Phylogenetic and Expression Analysis of CENH3 and APOLLO Genes in Sexual and Apomictic Boechera Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. APOLLO and CENH3 Genes Retrieval and Pre-Processing
2.3. General Characteristics of CENH3 Gene
2.4. Evolutionary Analysis of the CENH3 and APOLLO Genes
2.5. CENH3 and APOLLO Genes Expression Studies
3. Results
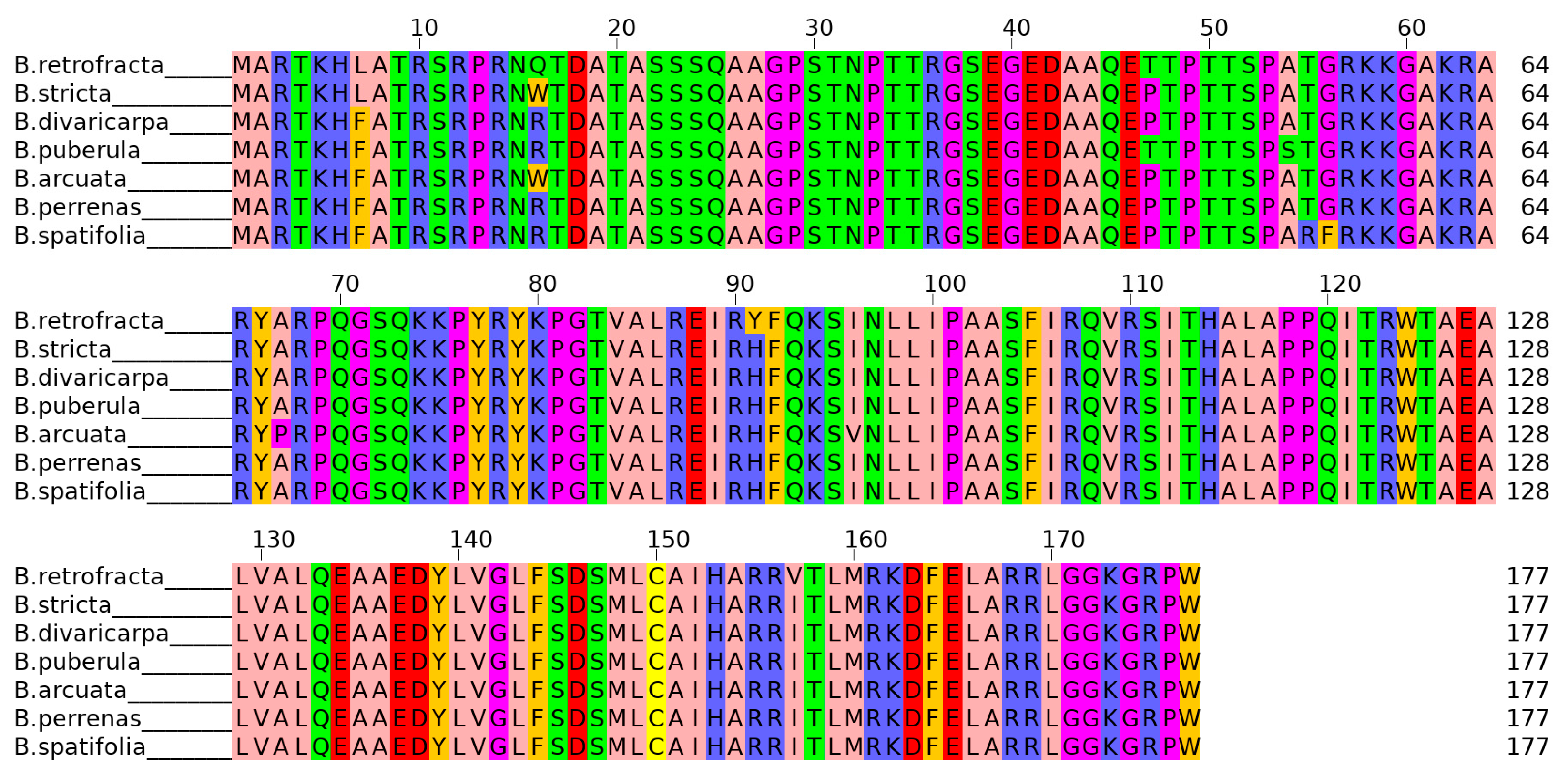
3.1. Characteristics of CENH3 Gene and Protein Isoforms
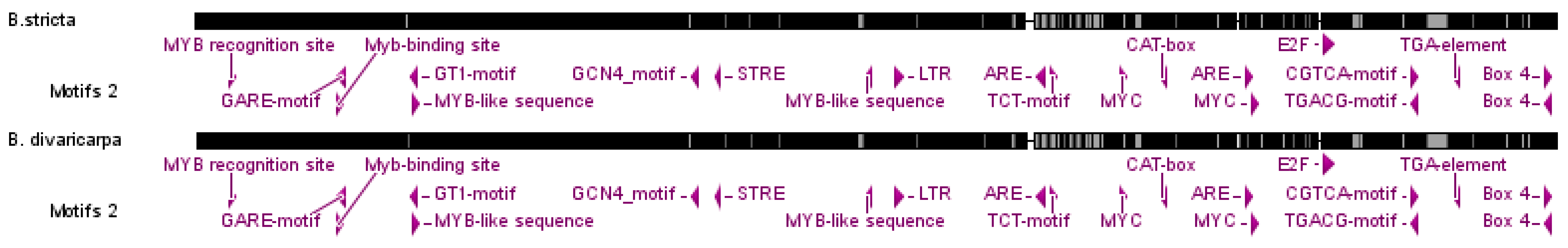
3.2. CENH3 Promoter Characteristic
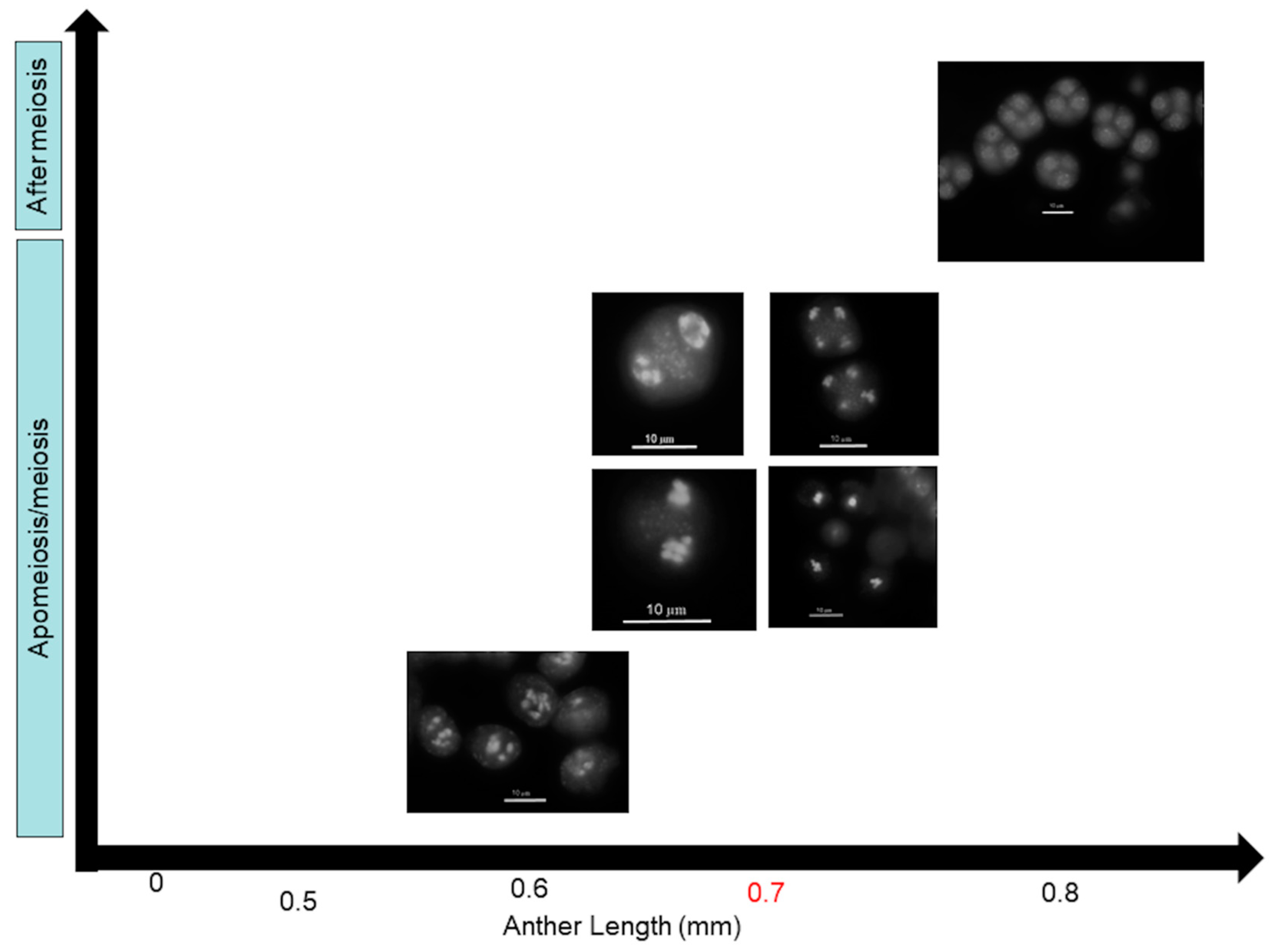
3.3. CENH3 Expression during and after Meiosis in Gynoecia/Siliques and Anthers
3.4. APOLLO Expression during and after Meiosis in Gynoecia/Siliques and Anthers
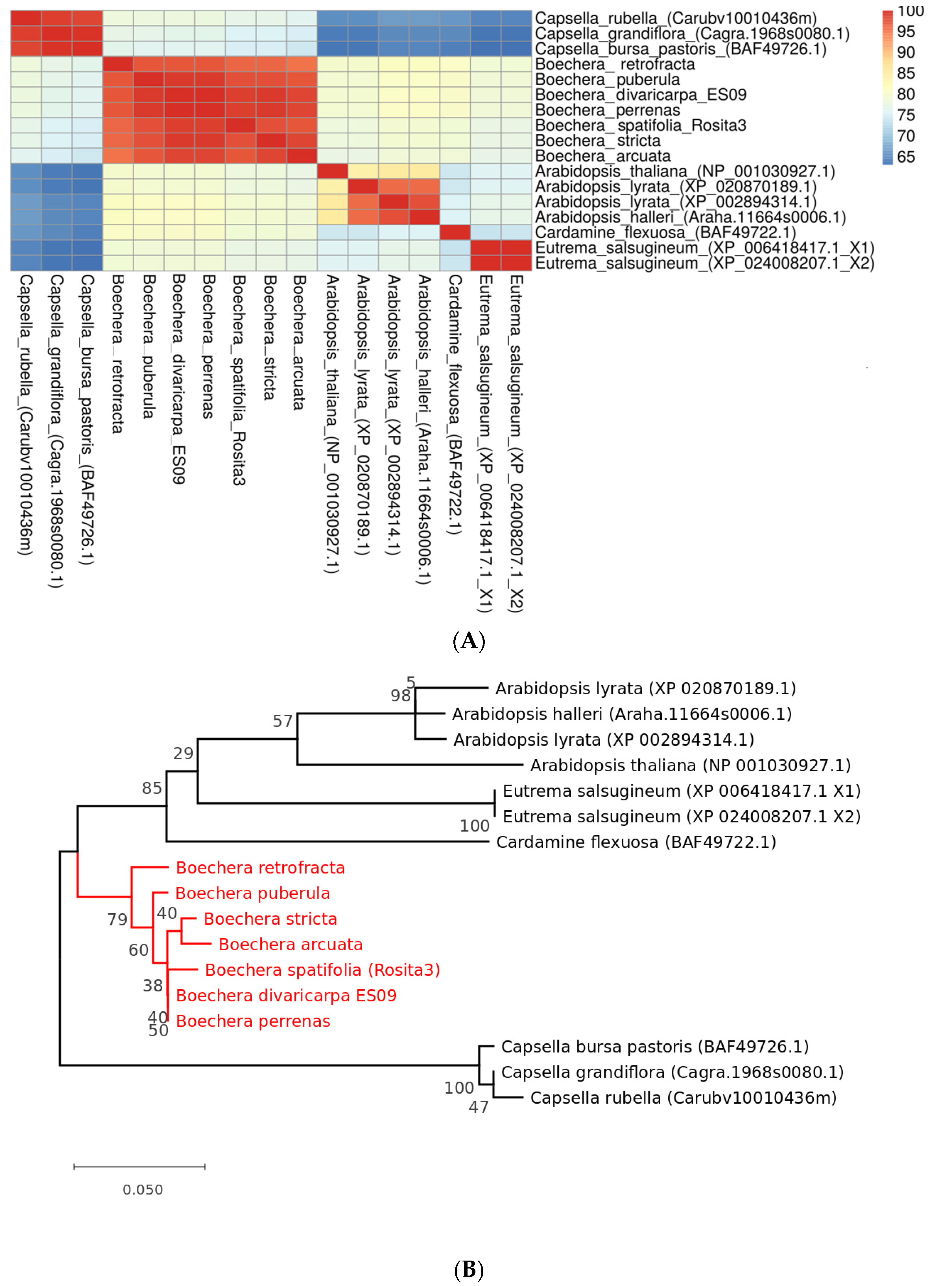
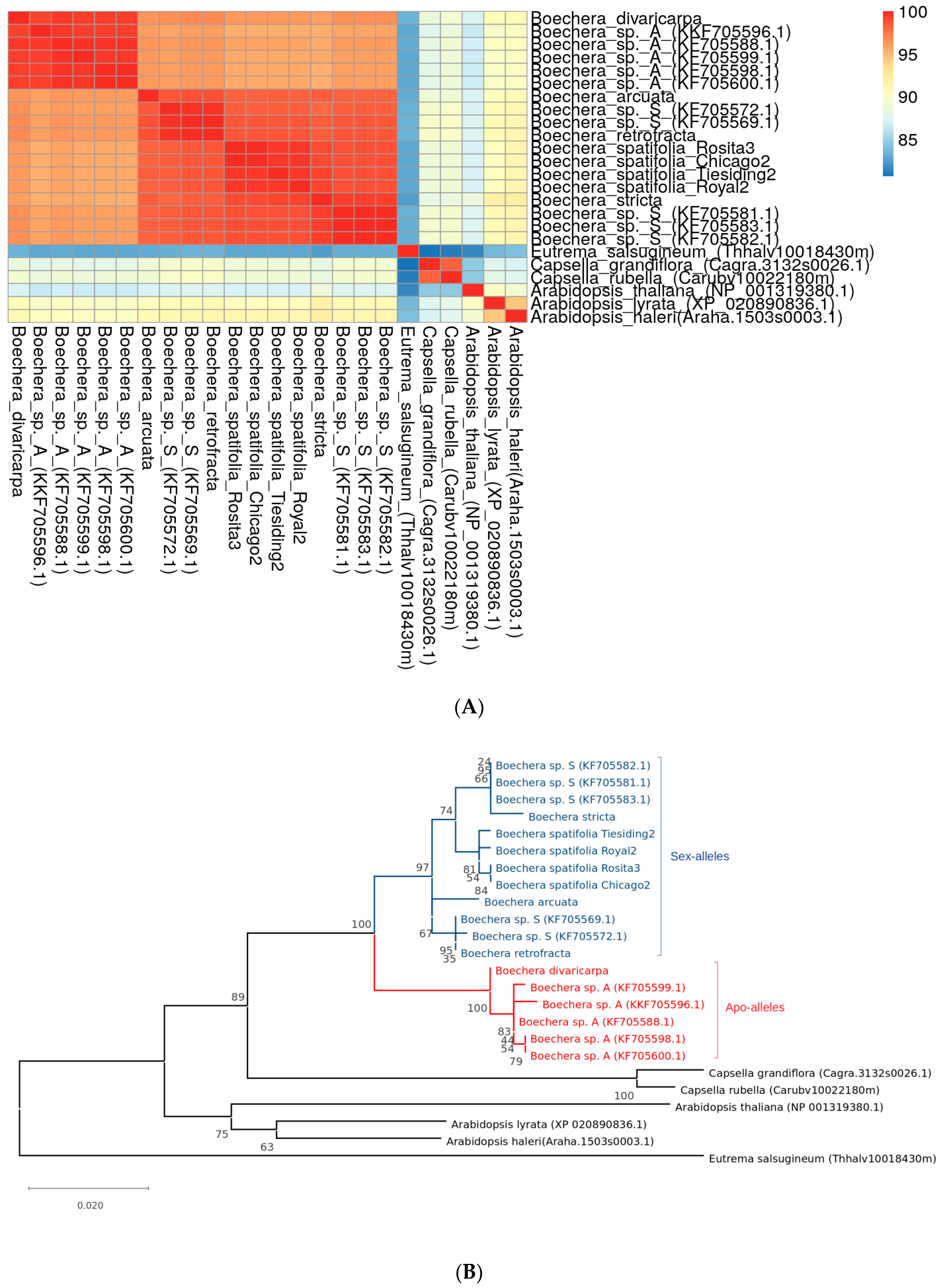
3.5. Phylogenetic Analysis of APOLLO Gene
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Muller, H.J. Some Genetic Aspects of Sex. Am. Nat. 1932, 66, 118–138. [Google Scholar] [CrossRef]
- Fisher, R.A. The Genetical Theory of Natural Selection: By R.A. Fisher; Bennett, J.H., Ed.; Oxford University Press: Oxford, UK, 1999; ISBN 978-0-19-850440-5. [Google Scholar]
- Hill, W.G.; Robertson, A. The Effect of Linkage on Limits to Artificial Selection. Genet. Res. 1966, 8, 269–294. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. The Evolutionary Advantage of Recombination. Genetics 1974, 78, 737–756. [Google Scholar] [CrossRef] [PubMed]
- Spillane, C.; Curtis, M.D.; Grossniklaus, U. Apomixis Technology Development—Virgin Births in Farmers’ Fields? Nat. Biotechnol. 2004, 22, 687–691. [Google Scholar] [CrossRef]
