A Point Mutation in Phytochromobilin synthase Alters the Circadian Clock and Photoperiodic Flowering of Medicago truncatula
Abstract
:1. Introduction
2. Results
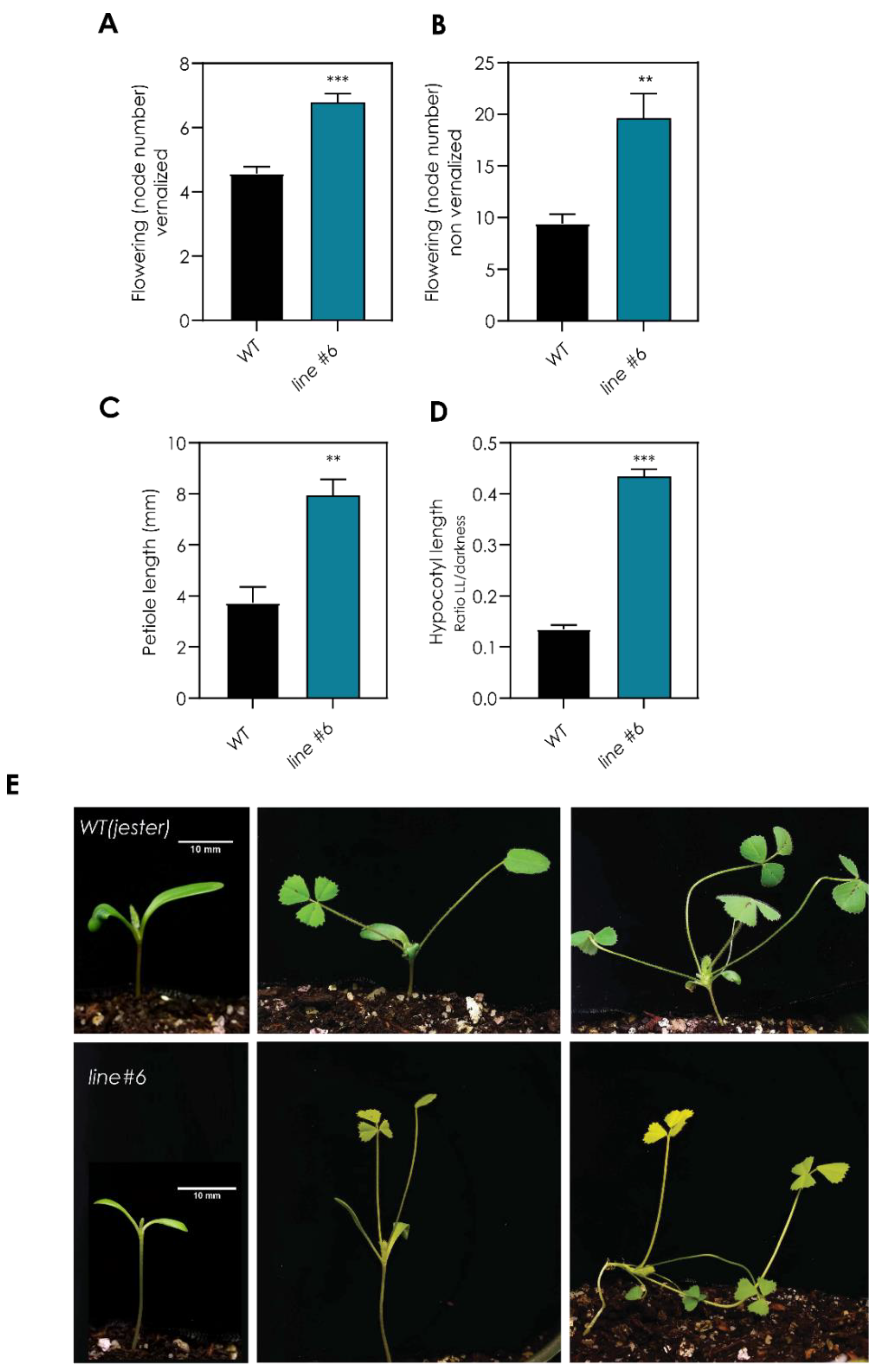
2.1. Screening of a TILLING Population Reveals a Late-Flowering Mutant with Light-Sensing Deficiency
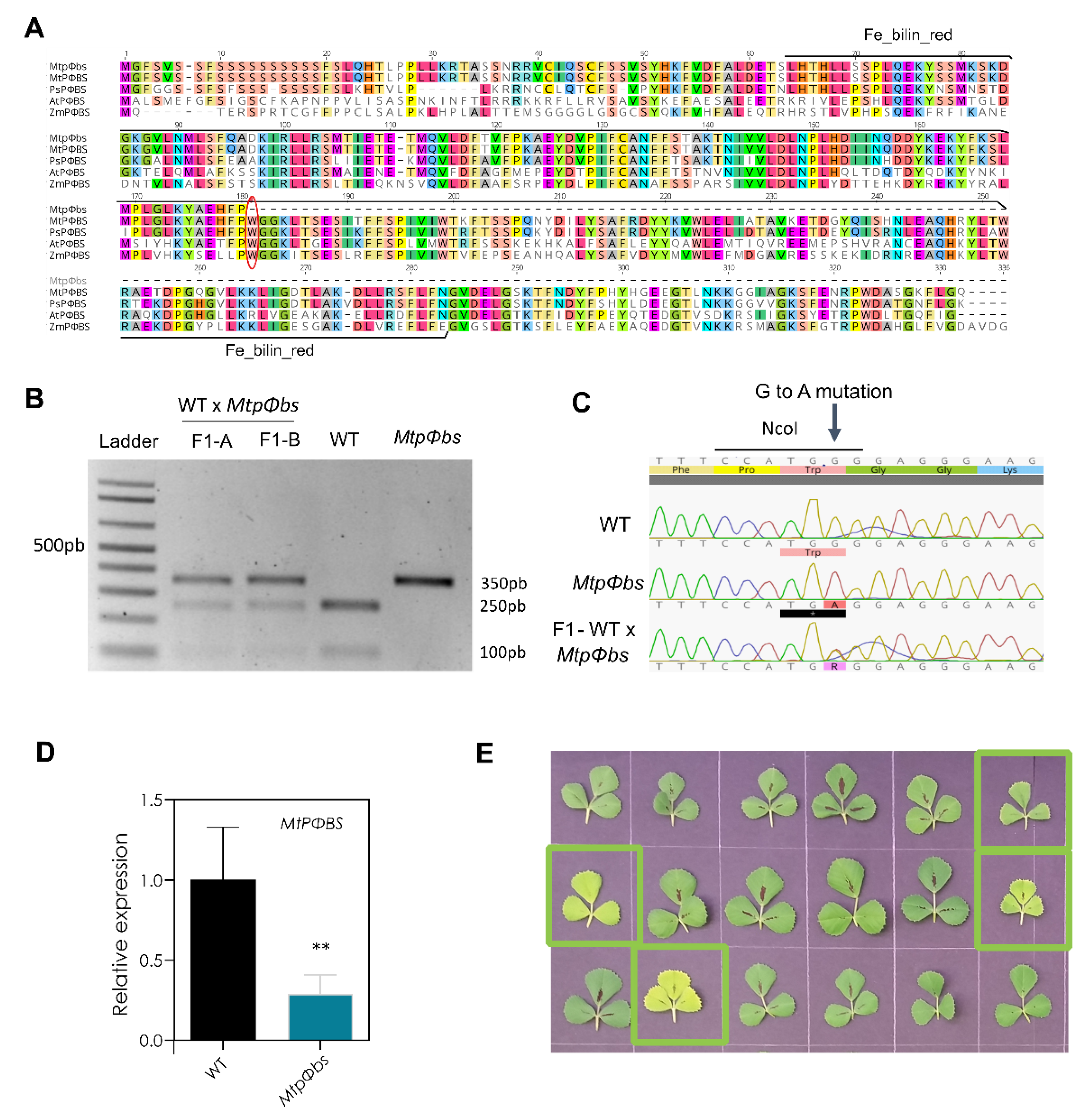
2.2. A Point Mutation in the MtPΦBS Gene Is Responsible for the Photo-Morphogenic and Late-Flowering Phenotypes
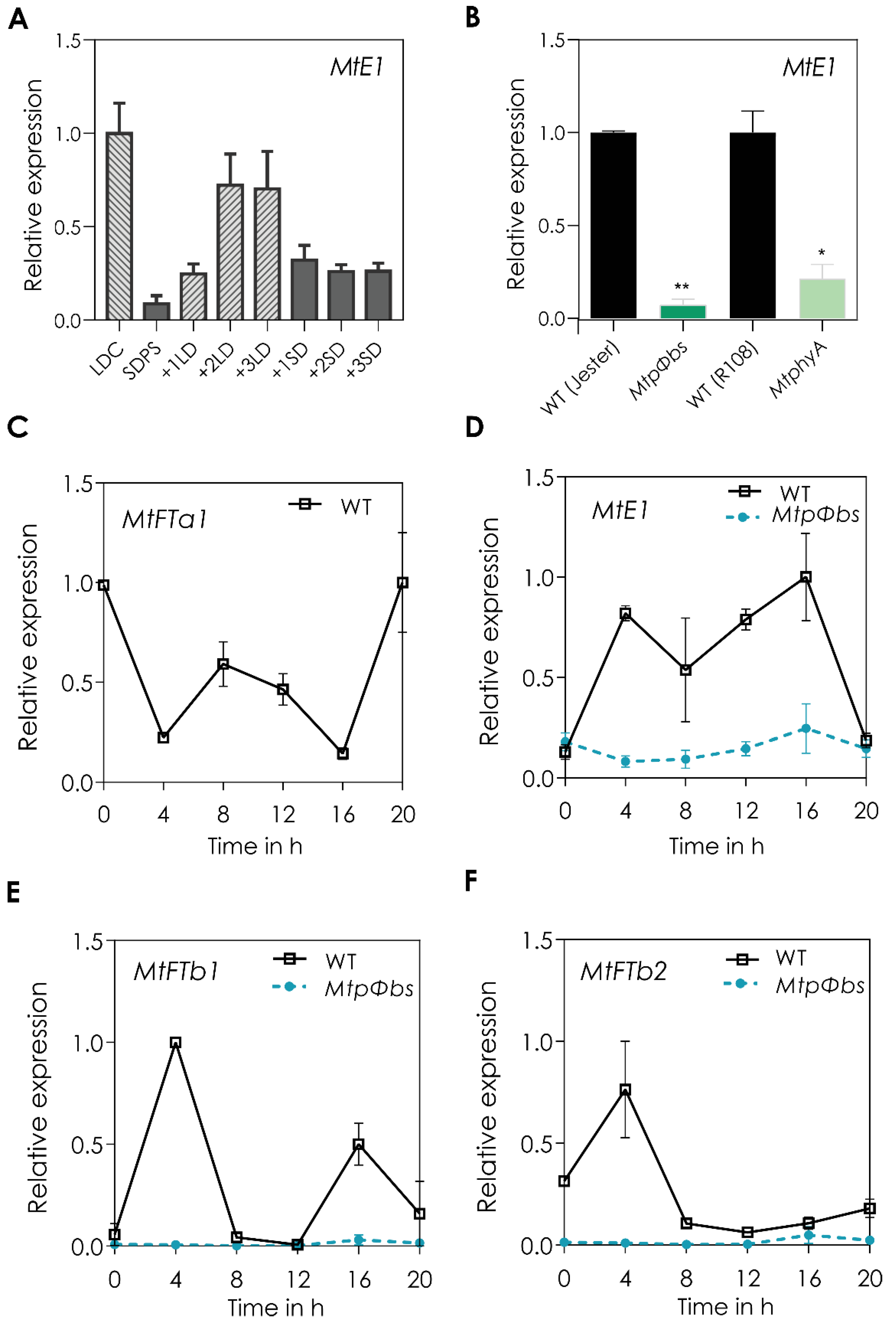
2.3. MtE1, a Legume Specific Flowering Gene, Is Only Expressed in LD Conditions and It Is Downregulated in the Mtpφbs Mutant
2.4. The MtpΦbs Mutant Has Altered Circadian Clock Function
2.5. The MtpΦbs Mutant Has a Shift in Expression of the Core Clock Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Growth Conditions
4.3. Hypocotyl Length
4.4. Flowering Time Analysis
4.5. Circadian Leaf Movement Analysis
4.6. Genotyping and DNA Sequencing, Protein Alignments and Phylogenetic Analysis
4.7. Generation of the F2 Population
4.8. RNA Extraction and RNA Sequencing
4.9. Processing of RNA Sequencing Reads
4.10. cDNA Synthesis and Quantitative RT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amasino, R.M.; Michaels, S.D. The timing of flowering. Plant Physiol. 2010, 154, 516–520. [Google Scholar] [CrossRef]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Suárez-López, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 2016, 173, 5–15. [Google Scholar] [CrossRef]
