Coordination of Chloroplast Activity with Plant Growth: Clues Point to TOR
Abstract
1. Why Plants Need to Regulate Chloroplasts Activity
2. Mechanisms Regulating Chloroplasts Activity
3. Chloroplastic Import: A Dynamic Gatekeeper of Coordination?
4. Nutrients/Metabolites Exchange as Signaling
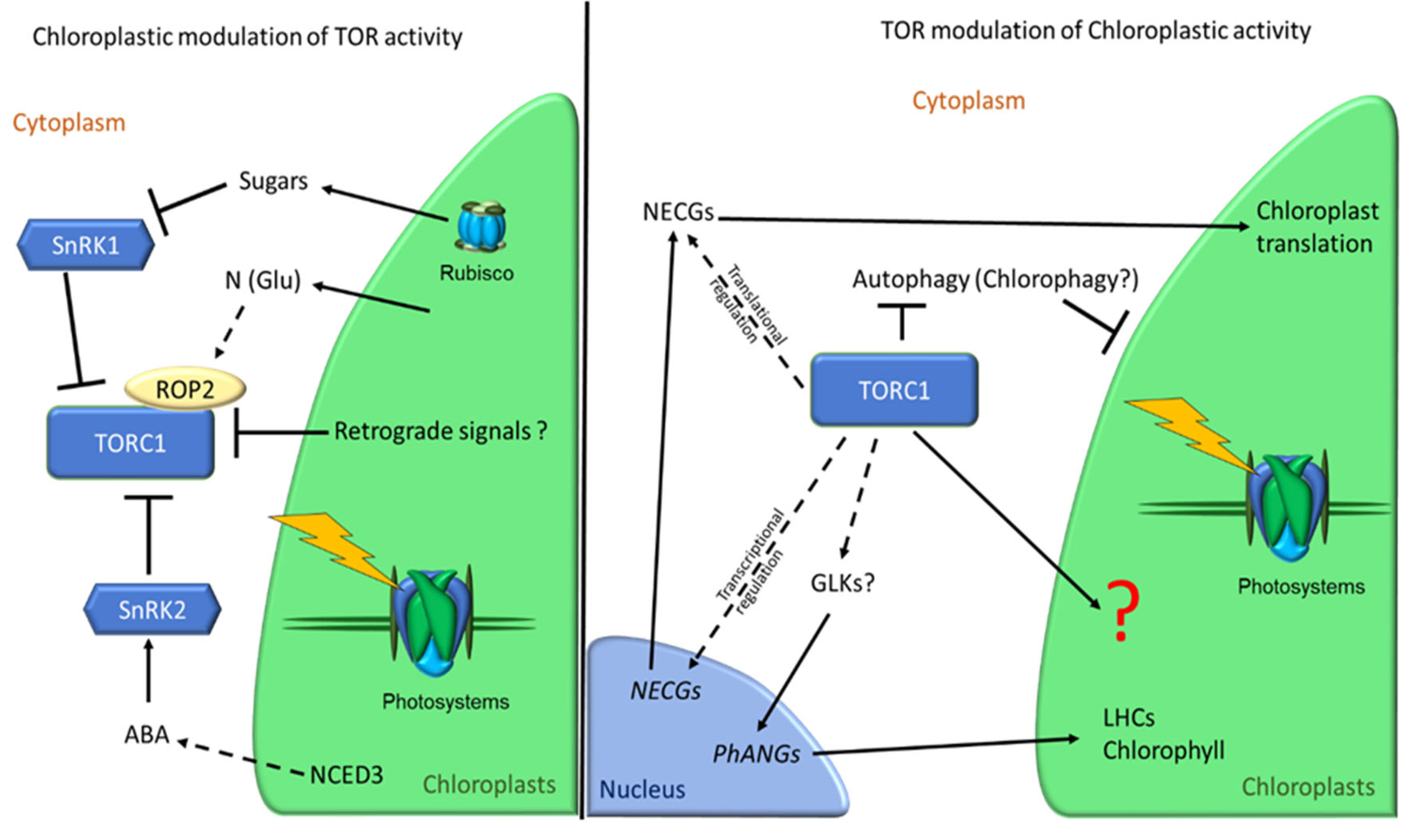
5. Mutual Regulation of TOR and Chloroplast Activity
6. Conclusions and Open Questions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arp, T.B.; Kistner-Morris, J.; Aji, V.; Cogdell, R.J.; van Grondelle, R.; Gabor, N.M. Quieting a Noisy Antenna Reproduces Photosynthetic Light-Harvesting Spectra. Science 2020, 368, 1490–1495. [Google Scholar] [CrossRef] [PubMed]
- Goral, T.K.; Johnson, M.P.; Duffy, C.D.P.; Brain, A.P.R.; Ruban, A.V.; Mullineaux, C.W. Light-Harvesting Antenna Composition Controls the Macrostructure and Dynamics of Thylakoid Membranes in Arabidopsis. Plant J. 2012, 69, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Suorsa, M.; Rossi, F.; Ferrari, R.; Tadini, L.; Barbato, R.; Pesaresi, P. Photosynthesis Control: An Underrated Short-Term Regulatory Mechanism Essential for Plant Viability. Plant Signal Behav. 2016, 11, e1165382. [Google Scholar] [CrossRef] [PubMed]
- Holdmann, C.; Schmid-Staiger, U.; Hirth, T. Outdoor Microalgae Cultivation at Different Biomass Concentrations—Assessment of Different Daily and Seasonal Light Scenarios by Modeling. Algal Res. 2019, 38, 101405. [Google Scholar] [CrossRef]
- Malnoë, A. Photoinhibition or Photoprotection of Photosynthesis? Update on the (Newly Termed) Sustained Quenching Component QH. Environ. Exp. Bot. 2018, 154, 123–133. [Google Scholar] [CrossRef]
- Ort, D.R. When There Is Too Much Light. Plant Physiol. 2001, 125, 29–32. [Google Scholar] [CrossRef]
- Shameer, S.; Ratcliffe, R.G.; Sweetlove, L.J. Leaf Energy Balance Requires Mitochondrial Respiration and Export of Chloroplast NADPH in the Light1. Plant Physiol. 2019, 180, 1947–1961. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Beaugelin, I.; Havaux, M. Tanned or Sunburned: How Excessive Light Triggers Plant Cell Death. Mol. Plant 2020, 13, 1545–1555. [Google Scholar] [CrossRef]
- Montané, M.-H.; Tardy, F.; Kloppstech, K.; Havaux, M. Differential Control of Xanthophylls and Light-Induced Stress Proteins, as Opposed to Light-Harvesting Chlorophyll a/b Proteins, during Photosynthetic Acclimation of Barley Leaves to Light Irradiance. Plant Physiol. 1998, 118, 227–235. [Google Scholar] [CrossRef]
- Asada, K. Production and Scavenging of Reactive Oxygen Species in Chloroplasts and Their Functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Li, Z.; Wakao, S.; Fischer, B.B.; Niyogi, K.K. Sensing and Responding to Excess Light. Annu. Rev. Plant Biol. 2009, 60, 239–260. [Google Scholar] [CrossRef]
