Inhibition of α-Glucosidase, Acetylcholinesterase, and Nitric Oxide Production by Phytochemicals Isolated from Millettia speciosa—In Vitro and Molecular Docking Studies
Abstract
:1. Introduction
2. Results
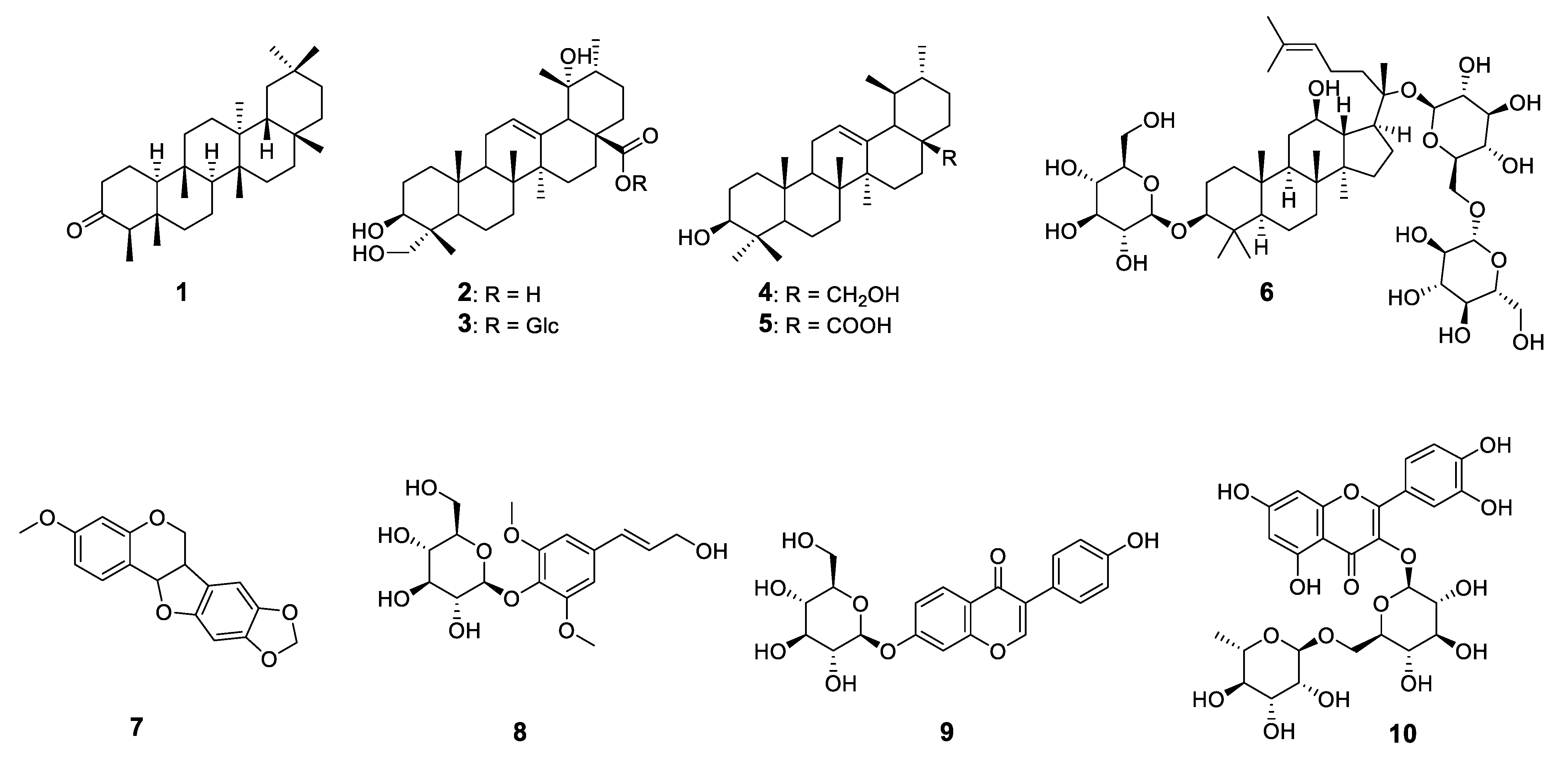
2.1. Structural Characterization of the Isolated Compounds
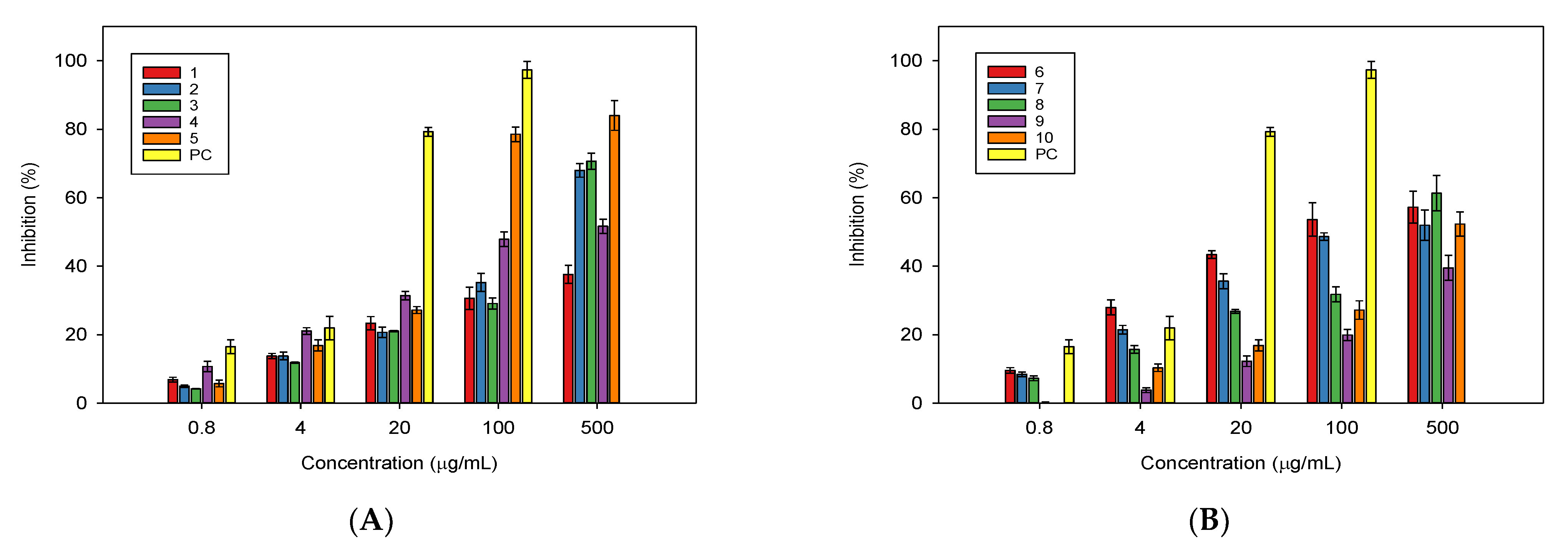
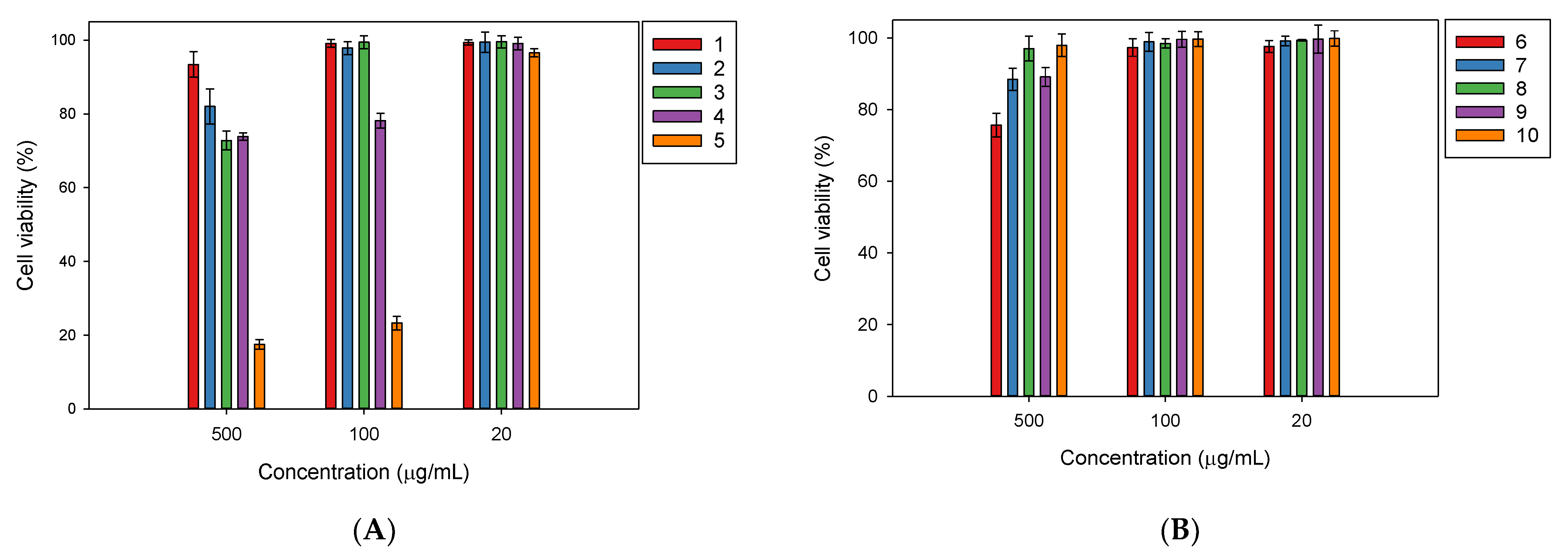
2.2. In Vitro Biological Activities of the Isolated Compounds from Millettia speciosa
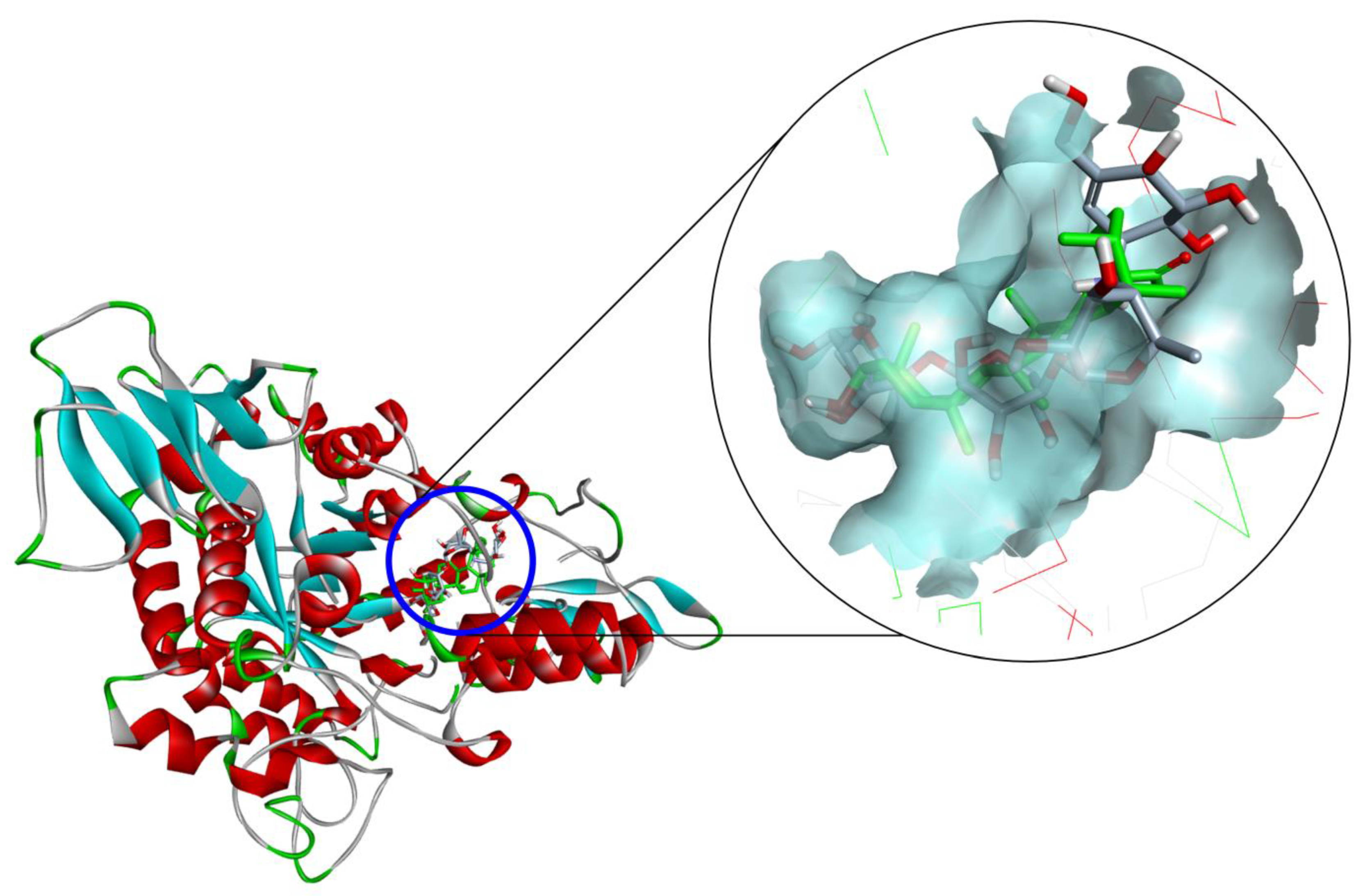
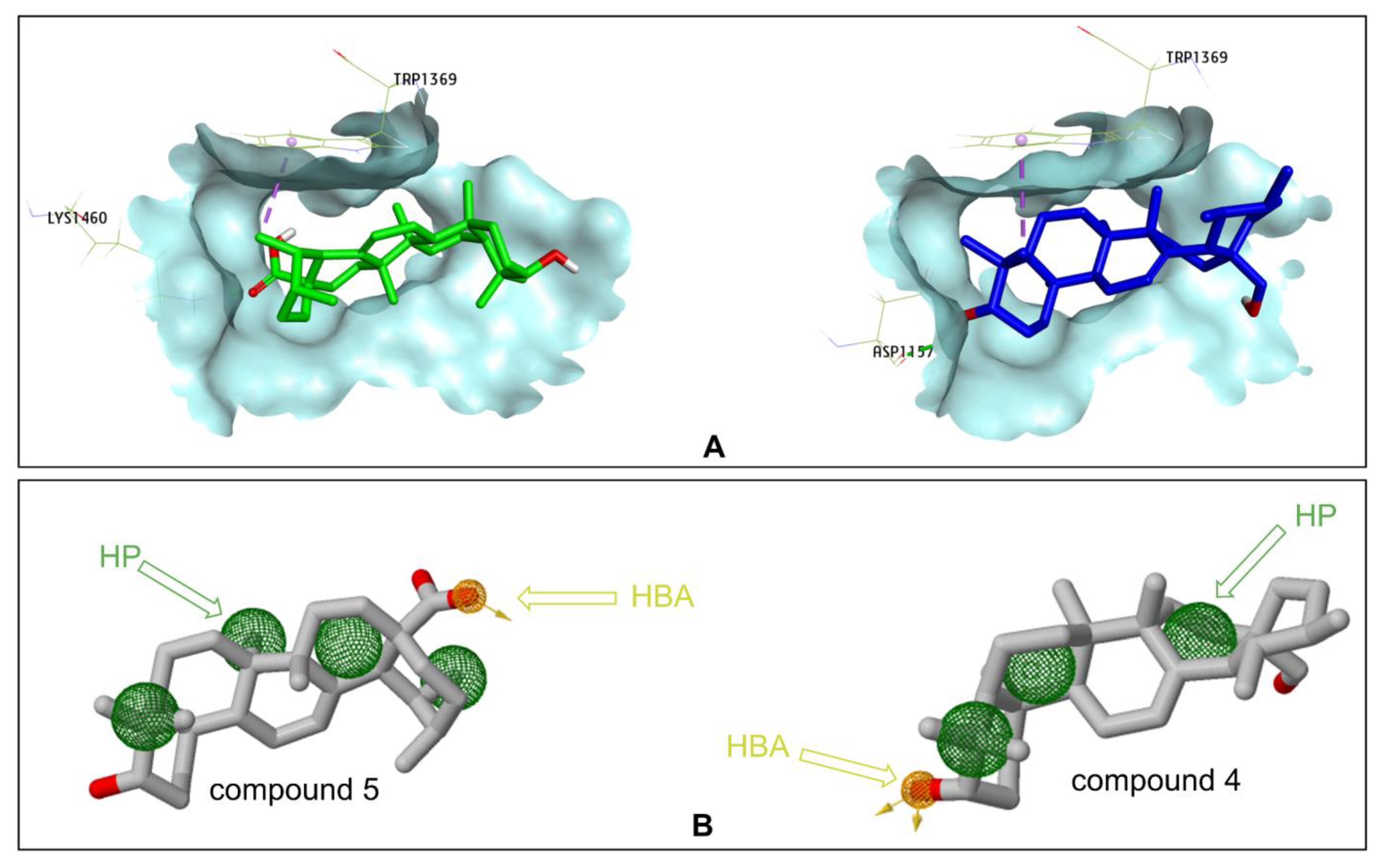
2.3. Docking Study for α-Glucosidase Inhibition by Compounds 3–5 and 10
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. General Procedures
4.3. Isolation and Characterization of Phytochemical Constituents
4.4. Structural Characterization of the Isolated Compounds
4.5. In Vitro Evaluation of NO Production Inhibitory Activity of the Isolated Compounds
4.6. Cell Viability Assay for the Evaluation of the Cytotoxicity of the Isolated Compounds
4.7. In Vitro Bioassay for α-Glucosidase Inhibition of the Isolated Compounds from the Roots of Milletia speciosa
4.8. In Vitro Bioassay for Acetylcholinesterase Inhibition of the Isolated Compounds
4.9. Molecular Docking Study for Anti-α-Glucosidase Inhibition
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Banzouzi, J.T.; Prost, A.; Rajemiarimiraho, M.; Ongoka, P. Traditional Uses of the African Millettia Species (Fabaceae). Int. J. Bot. 2008, 4, 406–420. [Google Scholar] [CrossRef] [Green Version]
- Deyou, T.; Gumula, I.; Pang, F.; Gruhonjic, A.; Mumo, M.; Holleran, J.; Duffy, S.; Fitzpatrick, P.A.; Heydenreich, M.; Landberg, G.; et al. Rotenoids, Flavonoids, and Chalcones from the Root Bark of Millettia Usaramensis. J. Nat. Prod. 2015, 78, 2932–2939. [Google Scholar] [CrossRef] [PubMed]