- Barak, S.; Tobin, E.M.; Green, R.M.; Andronis, C.; Sugano, S. All in good time: The Arabidopsis circadian clock. Trends Plant Sci. 2000, 5, 517–522. [Google Scholar] [CrossRef]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef]
- Sanchez, S.E.; Rugnone, M.L.; Kay, S.A. Light perception: A matter of time. Mol. Plant 2020, 13, 363–385. [Google Scholar] [CrossRef]
- Laurie, R.E.; Diwadkar, P.; Jaudal, M.; Zhang, L.; Hecht, V.; Wen, J.; Tadege, M.; Mysore, K.S.; Putterill, J.; Weller, J.L.; et al. The Medicago FLOWERING LOCUS T Homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 2011, 156, 2207–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putterill, J.; Varkonyi-Gasic, E. FT and florigen long-distance flowering control in plants. Curr. Opin. Plant Biol. 2016, 33, 77–82. [Google Scholar] [CrossRef]
- Jaudal, M.; Wen, J.; Mysore, K.S.; Putterill, J. Medicago PHYA promotes flowering, primary stem elongation and expression of flowering time genes in long days. BMC Plant Biol. 2020, 20, 329. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, A.; Thomson, G.; Kerr-Phillips, M.; Phan, C.; Krueger, T.; Jaudal, M.; Wen, J.; Mysore, K.S.; Putterill, J. Overexpression of Medicago MtCDFd1_1 causes delayed flowering in Medicago via repression of MtFTa1 but not MtCO-like genes. Front. Plant Sci. 2019, 10, 1148. [Google Scholar] [CrossRef] [PubMed]
- Thomson, G.; Zhang, L.; Wen, J.; Mysore, K.S.; Putterill, J. The candidate photoperiod gene MtFE promotes growth and flowering in Medicago truncatula. Front. Plant Sci. 2021, 12, 634091. [Google Scholar] [CrossRef]