- Tikkanen, M.; Mekala, N.R.; Aro, E.-M. Photosystem II Photoinhibition-Repair Cycle Protects Photosystem I from Irreversible Damage. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1837, 210–215. [Google Scholar] [CrossRef]
- Triantaphylidès, C.; Krischke, M.; Hoeberichts, F.A.; Ksas, B.; Gresser, G.; Havaux, M.; Breusegem, F.V.; Mueller, M.J. Singlet Oxygen Is the Major Reactive Oxygen Species Involved in Photooxidative Damage to Plants. Plant Physiol. 2008, 148, 960–968. [Google Scholar] [CrossRef]
- Bechtold, U.; Field, B. Molecular Mechanisms Controlling Plant Growth during Abiotic Stress. J. Exp. Bot. 2018, 69, 2753–2758. [Google Scholar] [CrossRef] [PubMed]
- Van den Ende, W.; Valluru, R. Sucrose, Sucrosyl Oligosaccharides, and Oxidative Stress: Scavenging and Salvaging? J. Exp. Bot. 2009, 60, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Mueller, M.J. ROS-Mediated Lipid Peroxidation and RES-Activated Signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.S.; Mano, J. Lipid Peroxide-Derived Short-Chain Carbonyls Mediate Hydrogen Peroxide-Induced and Salt-Induced Programmed Cell Death in Plants. Plant Physiol. 2015, 168, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Exposito-Rodriguez, M.; Laissue, P.P.; Yvon-Durocher, G.; Smirnoff, N.; Mullineaux, P.M. Photosynthesis-Dependent H2O2 Transfer from Chloroplasts to Nuclei Provides a High-Light Signalling Mechanism. Nat. Commun. 2017, 8, 49. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Havaux, M. Sensing β-Carotene Oxidation in Photosystem II to Master Plant Stress Tolerance. New Phytol. 2019, 223, 1776–1783. [Google Scholar] [CrossRef]
- Breeze, E.; Mullineaux, P.M. The Passage of H2O2 from Chloroplasts to Their Associated Nucleus during Retrograde Signalling: Reflections on the Role of the Nuclear Envelope. Plants 2022, 11, 552. [Google Scholar] [CrossRef]
- Dogra, V.; Singh, R.M.; Li, M.; Li, M.; Singh, S.; Kim, C. EXECUTER2 Modulates the EXECUTER1 Signalosome through Its Singlet Oxygen-Dependent Oxidation. Mol. Plant 2022, 15, 438–453. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. REACTIVE OXYGEN SPECIES: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Ramel, F.; Birtic, S.; Ginies, C.; Soubigou-Taconnat, L.; Triantaphylidès, C.; Havaux, M. Carotenoid Oxidation Products Are Stress Signals That Mediate Gene Responses to Singlet Oxygen in Plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5535–5540. [Google Scholar] [CrossRef]
- D’Alessandro, S.; Ksas, B.; Havaux, M. Decoding β-Cyclocitral-Mediated Retrograde Signaling Reveals the Role of a Detoxification Response in Plant Tolerance to Photooxidative Stress. Plant Cell 2018, 30, 2495–2511. [Google Scholar] [CrossRef]