- Derese, S.; Barasa, L.; Akala, H.M.; Yusuf, A.O.; Kamau, E.; Heydenreich, M.; Yenesew, A. 4′-Prenyloxyderrone from the Stem Bark of Millettia Oblata Ssp. Teitensis and the Antiplasmodial Activities of Isoflavones from Some Millettia Species. Phytochem. Lett. 2014, 8, 69–72. [Google Scholar] [CrossRef]
- Cheng, J.; Zhao, Y.-Y.; Wang, B.; Qiao, L.; Liang, H. Flavonoids from Millettia Nitida Var. Hirsutissima. Chem. Pharm. Bull. 2005, 53, 419–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, T.; Liang, H.; Wang, B.; Zhao, Y. A New Flavonol Glycoside from Millettia Speciosa. Fitoterapia 2010, 81, 274–275. [Google Scholar] [CrossRef] [PubMed]
- Yankep, E.; Njamen, D.; Fotsing, M.T.; Fomum, Z.T.; Mbanya, J.-C.; Giner, R.M.; Recio, M.C.; Máñez, S.; Ríos, J.L. Griffonianone D, an Isoflavone with Anti-Inflammatory Activity from the Root Bark of Millettia Griffoniana. J. Nat. Prod. 2003, 66, 1288–1290. [Google Scholar] [CrossRef] [PubMed]
- Ketcha Wanda, G.J.M.; Njamen, D.; Yankep, E.; Tagatsing Fotsing, M.; Tanee Fomum, Z.; Wober, J.; Starcke, S.; Zierau, O.; Vollmer, G. Estrogenic Properties of Isoflavones Derived from Millettia Griffoniana. Phytomedicine 2006, 13, 139–145. [Google Scholar] [CrossRef]
- Fu, M.; Xiao, G.; Xu, Y.; Wu, J.; Chen, Y.; Qiu, S.-X. Chemical Constituents from Roots of Millettia Speciosa. Chin. Herb. Med. 2016, 8, 385–389. [Google Scholar] [CrossRef]
- Ding, P.; Qiu, J.; Ying, G.; Dai, L. Chemical Constituents of Millettia Speciosa. Chin. Herb. Med. 2014, 6, 332–334. [Google Scholar] [CrossRef]
- Editorial Board of Chinese Materia Medica in Guangdong; Guangdong Science and Technology Press: Guangzhou, China, 1991.
- Vo, C.V. A Dictionary of the Medicinal Plants of Vietnam; Y Hoc Publisher: Ha Noi, Vietnam, 2012. [Google Scholar]
- Dao, D.T.; Nguyen, T.T.; Nguyen, T.H.A.; Dong, T.K.C.; Nguyen, T.T.M.; Tran, D.Q.; Nguyen, H.S.; Trinh, T.T. A New Oleanane Triterpenoid from the Roots of Callerya Speciosa. Lett. Org. Chem. 2020, 17, 388–392. [Google Scholar]