- Brukhin, V. Molecular and Genetic Regulation of Apomixis. Russ. J. Genet. 2017, 53, 943–964. [Google Scholar] [CrossRef]
- Carman, J.G. Asynchronous Expression of Duplicate Genes in Angiosperms May Cause Apomixis, Bispory, Tetraspory, and Polyembryony. Biol. J. Linn. Soc. 1997, 61, 51–94. [Google Scholar] [CrossRef]
- Johri, B.M. Embryology of Angiosperms; Springer: Berlin/Heidelberg, Germany, 1984; ISBN 978-3-642-69302-1. [Google Scholar]
- Asker, S.E.; Jerling, L. Apomixis in Plants; CRC Press: Boca Raton, FL, USA, 1992; ISBN 978-0-8493-4545-6. [Google Scholar]
- Savidan, Y.; Carman, J.G.; Dresselhaus, T.; International Maize and Wheat Improvement Center (Eds.) The Flowering of Apomixis: From Mechanisms to Genetic Engineering; International Maize and Wheat Improvement Center: Mexico City, Mexico; European Union: Luxembourg; Institut de Recherche pour le Développement: Paris, France, 2001; ISBN 978-970-648-074-3. [Google Scholar]
- Bicknell, R.A. Understanding Apomixis: Recent Advances and Remaining Conundrums. Plant Cell Online 2004, 16, S228–S245. [Google Scholar] [CrossRef] [Green Version]
- Schön, I.; Martens, K.; van Dijk, P. (Eds.) Lost Sex: The Evolutionary Biology of Parthenogenesis; Springer: Dordrecht, The Netherlands; New York, NY, USA, 2009; ISBN 978-90-481-2769-6. [Google Scholar]
- Kotani, Y.; Henderson, S.T.; Suzuki, G.; Johnson, S.D.; Okada, T.; Siddons, H.; Mukai, Y.; Koltunow, A.M.G. The Loss of Apomeiosis (LOA) Locus in Hieracium Praealtum Can Function Independently of the Associated Large-Scale Repetitive Chromosomal Structure. New Phytol. 2014, 201, 973–981. [Google Scholar] [CrossRef]