- Xiao, Y.; Savchenko, T.; Baidoo, E.E.K.; Chehab, W.E.; Hayden, D.M.; Tolstikov, V.; Corwin, J.A.; Kliebenstein, D.J.; Keasling, J.D.; Dehesh, K. Retrograde Signaling by the Plastidial Metabolite MEcPP Regulates Expression of Nuclear Stress-Response Genes. Cell 2012, 149, 1525–1535. [Google Scholar] [CrossRef]
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP Chloroplast Retrograde Pathway That Functions in Drought and High Light Signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef]
- Mano, J.; Biswas, M.S.; Sugimoto, K. Reactive Carbonyl Species: A Missing Link in ROS Signaling. Plants 2019, 8, 391. [Google Scholar] [CrossRef]
- Koschmieder, J.; Wüst, F.; Schaub, P.; Álvarez, D.; Trautmann, D.; Krischke, M.; Rustenholz, C.; Mano, J.; Mueller, M.J.; Bartels, D.; et al. Plant Apocarotenoid Metabolism Utilizes Defense Mechanisms against Reactive Carbonyl Species and Xenobiotics. Plant Physiol. 2021, 185, 331–351. [Google Scholar] [CrossRef]
- Sharkey, T.D. Photorespiration. In eLS; John Wiley & Sons: Hoboken, NJ, USA, 2001. [Google Scholar]
- Busch, F.A.; Sage, R.F.; Farquhar, G.D. Plants Increase CO2 Uptake by Assimilating Nitrogen via the Photorespiratory Pathway. Nat. Plants 2018, 4, 46–54. [Google Scholar] [CrossRef]
- Slesak, I.; Libik, M.; Karpinska, B.; Karpinski, S.; Miszalski, Z. The Role of Hydrogen Peroxide in Regulation of Plant Metabolism and Cellular Signalling in Response to Environmental Stresses. Acta Biochim. Pol. 2007, 54, 39–50. [Google Scholar] [CrossRef]
- Voon, C.P.; Guan, X.; Sun, Y.; Sahu, A.; Chan, M.N.; Gardeström, P.; Wagner, S.; Fuchs, P.; Nietzel, T.; Versaw, W.K.; et al. ATP Compartmentation in Plastids and Cytosol of Arabidopsis Thaliana Revealed by Fluorescent Protein Sensing. Proc. Natl. Acad. Sci. USA 2018, 115, E10778–E10787. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Milo, R. The Global Mass and Average Rate of Rubisco. Proc. Natl. Acad. Sci. USA 2019, 116, 4738–4743. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.-Z.; Bock, R. GUN Control in Retrograde Signaling: How GENOMES UNCOUPLED Proteins Adjust Nuclear Gene Expression to Plastid Biogenesis. Plant Cell 2021, 33, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S. The Economics of Organellar Gene Loss and Endosymbiotic Gene Transfer. Genome Biol. 2021, 22, 345. [Google Scholar] [CrossRef]
- Berry, J.O.; Yerramsetty, P.; Zielinski, A.M.; Mure, C.M. Photosynthetic Gene Expression in Higher Plants. Photosynth. Res. 2013, 117, 91–120. [Google Scholar] [CrossRef]
- Patel, M.; Berry, J.O. Rubisco Gene Expression in C4 Plants. J. Exp. Bot. 2008, 59, 1625–1634. [Google Scholar] [CrossRef]
- Smith, C.W.; Scadden, A. RNA Processing. In eLS; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Tyagi, A.K.; Gaur, T. Light Regulation of Nuclear Photosynthetic Genes in Higher Plants. Crit. Rev. Plant Sci. 2003, 22, 417–452. [Google Scholar] [CrossRef]