- Liew, L.C.; Hecht, V.; Laurie, R.E.; Knowles, C.L.C.; Vander Schoor, J.K.; Macknight, R.C.; Weller, J.L. DIE NEUTRALIS and LATE BLOOMER 1 contribute to regulation of the pea circadian clock. Plant Cell 2009, 21, 3198–3211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, J.L.; Liew, L.C.; Hecht, V.F.G.; Rajandran, V.; Laurie, R.E.; Ridge, S.; Wenden, B.; Vander Schoor, J.K.; Jaminon, O.; Blassiau, C.; et al. A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. USA 2012, 109, 21158–22163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liew, L.C.; Hecht, V.; Sussmilch, F.C.; Weller, J.L. The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol. 2014, 165, 648–657. [Google Scholar] [CrossRef] [Green Version]
- Rubenach, A.J.S.; Hecht, V.F.; Vander Schoor, J.K.; Liew, L.C.; Aubert, G.; Burstin, J.; Weller, J.L. 2017. EARLY FLOWERING 3 redundancy fine-tunes photoperiod sensitivity. Plant Physiol. 2017, 173, 2253–2264. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.; Hecht, V.F.; Picard, K.; Diwadkar, P.; Laurie, R.E.; Wen, J.; Mysore, K.; Macknight, R.C.; Weller, J.L. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 2014, 5, 486. [Google Scholar] [CrossRef]
- Hecht, V.; Foucher, F.; Ferrándiz, C.; Macknight, R.C.; Navarro, C.; Morin, J.; Vardy, M.E.; Ellis, N.; Beltrán, J.P.; Rameau, C.; et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005, 137, 1420–1434. [Google Scholar] [CrossRef] [Green Version]