- Aliyu, O.M.; Schranz, M.E.; Sharbel, T.F. Quantitative Variation for Apomictic Reproduction in the Genus Boechera (Brassicaceae). Am. J. Bot. 2010, 97, 1719–1731. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Klatt, S.; Baier, R.; Carman, J.G.; Hörandl, E. Taxonomy and Biogeography of Apomixis in Angiosperms and Associated Biodiversity Characteristics. Crit. Rev. Plant Sci. 2014, 33, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Hojsgaard, D. Transient Activation of Apomixis in Sexual Neotriploids May Retain Genomically Altered States and Enhance Polyploid Establishment. Front. Plant Sci. 2018, 9, 230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brukhin, V.; Baskar, R. A Brief Note on Genes That Trigger Components of Apomixis. J. Biosci. 2019, 44, 45. [Google Scholar] [CrossRef] [PubMed]
- Corral, J.M.; Vogel, H.; Aliyu, O.M.; Hensel, G.; Thiel, T.; Kumlehn, J.; Sharbel, T.F. A Conserved Apomixis-Specific Polymorphism Is Correlated with Exclusive Exonuclease Expression in Premeiotic Ovules of Apomictic Boechera Species. Plant Physiol. 2013, 163, 1660–1672. [Google Scholar] [CrossRef] [Green Version]
- Kliver, S.; Rayko, M.; Komissarov, A.; Bakin, E.; Zhernakova, D.; Prasad, K.; Rushworth, C.; Baskar, R.; Smetanin, D.; Schmutz, J.; et al. Assembly of the Boechera Retrofracta Genome and Evolutionary Analysis of Apomixis-Associated Genes. Genes 2018, 9, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lermontova, I.; Koroleva, O.; Rutten, T.; Fuchs, J.; Schubert, V.; Moraes, I.; Koszegi, D.; Schubert, I. Knockdown of CENH3 in Arabidopsis Reduces Mitotic Divisions and Causes Sterility by Disturbed Meiotic Chromosome Segregation: Consequences of AtCENH3 Depletion. Plant J. 2011, 68, 40–50. [Google Scholar] [CrossRef]