- López-Ochoa, L.; Acevedo-Hernández, G.; Martínez-Hernández, A.; Argüello-Astorga, G.; Herrera-Estrella, L. Structural Relationships between Diverse Cis-Acting Elements Are Critical for the Functional Properties of a RbcS Minimal Light Regulatory Unit. J. Exp. Bot. 2007, 58, 4397–4406. [Google Scholar] [CrossRef]
- Dubreuil, C.; Ji, Y.; Strand, Å.; Grönlund, A. A Quantitative Model of the Phytochrome-PIF Light Signalling Initiating Chloroplast Development. Sci. Rep. 2017, 7, 13884. [Google Scholar] [CrossRef]
- Stephenson, P.G.; Fankhauser, C.; Terry, M.J. PIF3 Is a Repressor of Chloroplast Development. Proc. Natl. Acad. Sci. USA 2009, 106, 7654–7659. [Google Scholar] [CrossRef]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and Phytochrome Interacting Factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef]
- Griffin, J.H.C.; Prado, K.; Sutton, P.; Toledo-Ortiz, G. Coordinating Light Responses between the Nucleus and the Chloroplast, a Role for Plant Cryptochromes and Phytochromes. Physiol. Plant. 2020, 169, 515–528. [Google Scholar] [CrossRef]
- Oh, S.; Montgomery, B.L. Mesophyll-Specific Phytochromes Impact Chlorophyll Light-Harvesting Complexes (LHCs) and Non-Photochemical Quenching. Plant Signal Behav. 2019, 14, 1609857. [Google Scholar] [CrossRef]
- Devireddy, A.R.; Liscum, E.; Mittler, R. Phytochrome B Is Required for Systemic Stomatal Responses and Reactive Oxygen Species Signaling during Light Stress. Plant Physiol. 2020, 184, 1563–1572. [Google Scholar] [CrossRef]
- Martín, G.; Leivar, P.; Ludevid, D.; Tepperman, J.M.; Quail, P.H.; Monte, E. Phytochrome and Retrograde Signalling Pathways Converge to Antagonistically Regulate a Light-Induced Transcriptional Network. Nat. Commun. 2016, 7, 11431. [Google Scholar] [CrossRef]
- Ushijima, T.; Hanada, K.; Gotoh, E.; Yamori, W.; Kodama, Y.; Tanaka, H.; Kusano, M.; Fukushima, A.; Tokizawa, M.; Yamamoto, Y.Y.; et al. Light Controls Protein Localization through Phytochrome-Mediated Alternative Promoter Selection. Cell 2017, 171, 1316–1325.e12. [Google Scholar] [CrossRef]
- Inoue, H.; Rounds, C.; Schnell, D.J. The Molecular Basis for Distinct Pathways for Protein Import into Arabidopsis Chloroplasts. Plant Cell 2010, 22, 1947–1960. [Google Scholar] [CrossRef]
- Watson, S.J.; Sowden, R.G.; Jarvis, P. Abiotic Stress-Induced Chloroplast Proteome Remodelling: A Mechanistic Overview. J. Exp. Bot. 2018, 69, 2773–2781. [Google Scholar] [CrossRef]
- Börner, T.; Aleynikova, A.Y.; Zubo, Y.O.; Kusnetsov, V.V. Chloroplast RNA Polymerases: Role in Chloroplast Biogenesis. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1847, 761–769. [Google Scholar] [CrossRef]
- Macadlo, L.A.; Ibrahim, I.M.; Puthiyaveetil, S. Sigma Factor 1 in Chloroplast Gene Transcription and Photosynthetic Light Acclimation. J. Exp. Bot. 2020, 71, 1029–1038. [Google Scholar] [CrossRef]