- Hecht, V.; Knowles, C.L.; Vander Schoor, J.K.; Liew, L.C.; Jones, S.E.; Lambert, M.J.M.; Weller, J.L. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 2007, 144, 648–661. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lu, S.; et al. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2012, 109, E2155–E2164. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Jiang, B.; Ma, L.; Zhang, S.; Zhai, H.; Xu, X.; Hou, W.; Xia, Z.; Wu, C.; Sun, S.; et al. Functional diversification of flowering locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 2018, 217, 1335–1345. [Google Scholar] [CrossRef] [Green Version]
- Thakare, D.; Kumudini, S.; Dinkins, R.D. The alleles at the E1 locus impact the expression pattern of two soybean FT-like genes shown to induce flowering in Arabidopsis. Planta 2011, 234, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yamagishi, N.; Zhao, C.; Takeshima, R.; Kasai, M.; Watanabe, S.; Kanazawa, A.; Yoshikawa, N.; Liu, B.; Yamada, T.; et al. The soybean-specific maturity gene E1 family of floral repressors controls night-break responses through down-regulation of FLOWERING LOCUS T orthologs. Plant Physiol. 2015, 168, 1735–1746. [Google Scholar] [CrossRef] [Green Version]
- Zhai, H.; Lü, S.; Liang, S.; Wu, H.; Zhang, X.; Liu, B.; Kong, F.; Yuan, X.; Li, J.; Xia, Z. GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS ONE 2014, 9, e89030. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhai, H.; Wang, Y.; Tian, X.; Zhang, Y.; Wu, H.; Lü, S.; Yang, G.; Li, Y.; Wang, L.; et al. Functional conservation and diversification of the soybean maturity gene E1 and its homologs in legumes. Sci. Rep. 2016, 6, 29548. [Google Scholar] [CrossRef] [Green Version]