- Lermontova, I.; Sandmann, M.; Mascher, M.; Schmit, A.-C.; Chabouté, M.-E. Centromeric Chromatin and Its Dynamics in Plants. Plant J. 2015, 83, 4–17. [Google Scholar] [CrossRef] [Green Version]
- Talbert, P.B.; Henikoff, S. Histone Variants—Ancient Wrap Artists of the Epigenome. Nat. Rev. Mol. Cell Biol. 2010, 11, 264–275. [Google Scholar] [CrossRef]
- Maheshwari, S.; Tan, E.H.; West, A.; Franklin, F.C.H.; Comai, L.; Chan, S.W.L. Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids. PLoS Genet. 2015, 11, e1004970. [Google Scholar] [CrossRef] [Green Version]
- Ravi, M.; Shibata, F.; Ramahi, J.S.; Nagaki, K.; Chen, C.; Murata, M.; Chan, S.W.L. Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in Arabidopsis Thaliana. PLoS Genet. 2011, 7, e1002121. [Google Scholar] [CrossRef] [Green Version]
- Ravi, M.; Chan, S.W.L. Haploid Plants Produced by Centromere-Mediated Genome Elimination. Nature 2010, 464, 615–618. [Google Scholar] [CrossRef]
- Marimuthu, M.P.A.; Jolivet, S.; Ravi, M.; Pereira, L.; Davda, J.N.; Cromer, L.; Wang, L.; Nogue, F.; Chan, S.W.L.; Siddiqi, I.; et al. Synthetic Clonal Reproduction Through Seeds. Science 2011, 331, 876. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Ashtiyani, R.; Ishii, T.; Niessen, M.; Stein, N.; Heckmann, S.; Gurushidze, M.; Banaei-Moghaddam, A.M.; Fuchs, J.; Schubert, V.; Koch, K.; et al. Point Mutation Impairs Centromeric CENH3 Loading and Induces Haploid Plants. Proc. Natl. Acad. Sci. USA 2015, 112, 11211–11216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evtushenko, E.V.; Lipikhina, Y.A.; Stepochkin, P.I.; Vershinin, A.V. Cytogenetic and Molecular Characteristics of Rye Genome in Octoploid Triticale (× Triticosecale Wittmack). Comp. Cytogenet. 2019, 13, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Haubold, B.; Mitchell-Olds, T. Molecular Systematics of the Brassicaceae: Evidence from Coding Plastidic MatK and Nuclear Chs Sequences. Am. J. Bot. 2001, 88, 534–544. [Google Scholar] [CrossRef]
- Al-Shehbaz, I.A. Transfer of Most North American Species of Arabis to Boechera (Brassicaceae). Novon 2003, 13, 381. [Google Scholar] [CrossRef]
- Brukhin, V.; Osadtchiy, J.V.; Florez-Rueda, A.M.; Smetanin, D.; Bakin, E.; Nobre, M.S.; Grossniklaus, U. The Boechera Genus as a Resource for Apomixis Research. Front. Plant Sci. 2019, 10, 392. [Google Scholar] [CrossRef] [Green Version]
- Böcher, T. Experimental Taxonomical Studies in the Arabis Holboellii Complex. Sven. Bot. Tidskr. 1954, 48, 31–44. [Google Scholar]
- Naumova, T.N.; van der Laak, J.; Osadtchiy, J.; Matzk, F.; Kravtchenko, A.; Bergervoet, J.; Ramulu, K.S.; Boutilier, K. Reproductive Development in Apomictic Populations of Arabis Holboellii (Brassicaceae). Sex Plant Reprod. 2001, 14, 195–200. [Google Scholar] [CrossRef]
- Taskin, K.M.; Turgut, K.; Scott, R.J. Apomictic Development in Arabis Gunnisoniana. Isr. J. Plant Sci. 2004, 52, 155–160. [Google Scholar] [CrossRef]