- Chen, X.; Sun, W.; Xu, B.; Wu, E.; Cui, Y.; Hao, K.; Zhang, G.; Zhou, C.; Xu, Y.; Li, J.; et al. Polysaccharides From the Roots of Millettia Speciosa Champ Modulate Gut Health and Ameliorate Cyclophosphamide-Induced Intestinal Injury and Immunosuppression. Front. Immunol. 2021, 12, 766296. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zeng, Y.-J.; Chen, X.; Luo, S.-Y.; Pu, L.; Li, F.-Z.; Zong, M.-H.; Lou, W.-Y. A Novel Polysaccharide from the Roots of Millettia Speciosa Champ: Preparation, Structural Characterization and Immunomodulatory Activity. Int. J. Biol. Macromol. 2020, 145, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Jena, R.; Rath, D.; Rout, S.S.; Kar, D.M. A Review on Genus Millettia: Traditional Uses, Phytochemicals and Pharmacological Activities. Saudi Pharm. J. 2020, 28, 1686–1703. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, P.; Ma, S.; Wang, S.; Ang, L.; Liu, J.; Wang, M. Botanical Characteristics, Chemical and Nutritional Composition and Pharmacological and Toxicological Effects of Medicinal and Edible Plant Millettia Speciosa Champ. Food Sci. 2017, 9, 293–306. [Google Scholar] [CrossRef]
- Zhang, M.; Cui, C.; Lin, Y.; Cai, J. Ameliorating Effect on Glycolipid Metabolism and Chemical Profile of Millettia Speciosa Champ. Extract. J. Ethnopharmacol. 2021, 279, 114360. [Google Scholar] [CrossRef]
- Uchiyama, T.; Furukawa, M.; Isobe, S. New Oleanane-Type Triterpene Saponins from Millettia Speciosa. Heterocycles 2003, 60, 655–661. [Google Scholar]
- Wang, C.H.; Wang, Y.; Wang, G.C.; Ya, J.; Zhang, X.; Ye, W.C. Chemical Constituents from Roots of Millettia Speciosa. Chin. Tradit. Herb. Drugs 2008, 39, 972–975. [Google Scholar]
- Yin, T.; Tu, G.; Zhang, Q.; Wang, B.; Zhao, Y. Three New Phenolic Glycosides from the Caulis of Millettia Speciosa. Magn. Reson. Chem. 2008, 46, 387–391. [Google Scholar] [CrossRef]