- Weller, J.; Murfet, I.C.; Reid, J.B. Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol. 1997, 114, 1225–1236. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Hideshima, R.; Xia, Z.; Tsubokura, Y.; Sato, S.; Nakamoto, Y.; Yamanaka, N.; Takahashi, R.; Ishimoto, M.; Anai, T.; et al. Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 2009, 182, 1251–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terry, M.J.; Wahleithner, J.A.; Lagarias, J.C. Biosynthesis of the plant photoreceptor phytochrome. Arch. Biochem. Biophys. 1993, 306, 1–15. [Google Scholar] [CrossRef]
- Weller, J.L.; Terry, M.J.; Rameau, C.; Reid, J.B.; Kendrick, R.E. The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXalpha. Plant Cell 1996, 8, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J.; Kurepa, J.; Vierstra, R.D. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc. Natl. Acad. Sci. USA 2002, 96, 6541–6546. [Google Scholar] [CrossRef] [Green Version]
- Weller, J.L.; Terry, M.J.; Reid, J.B.; Kendrick, R.E. The phytochrome-deficient pcd2 mutant of pea is unable to convert biliverdin IXalpha to 3(Z)-phytochromobilin. Plant J. 1997, 11, 1177–1186. [Google Scholar] [CrossRef]
- Kohchi, T.; Mukougawa, K.; Frankenberg, N.; Masuda, M.; Yokota, A.; Lagarias, J.C. The Arabidopsis HY2 gene encodes Phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 2007, 13, 425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- RevGenUK [WWW Document]. Available online: https://www.jic.ac.uk/research-impact/technology-research-platforms/reverse-genetics/ (accessed on 6 December 2021).