- Carman, J.G.; De Arias, M.M.; Gao, L.; Zhao, X.; Kowallis, B.M.; Sherwood, D.A.; Srivastava, M.K.; Dwivedi, K.K.; Price, B.J.; Watts, L.; et al. Apospory and diplospory in diploid Boechera (Brassicaceae) may facilitate speciation by recombination-driven apomixis-to-sex reversals. Front. Plant Sci. 2019, 10, 724. [Google Scholar] [CrossRef]
- Koch, M.A. Multiple Hybrid Formation in Natural Populations: Concerted Evolution of the Internal Transcribed Spacer of Nuclear Ribosomal DNA (ITS) in North American Arabis Divaricarpa (Brassicaceae). Mol. Biol. Evol. 2003, 20, 338–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voigt-Zielinski, M.-L.; Piwczyński, M.; Sharbel, T.F. Differential Effects of Polyploidy and Diploidy on Fitness of Apomictic Boechera. Sex Plant Reprod. 2012, 25, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Dobes, C.; Koch, M.A.; Mitchell-Olds, T. Sexual Reproduction, Hybridization, Apomixis, and Polyploidization in the Genus Boechera (Brassicaceae). Am. J. Bot. 2005, 92, 1797–1810. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Kantama, L.; de Jong, H.; Mitchell-Olds, T. Asexual Reproduction in a Close Relative of Arabidopsis: A Genetic Investigation of Apomixis in Boechera (Brassicaceae). New Phytol. 2006, 171, 425–438. [Google Scholar] [CrossRef]
- Taskin, K.M.; Turgut, K.; Scott, R.J. Apomeiotic Pollen Mother Cell Development in the Apomictic Boechera Species. Biologia Plant. 2009, 53, 468–474. [Google Scholar] [CrossRef]