- Allison, L.A. The Role of Sigma Factors in Plastid Transcription. Biochimie 2000, 82, 537–548. [Google Scholar] [CrossRef]
- Chi, W.; He, B.; Mao, J.; Jiang, J.; Zhang, L. Plastid Sigma Factors: Their Individual Functions and Regulation in Transcription. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1847, 770–778. [Google Scholar] [CrossRef]
- Field, B. Green Magic: Regulation of the Chloroplast Stress Response by (p)PpGpp in Plants and Algae. J. Exp. Bot. 2018, 69, 2797–2807. [Google Scholar] [CrossRef]
- Steinchen, W.; Zegarra, V.; Bange, G. (P)PpGpp: Magic Modulators of Bacterial Physiology and Metabolism. Front. Microbiol. 2020, 11, 2072. [Google Scholar] [CrossRef]
- Sugliani, M.; Abdelkefi, H.; Ke, H.; Bouveret, E.; Robaglia, C.; Caffarri, S.; Field, B. An Ancient Bacterial Signaling Pathway Regulates Chloroplast Function to Influence Growth and Development in Arabidopsis. Plant Cell 2016, 28, 661–679. [Google Scholar] [CrossRef]
- Atkinson, G.C.; Tenson, T.; Hauryliuk, V. The RelA/SpoT Homolog (RSH) Superfamily: Distribution and Functional Evolution of PpGpp Synthetases and Hydrolases across the Tree of Life. PLoS ONE 2011, 6, e23479. [Google Scholar] [CrossRef]
- Honoki, R.; Ono, S.; Oikawa, A.; Saito, K.; Masuda, S. Significance of Accumulation of the Alarmone (p)PpGpp in Chloroplasts for Controlling Photosynthesis and Metabolite Balance during Nitrogen Starvation in Arabidopsis. Photosynth. Res. 2018, 135, 299–308. [Google Scholar] [CrossRef]
- Abdelkefi, H.; Sugliani, M.; Ke, H.; Harchouni, S.; Soubigou-Taconnat, L.; Citerne, S.; Mouille, G.; Fakhfakh, H.; Robaglia, C.; Field, B. Guanosine Tetraphosphate Modulates Salicylic Acid Signalling and the Resistance of Arabidopsis Thaliana to Turnip Mosaic Virus. Mol. Plant Pathol. 2017, 19, 634–646. [Google Scholar] [CrossRef]
- Romand, S.; Abdelkefi, H.; Lecampion, C.; Belaroussi, M.; Dussenne, M.; Ksas, B.; Citerne, S.; Caius, J.; D’Alessandro, S.; Fakhfakh, H.; et al. A Guanosine Tetraphosphate (PpGpp) Mediated Brake on Photosynthesis Is Required for Acclimation to Nitrogen Limitation in Arabidopsis. Elife 2022, 11, e75041. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Small, I. Pentatricopeptide Repeat Proteins: A Socket Set for Organelle Gene Expression. Trends Plant Sci. 2008, 13, 663–670. [Google Scholar] [CrossRef]
- Asakura, Y.; Barkan, A. A CRM Domain Protein Functions Dually in Group I and Group II Intron Splicing in Land Plant Chloroplasts. Plant Cell 2007, 19, 3864–3875. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.B.; Goldschmidt-Clermont, M.; Hanson, M.R. Chloroplast RNA Metabolism. Annu. Rev. Plant Biol. 2010, 61, 125–155. [Google Scholar] [CrossRef] [PubMed]