- Tang, H.; Krishnakumar, V.; Bidwell, S.; Rosen, B.; Chan, A.; Zhou, S.; Gentzbittel, L.; Childs, K.L.; Yandell, M.; Gundlach, H. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genom. 2014, 15, 312. [Google Scholar] [CrossRef] [Green Version]
- Nickless, A.; Bailis, J.M.; You, Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci. 2017, 7, 26. [Google Scholar] [CrossRef]
- Weller, J.L.; Reid, J.B.; Taylor, S.A.; Murfet, I.C. The genetic control of flowering in pea. Trends Plant Sci. 1997, 2, 412–418. [Google Scholar] [CrossRef]
- Legris, M.; Ince, Y.Ç.; Fankhauser, C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat Commun 2019, 10, 5219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, G.; Taylor, J.; Putterill, J. The transcriptomic response to a short day to long day shift in leaves of the reference legume Medicago truncatula. PeerJ 2019, 7, e6626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elich, T.D.; McDonagh, A.F.; Palma, L.A.; Lagarias, J.C. Phytochrome chromophore biosynthesis. Treatment of tetrapyrrole-deficient Avena explants with natural and non-natural bilatrienes leads to formation of spectrally active holoproteins. J. Biol. Chem. 1989, 264, 183–189. [Google Scholar] [CrossRef]
- Bhaskara, G.B.; Wen, T.-N.; Nguyen, T.T.; Verslues, P.E. Protein phosphatase 2Cs and microtubule-associated stress protein 1 control microtubule stability, plant growth, and drought response. Plant Cell 2017, 29, 169–191. [Google Scholar] [CrossRef] [Green Version]
- Weller, J.L.; Ortega, R. Genetic control of flowering time in legumes. Front. Plant Sci. 2015, 6, 207. [Google Scholar] [CrossRef] [Green Version]
- Somers, D.E.; Devlin, P.F.; Kay, S.A. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 1998, 282, 1488–1490. [Google Scholar] [CrossRef] [Green Version]
- Johnson, E.; Bradley, M.; Harberd, N.P.; Whitelam, G.C. Photoresponses of light-grown phyA mutants of Arabidopsis (phytochrome A is required for the perception of daylength extensions). Plant Physiol. 1994, 105, 141–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, J.L.; Batge, S.L.; Smith, J.J.; Kerckhoffs, L.H.J.; Sineshchekov, V.A.; Murfet, I.C.; Reid, J.B. A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol. 2004, 135, 2186–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, J.W.; Nagatani, A.; Elich, T.D.; Fagan, M.; Chory, J. Phytochrome A and Phytochrome B Have Overlapping but Distinct Functions in Arabidopsis Development. Plant Physiol. 1994, 104, 1139–1149. [Google Scholar] [CrossRef]
- Weller, J.L.; Nagatani, A.; Kendrick, R.E.; Murfet, I.C.; Reid, J.B. New lv Mutants of Pea Are Deficient in Phytochrome B. Plant Physiol. 1995, 108, 525–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.; Wang, M.; Lee, H.K.; Tadege, M.; Ratet, P.; Udvardi, M.; Mysore, K.S.; Wen, J. An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol. 2014, 201, 1065–1076. [Google Scholar] [CrossRef]
- Iserte, J.; Yanovsky, M.J. Leaftion 2016 [Computer software]. Available online: https://gitlab.com/javieriserte/leaftion (accessed on 6 December 2021).
- Greenham, K.; Lou, P.; Remsen, S.E.; Farid, H.; McClung, C.R. TRiP: Tracking rhythms in plants, an automated leaf movement analysis program for circadian period estimation. Plant Methods 2015, 11, 33. [Google Scholar] [CrossRef] [Green Version]
- Zielinski, T.; Moore, A.M.; Troup, E.; Halliday, K.J.; Millar, A.J. Strengths and limitations of period estimation methods for circadian data. PLoS ONE 2014, 9, e96462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raw Sequencing Data. Available online: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA794171 (accessed on 6 December 2021).
- Kakar, K.; Wandrey, M.; Czechowski, T.; Gaertner, T.; Scheible, W.R.; Stitt, M.; Torres-Jerez, I.; Xiao, Y.; Redman, J.C.; Wu, H.C.; et al. A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods. 2008, 4, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bookout, A.L.; Mangelsdorf, D.J. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Recept. Signal. 2003, 1, nrs-01012. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perez-Santangelo, S.; Napier, N.; Robson, F.; Weller, J.L.; Bond, D.M.; Macknight, R.C. A Point Mutation in Phytochromobilin synthase Alters the Circadian Clock and Photoperiodic Flowering of Medicago truncatula. Plants 2022, 11, 239. https://doi.org/10.3390/plants11030239
Perez-Santangelo S, Napier N, Robson F, Weller JL, Bond DM, Macknight RC. A Point Mutation in Phytochromobilin synthase Alters the Circadian Clock and Photoperiodic Flowering of Medicago truncatula. Plants. 2022; 11(3):239. https://doi.org/10.3390/plants11030239
Chicago/Turabian StylePerez-Santangelo, Soledad, Nathanael Napier, Fran Robson, James L. Weller, Donna M. Bond, and Richard C. Macknight. 2022. "A Point Mutation in Phytochromobilin synthase Alters the Circadian Clock and Photoperiodic Flowering of Medicago truncatula" Plants 11, no. 3: 239. https://doi.org/10.3390/plants11030239