- Zhang, H.; Ding, G.; Li, R.; Wei, J.; Zou, Z. Isolation, Identification and Quantitative Analysis of Hypaphorine in the Root of Millettia Speciosa Champ. Chin. J. Pharm. Anal. 2011, 31, 1024–1026. [Google Scholar]
- Zong, X.-K.; Lai, F.-L.; Wang, Z.-N.; Wang, J.-R. Studies on chemical constituents of root of Millettia speciosa. Zhong Yao Cai 2009, 32, 520–521. [Google Scholar]
- Dandan, Y.; Xianrui, L. Characterization and Identification of Isoflavonoids in the Roots of Millettia Speciosa Champ. by UPLC-Q-TOF-MS/MS. Curr. Pharm. Anal. 2019, 15, 580–591. [Google Scholar]
- Cheng, L.-Q.; Na, J.-R.; Kim, M.K.; Bang, M.-H.; Yang, D.-C. Microbial Conversion of Ginsenoside Rb1 to Minor Ginsenoside F2 and Gypenoside XVII by Intrasporangium Sp. GS603 Isolated from Soil. J. Microbiol. Biotechnol. 2007, 17, 1937–1943. [Google Scholar] [PubMed]
- Wang, C.; Chao, Z.; Sun, W.; Wu, X.; Ito, Y. Isolation of Five Glycosides from the Barks of Ilex Rotunda by High-Speed Counter-Current Chromatography. J. Liq. Chromatogr. Relat. Technol. 2014, 37, 2363–2376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedoreyev, S.A.; Bulgakov, V.P.; Grishchenko, O.V.; Veselova, M.V.; Krivoschekova, O.E.; Kulesh, N.I.; Denisenko, V.A.; Tchernoded, G.K.; Zhuravlev, Y.N. Isoflavonoid Composition of a Callus Culture of the Relict Tree Maackia Amurensis Rupr. et Maxim. J. Agric. Food Chem. 2008, 56, 7023–7031. [Google Scholar] [CrossRef] [PubMed]
- Zor, M.; Aydin, S.; Güner, N.D.; Başaran, N.; Başaran, A.A. Antigenotoxic Properties of Paliurus Spina-Christi Mill Fruits and Their Active Compounds. BMC Complementary Altern. Med. 2017, 17, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Crystal Structures of Isomaltase from Saccharomyces Cerevisiae and in Complex with Its Competitive Inhibitor Maltose. FEBS J. 2010, 277, 4205–4214. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Qin, X.; Cao, X.; Wang, L.; Bai, F.; Bai, G.; Shen, Y. Structural Insight into Substrate Specificity of Human Intestinal Maltase-Glucoamylase. Protein Cell 2011, 2, 827–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koes, D.R.; Camacho, C.J. ZINCPharmer: Pharmacophore Search of the ZINC Database. Nucleic Acids Res. 2012, 40, W409–W414. [Google Scholar] [CrossRef]
- Chen, D.-L.; Liu, Y.-Y.; Ma, G.-X.; Zhu, N.-L.; Wu, H.-F.; Wang, D.-L.; Xu, X.-D. Two New Rotenoids from the Roots of Millettia Speciosa. Phytochem. Lett. 2015, 12, 196–199. [Google Scholar] [CrossRef]
- You, H.J.; Choi, C.Y.; Kim, J.Y.; Park, S.J.; Hahm, K.-S.; Jeong, H.G. Ursolic Acid Enhances Nitric Oxide and Tumor Necrosis Factor-α Production via Nuclear Factor-ΚB Activation in the Resting Macrophages. FEBS Lett. 2001, 509, 156–160. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.-W.; Chen, T.-L.; Shih, C.-M.; Huang, C.-Y.; Tsao, N.-W.; Chang, N.-C.; Chen, Y.-H.; Fong, T.-H.; Lin, F.-Y. Ursolic Acid Induces Allograft Inflammatory Factor-1 Expression via a Nitric Oxide-Related Mechanism and Increases Neovascularization. J. Agric. Food Chem. 2010, 58, 12941–12949. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, S.K.; Park, K.-S.; Kwon, N.-Y.; Park, H.-J. Isolation of Constituents with Nitric Oxide Synthase Inhibition Activity from Phryma leptostachya var. asiatica. Nat. Prod. Sci. 2019, 25, 34–37. [Google Scholar] [CrossRef]
- Cho, J.Y.; Nam, K.H.; Kim, A.R.; Park, J.; Yoo, E.S.; Baik, K.U.; Yu, Y.H.; Park, M.H. In-Vitro and in-Vivo Immunomodulatory Effects of Syringin. J. Pharm. Pharmacol. 2001, 53, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Ugusman, A.; Zakaria, Z.; Chua, K.H.; Megat Mohd Nordin, N.A.; Abdullah Mahdy, Z. Role of Rutin on Nitric Oxide Synthesis in Human Umbilical Vein Endothelial Cells. Sci. World J. 2014, 2014, e169370. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.; Ganeshpurkar, A.; Ganeshpurkar, A.; Bansal, D.; Dubey, N. Glycolytic Enzyme Inhibitory and Antiglycation Potential of Rutin. Future J. Pharm. Sci. 2017, 3, 158–162. [Google Scholar] [CrossRef]
