5D | 5D | 451 | 49.77 | 4.6 | −0.22 | cytosol | WGD/segmental |
| TraesCS1A02G101000 | TaTUB-1A1 | 1A | 232 | 25.06 | 7.31 | −0.23 | plastid | dispersed |
| TraesCS1A02G142800 | TaTUB-1A2 | 1A | 243 | 26.04 | 8.68 | −0.33 | plastid | dispersed |
| TraesCS1A02G258800 | TaTUB-1A3 | 1A | 445 | 50.14 | 4.48 | −0.36 | GA | WGD/segmental |
| TraesCS1A02G309700 | TaTUB-1A4 | 1A | 447 | 50.31 | 4.45 | −0.34 | GA | WGD/segmental |
| TraesCS1A02G437700 | TaTUB-1A5 | 1A | 239 | 25.69 | 6.5 | −0.33 | plastid | dispersed |
| TraesCS1B02G269400 | TaTUB-1B1 | 1B | 445 | 50.15 | 4.49 | −0.36 | cytosol | WGD/segmental |
| TraesCS1B02G320800 | TaTUB-1B2 | 1B | 447 | 50.31 | 4.45 | −0.34 | cytosol | WGD/segmental |
| TraesCS1D02G258800 | TaTUB-1D1 | 1D | 445 | 50.15 | 4.49 | −0.36 | cytosol | WGD/segmental |
| TraesCS1D02G309200 | TaTUB-1D2 | 1D | 447 | 50.31 | 4.45 | −0.34 | cytosol | WGD/segmental |
| TraesCS3A02G333300 | TaTUB-3A | 3A | 451 | 50.68 | 4.45 | −0.36 | cytosol | WGD/segmental |
| TraesCS3B02G363500 | TaTUB-3B | 3B | 447 | 50.21 | 4.42 | −0.37 | GA | WGD/segmental |
| TraesCS3D02G326900 | TaTUB-3D | 3D | 499 | 55.81 | 4.8 | −0.36 | plastid | WGD/segmental |
| TraesCS4A02G296600 | TaTUB-4A1 | 4A | 445 | 49.73 | 4.46 | −0.35 | cytosol | WGD/segmental |
| TraesCS4B02G017100 | TaTUB-4B1 | 4B | 445 | 49.76 | 4.47 | −0.36 | cytosol | WGD/segmental |
| TraesCS4B02G366500 | TaTUB-4B2 | 4B | 449 | 50.24 | 4.41 | −0.34 | cytosol | WGD/segmental |
| TraesCS4D02G015400 | TaTUB-4D1 | 4D | 445 | 49.76 | 4.47 | −0.35 | cytosol | WGD/segmental |
| TraesCS5A02G069700 | TaTUB-5A1 | 5A | 243 | 26.01 | 8.68 | −0.30 | plastid | tandem |
| TraesCS5A02G069800 | TaTUB-5A2 | 5A | 243 | 26.02 | 8.49 | −0.28 | plastid | tandem |
| TraesCS5A02G416400 | TaTUB-5A3 | 5A | 444 | 49.84 | 4.53 | −0.36 | cytosol | WGD/segmental |
| TraesCS5A02G443500 | TaTUB-5A4 | 5A | 239 | 25.53 | 6.88 | −0.34 | plastid | dispersed |
| TraesCS5A02G534300 | TaTUB-5A5 | 5A | 447 | 50.10 | 4.45 | −0.35 | cytosol | WGD/segmental |
| TraesCS5B02G418700 | TaTUB-5B | 5B | 445 | 49.97 | 4.5 | −0.37 | GA | WGD/segmental |
| TraesCS5D02G424100 | TaTUB-5D | 5D | 445 | 49.97 | 4.5 | −0.37 | cytosol | WGD/segmental |
| TraesCS6B02G169300 | TaTUB-6B | 6B | 444 | 49.87 | 4.4 | −0.38 | cytosol | WGD/segmental |
| TraesCS6D02G130500 | TaTUB-6D | 6D | 445 | 49.96 | 4.4 | −0.37 | GA | WGD/segmental |
| TraesCS7A02G466600 | TaTUB-7A | 7A | 446 | 50.21 | 4.43 | −0.39 | GA | WGD/segmental |
| TraesCS7D02G454200 | TaTUB-7D | 7D | 446 | 50.21 | 4.43 | −0.39 | cytosol | WGD/segmental |
| TraesCSU02G137900 | TaTUB-Un | Un | 448 | 50.21 | 4.45 | −0.34 | cytosol | WGD/segmental |
| Number of Homologous Loci | Distribution of Genome | Number of Genes | Total Number of Genes |
|---|---|---|---|
| Three homeologs | A, B, D | 10 | 30 |
| Two homeologs | A, D | 1 | 2 |
| B, D | 1 | 2 | |
| One homeolog | A | 7 | 14 |
| B | 1 | 2 | |
| Un | 1 | 2 | |
| Total | 43 |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| TaActin | ACCTTCAGTTGCCCAGCAAT | CAGAGTCGAGCACAATACCAGTTG |
| TUB-1B1 | AGCACCAAGGAGGTTGATGAACA | GTTGCCGATGAAGGTGGACG |
| TUA-2B | AGCTCATCTCTGGGAAGGAGG | GATCCAGTTCCACCACCAACA |
| TUB-3B | ACAAAGGAGGTGGACGAGCAG | TGGAGGTCGAGTTGCCAACAA |
| TUB-4A1 | GACGCCAAGAACATGATGTGTG | TCTGCTCATCCACCTCCTTTGT |
| TUA-4B2 | TTTGTTGATCTTGAGCCCACTGT | TGATACGGTCCAGGCATAGGTC |
| TUB-6D | GTCAGCTGAACTCCGACCTC | GGGTCAACTCAGGAACAGTGA |
| TUB-Un | CTCCACCTTCATCGGCAACTC | GGCCTCGGTGAACTCCATC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Y.; Song, Q.; Shan, S.; Wang, J.; Ma, S.; Song, Y.; Ma, L.; Zhang, G.; Niu, N. Genome-Wide Identification and Expression Analysis of TUA and TUB Genes in Wheat (Triticum aestivum L.) during Its Development. Plants 2022, 11, 3495. https://doi.org/10.3390/plants11243495
Ren Y, Song Q, Shan S, Wang J, Ma S, Song Y, Ma L, Zhang G, Niu N. Genome-Wide Identification and Expression Analysis of TUA and TUB Genes in Wheat (Triticum aestivum L.) during Its Development. Plants. 2022; 11(24):3495. https://doi.org/10.3390/plants11243495
Chicago/Turabian StyleRen, Yang, Qilu Song, Sicong Shan, Junwei Wang, Shoucai Ma, Yulong Song, Lingjian Ma, Gaisheng Zhang, and Na Niu. 2022. "Genome-Wide Identification and Expression Analysis of TUA and TUB Genes in Wheat (Triticum aestivum L.) during Its Development" Plants 11, no. 24: 3495. https://doi.org/10.3390/plants11243495
APA StyleRen, Y., Song, Q., Shan, S., Wang, J., Ma, S., Song, Y., Ma, L., Zhang, G., & Niu, N. (2022). Genome-Wide Identification and Expression Analysis of TUA and TUB Genes in Wheat (Triticum aestivum L.) during Its Development. Plants, 11(24), 3495. https://doi.org/10.3390/plants11243495