- Lee, C.-R.; Wang, B.; Mojica, J.P.; Mandáková, T.; Prasad, K.V.S.K.; Goicoechea, J.L.; Perera, N.; Hellsten, U.; Hundley, H.N.; Johnson, J.; et al. Young Inversion with Multiple Linked QTLs under Selection in a Hybrid Zone. Nat. Ecol. Evol. 2017, 1, 0119. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, C.; Willing, E.-M.; Jiao, W.-B.; Sun, H.; Piednoël, M.; Hümann, U.; Hartwig, B.; Koch, M.A.; Schneeberger, K. Interspecies Association Mapping Links Reduced CG to TG Substitution Rates to the Loss of Gene-Body Methylation. Nat. Plants 2019, 5, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Lovell, J.T.; Aliyu, O.M.; Mau, M.; Schranz, M.E.; Koch, M.; Kiefer, C.; Song, B.-H.; Mitchell-Olds, T.; Sharbel, T.F. On the Origin and Evolution of Apomixis in Boechera. Plant Reprod. 2013, 26, 309–315. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, T.W.; Chou, H.; Brendel, V.P. SRAssembler: Selective Recursive Local Assembly of Homologous Genomic Regions. BMC Bioinform. 2019, 20, 371. [Google Scholar] [CrossRef]
- Kajitani, R.; Toshimoto, K.; Noguchi, H.; Toyoda, A.; Ogura, Y.; Okuno, M.; Yabana, M.; Harada, M.; Nagayasu, E.; Maruyama, H.; et al. Efficient de Novo Assembly of Highly Heterozygous Genomes from Whole-Genome Shotgun Short Reads. Genome Res. 2014, 24, 1384–1395. [Google Scholar] [CrossRef] [Green Version]
- Slater, G.; Birney, E. Automated Generation of Heuristics for Biological Sequence Comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-58829-343-5. [Google Scholar]
- Yu, C.-S.; Chen, Y.-C.; Lu, C.-H.; Hwang, J.-K. Prediction of Protein Subcellular Localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s Conserved Domain Database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 15 December 2021).