- Ding, H.; Hu, X.; Xu, X.; Zhang, G.; Gong, D. Inhibitory Mechanism of Two Allosteric Inhibitors, Oleanolic Acid and Ursolic Acid on α-Glucosidase. Int. J. Biol. Macromol. 2018, 107, 1844–1855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-W.; Xing, Y.; Wen, C.; Yu, X.-X.; Sun, W.-L.; Xiu, Z.-L.; Dong, Y.-S. Pentacyclic Triterpenes as α-Glucosidase and α-Amylase Inhibitors: Structure-Activity Relationships and the Synergism with Acarbose. Bioorg. Med. Chem. Lett. 2017, 27, 5065–5070. [Google Scholar] [CrossRef] [PubMed]
- Kalaycıoğlu, Z.; Uzaşçı, S.; Dirmenci, T.; Erim, F.B. α-Glucosidase Enzyme Inhibitory Effects and Ursolic and Oleanolic Acid Contents of Fourteen Anatolian Salvia Species. J. Pharm. Biomed. Anal. 2018, 155, 284–287. [Google Scholar] [CrossRef]
- Morocho, V.; Valle, A.; García, J.; Gilardoni, G.; Cartuche, L.; Suárez, A.I. α-Glucosidase Inhibition and Antibacterial Activity of Secondary Metabolites from the Ecuadorian Species Clinopodium Taxifolium (Kunth) Govaerts. Molecules 2018, 23, 146. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhao, J.; Yan, Y.; Liu, D.; Wang, C.; Wang, H. Inhibition of Glycosidase by Ursolic Acid: In Vitro, in Vivo and in Silico Study. J. Sci. Food Agric. 2020, 100, 986–994. [Google Scholar] [CrossRef]
- Peytam, F.; Takalloobanafshi, G.; Saadattalab, T.; Norouzbahari, M.; Emamgholipour, Z.; Moghimi, S.; Firoozpour, L.; Bijanzadeh, H.R.; Faramarzi, M.A.; Mojtabavi, S.; et al. Design, Synthesis, Molecular Docking, and in Vitro α-Glucosidase Inhibitory Activities of Novel 3-Amino-2,4-Diarylbenzo[4,5]Imidazo[1,2-a]Pyrimidines against Yeast and Rat α-Glucosidase. Sci. Rep. 2021, 11, 11911. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Wang, G.; Wang, J.; Chen, M.; Peng, Y.; Li, L.; Deng, B.; Chen, S.; Li, W. Synthesis, Biological Evaluation, and Molecular Docking Studies of Novel Isatin-Thiazole Derivatives as α-Glucosidase Inhibitors. Molecules 2017, 22, E659. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, B.; Tan, C.; Huang, Q. α-Glucosidase Inhibitors: Consistency of in Silico Docking Data with in Vitro Inhibitory Data and Inhibitory Effect Prediction of Quercetin Derivatives. Food Funct. 2019, 10, 6312–6321. [Google Scholar] [CrossRef] [PubMed]
- Ting, L.; Xiao-dong, Z.; Yu-wen, S.; Jian-wen, L.I.U. A Microplate-Based Screening Method for Alpha-Glucosidase Inhibitors. Chin. J. Clin. Pharmacol. Ther. 2020, 10, 1128. [Google Scholar]
- Wang, G.; Peng, Y.; Xie, Z.; Wang, J.; Chen, M. Synthesis, α-Glucosidase Inhibition and Molecular Docking Studies of Novel Thiazolidine-2,4-Dione or Rhodanine Derivatives. Med. Chem. Commun. 2017, 8, 1477–1484. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.; Trucks, G.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A. Gaussian 09, Revision d. 01, Gaussian. Inc. Wallingford CT 2009, 201, 6–10. [Google Scholar]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Chemical Biology: Methods and Protocols; Hempel, J.E., Williams, C.H., Hong, C.C., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; pp. 243–250. ISBN 978-1-4939-2269-7. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]

| Compound 1 | IC50 (µg/mL) 2 |
|---|---|
| 1 | >500 |
| 2 | 241.3 ± 8.2 |
| 3 | 273.1 ± 8.2 |
| 4 | 246.5 ± 18.7 |
| 5 | 43.9 ± 3.7 |
| 6 | 93.9 ± 5.4 |
| 7 | 228.9 ± 18.6 |
| 8 | 303.1 ± 11.0 |
| 9 | >500 |
| 10 | 449.5 ± 5.2 |
| PC | 8.6 ± 0.9 |
| Compound 2 | IC50 (µg/mL) 1 | Fold Change |
|---|---|---|
| 1 | >256 | <0.663 |
| 2 | >256 | <0.663 |
| 3 | 184.9 ± 10.05 | 0.918 |
| 4 | 1.96 ± 0.09 | 86.632 |
| 5 | 1.1 ± 0.05 | 154.363 |
| 6 | >256 | <0.663 |
| 7 | >256 | <0.663 |
| 8 | >256 | <0.663 |