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB Transcription Factor Genes as Regulators for Plant Responses: An Overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mieulet, D.; Jolivet, S.; Rivard, M.; Cromer, L.; Vernet, A.; Mayonove, P.; Pereira, L.; Droc, G.; Courtois, B.; Guiderdoni, E.; et al. Turning Rice Meiosis into Mitosis. Cell Res. 2016, 26, 1242–1254. [Google Scholar] [CrossRef] [PubMed]
- Spielman, M.; Vinkenoog, R.; Scott, R.J. Genetic Mechanisms of Apomixis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003, 358, 1095–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, R.J. Polyspermy in Apomictic Crataegus: Yes and No. New Phytol. 2007, 173, 227–229. [Google Scholar] [CrossRef] [Green Version]
- Grossniklaus, U.; Spillane, C.; Page, D.R.; Köhler, C. Genomic Imprinting and Seed Development: Endosperm Formation with and without Sex. Curr. Opin. Plant Biol. 2001, 4, 21–27. [Google Scholar] [CrossRef]
- Scott, R.J.; Armstrong, S.J.; Doughty, J.; Spielman, M. Double Fertilization in Arabidopsis Thaliana Involves a Polyspermy Block on the Egg but Not the Central Cell. Mol. Plant 2008, 1, 611–619. [Google Scholar] [CrossRef] [Green Version]

| Species | Reproduction Mode | Raw Data NCBI Accession | Genome Assembly | Genome Annotation | Reference |
|---|---|---|---|---|---|
| B. stricta | Sexual | SRR396760 SRR396762 SRR396756 | Yes | Yes | [41] |
| B. retrofracta | Sexual | SRR3929707 | Yes | Yes | [19] |
| B. puberula | Sexual | ERX2578777 ERX2578776 | Yes | No | [42] |
| B. spatifolia (Rosita3) | Sexual | SRR5116719 | No | No | [43] |
| B. spatifolia (Tiesiding2) | Sexual | SRR5116723 | No | No | [43] |
| B. spatifolia (Chicago2) | Sexual | SRR5116732 | No | No | [43] |
| B. spatifolia (Royal2) | Sexual | SRR5116730 | No | No | [43] |
| B. arcuata | Sexual | SRR6448790 | No | No | n/a |
| B. divaricarpa | Apomictic | SRR3500627 SRR3500628 | No | No | n/a |
| B. perennas | Apomictic | SRR6448882 | No | No | n/a |
| GenBank Accession | Sample ID | Allele Type |
|---|---|---|
| KF705583.1 | 369S2_S3 | Sex-allele |
| KF705582.1 | 376S2_S5 | Sex-allele |
| KF705581.1 | 355S2_S3 | Sex-allele |
| KF705569.1 | 329S2_S1 | Sex-allele |
| KF705572.1 | 385S2_S11 | Sex-allele |
| KF705596.1 | 43A3_A3 | Apo-allele |
| KF705598.1 | 1A2_A6 | Apo-allele |
| KF705600.1 | 11A2_A1 | Apo-allele |
| KF705599.1 | 11A2_A3 | Apo-allele |
| KF705588.1 | 33A2_A5 | Apo-allele |
| Parameter | Value |
|---|---|
| Gene length | 2231–2298 b.p. |
| Number of exons | 10 |
| Protein length | 177 a.a. |
| Molecular weight | 19,616.82 ± 60.4 |
| Theoretical pI | 11.25 ± 0.10 |
| Subcellular Localization | Nuclear |
| Conserved Domains | Histone H3/CENP-A (from 57 a.a. to 172 a.a.) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakin, E.; Sezer, F.; Özbilen, A.; Kilic, I.; Uner, B.; Rayko, M.; Taskin, K.M.; Brukhin, V. Phylogenetic and Expression Analysis of CENH3 and APOLLO Genes in Sexual and Apomictic Boechera Species. Plants 2022, 11, 387. https://doi.org/10.3390/plants11030387
Bakin E, Sezer F, Özbilen A, Kilic I, Uner B, Rayko M, Taskin KM, Brukhin V. Phylogenetic and Expression Analysis of CENH3 and APOLLO Genes in Sexual and Apomictic Boechera Species. Plants. 2022; 11(3):387. https://doi.org/10.3390/plants11030387
Chicago/Turabian StyleBakin, Evgeny, Fatih Sezer, Aslıhan Özbilen, Irem Kilic, Buket Uner, Mike Rayko, Kemal Melih Taskin, and Vladimir Brukhin. 2022. "Phylogenetic and Expression Analysis of CENH3 and APOLLO Genes in Sexual and Apomictic Boechera Species" Plants 11, no. 3: 387. https://doi.org/10.3390/plants11030387