| 9 | >256 | <0.663 |
| 10 | 2.2 ± 0.09 | 77.534 |
| Positive control 3 | 169.8 ± 7.05 | 1.000 |
| Compound 1 | Hydrogen Bond Interacting Residues 2 |
|---|---|
| 3 | Ser157, Tyr158 (unfavorable bump), Asp242, His280, Asp307 (unfavorable bump), Pro312, Phe314, Arg315, Glu411 (unfavorable bump). |
| 4 | Leu313 (unfavorable bump), Arg315, Asp352, Gln353. |
| 5 | Tyr158 (pi–alkyl), Asp215, Val216 (alkyl), Glu277, Phe303 (pi–alkyl), Arg315, Glu411 (unfavorable bump). |
| 10 | Ser157, Ser240, Asp242, Phe303 (pi–pi stacked), Asp307 (pi–anion), Phe314, Ser311, Agr315, Asp352, Gln353, Glu411, Arg442. |
| Acarbose | Asp69, Asp215, Ser240, Asp242, His280, Phe303, Pro312, Arg315 (unfavorable bump), Arg442 (unfavorable bump). |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuan, N.N.; Thi, H.N.; My, C.L.T.; Hai, T.X.; Trung, H.T.; Kim, A.N.T.; Tan, T.N.; Van, T.L.; Nguyen, C.Q.; Tran, Q.D.; et al. Inhibition of α-Glucosidase, Acetylcholinesterase, and Nitric Oxide Production by Phytochemicals Isolated from Millettia speciosa—In Vitro and Molecular Docking Studies. Plants 2022, 11, 388. https://doi.org/10.3390/plants11030388
Tuan NN, Thi HN, My CLT, Hai TX, Trung HT, Kim ANT, Tan TN, Van TL, Nguyen CQ, Tran QD, et al. Inhibition of α-Glucosidase, Acetylcholinesterase, and Nitric Oxide Production by Phytochemicals Isolated from Millettia speciosa—In Vitro and Molecular Docking Studies. Plants. 2022; 11(3):388. https://doi.org/10.3390/plants11030388
Chicago/Turabian StyleTuan, Nguyen Ngoc, Huong Nguyen Thi, Chau Le Thi My, Tang Xuan Hai, Hieu Tran Trung, Anh Nguyen Thi Kim, Thanh Nguyen Tan, Tan Le Van, Cuong Quoc Nguyen, Quang De Tran, and et al. 2022. "Inhibition of α-Glucosidase, Acetylcholinesterase, and Nitric Oxide Production by Phytochemicals Isolated from Millettia speciosa—In Vitro and Molecular Docking Studies" Plants 11, no. 3: 388. https://doi.org/10.3390/plants11030388