- Flores-Pérez, Ú.; Jarvis, P. Molecular Chaperone Involvement in Chloroplast Protein Import. Biochim. Biophys. Acta 2013, 1833, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Fellerer, C.; Schweiger, R.; Schöngruber, K.; Soll, J.; Schwenkert, S. Cytosolic HSP90 Cochaperones HOP and FKBP Interact with Freshly Synthesized Chloroplast Preproteins of Arabidopsis. Mol. Plant 2011, 4, 1133–1145. [Google Scholar] [CrossRef]
- Lamberti, G.; Gügel, I.L.; Meurer, J.; Soll, J.; Schwenkert, S. The Cytosolic Kinases STY8, STY17, and STY46 Are Involved in Chloroplast Differentiation in Arabidopsis. Plant Physiol. 2011, 157, 70–85. [Google Scholar] [CrossRef]
- Nickel, C.; Soll, J.; Schwenkert, S. Phosphomimicking within the Transit Peptide of PHCF136 Leads to Reduced Photosystem II Accumulation in Vivo. FEBS Lett. 2015, 589, 1301–1307. [Google Scholar] [CrossRef]
- Jarvis, P. Targeting of Nucleus-Encoded Proteins to Chloroplasts in Plants. New Phytol. 2008, 179, 257–285. [Google Scholar] [CrossRef]
- Heckathorn, S.A.; Downs, C.A.; Coleman, J.S. Nuclear-Encoded Chloroplast Proteins Accumulate in the Cytosol During Severe Heat Stress. Int. J. Plant Sci. 1998, 159, 39–45. [Google Scholar] [CrossRef]
- Ling, Q.; Huang, W.; Baldwin, A.; Jarvis, P. Chloroplast Biogenesis Is Regulated by Direct Action of the Ubiquitin-Proteasome System. Science 2012. [Google Scholar] [CrossRef]
- Sjuts, I.; Soll, J.; Bölter, B. Import of Soluble Proteins into Chloroplasts and Potential Regulatory Mechanisms. Front. Plant Sci. 2017, 8, 168. [Google Scholar] [CrossRef]
- Ling, Q.; Jarvis, P. Regulation of Chloroplast Protein Import by the Ubiquitin E3 Ligase SP1 Is Important for Stress Tolerance in Plants. Curr. Biol. 2015, 25, 2527–2534. [Google Scholar] [CrossRef]
- Thomson, S.M.; Pulido, P.; Jarvis, R.P. Protein Import into Chloroplasts and Its Regulation by the Ubiquitin-Proteasome System. Biochem. Soc. Trans. 2020, 48, 71–82. [Google Scholar] [CrossRef]
- Lee, S.; Lee, D.W.; Lee, Y.; Mayer, U.; Stierhof, Y.-D.; Lee, S.; Jürgens, G.; Hwang, I. Heat Shock Protein Cognate 70-4 and an E3 Ubiquitin Ligase, CHIP, Mediate Plastid-Destined Precursor Degradation through the Ubiquitin-26S Proteasome System in Arabidopsis. Plant Cell 2009, 21, 3984–4001. [Google Scholar] [CrossRef]
- Tokumaru, M.; Adachi, F.; Toda, M.; Ito-Inaba, Y.; Yazu, F.; Hirosawa, Y.; Sakakibara, Y.; Suiko, M.; Kakizaki, T.; Inaba, T. Ubiquitin-Proteasome Dependent Regulation of the GOLDEN2-LIKE 1 Transcription Factor in Response to Plastid Signals. Plant Physiol. 2017, 173, 524–535. [Google Scholar] [CrossRef]
- Blazek, M.; Silva Santisteban, T.; Zengerle, R.; Meier, M. Analysis of Fast Protein Phosphorylation Kinetics in Single Cells on a Microfluidic Chip. Lab Chip 2015, 15, 726–734. [Google Scholar] [CrossRef]
- Salazar, C.; Höfer, T. Multisite Protein Phosphorylation—From Molecular Mechanisms to Kinetic Models. FEBS J. 2009, 276, 3177–3198. [Google Scholar] [CrossRef]
- Eisa, A.; Bölter, B.; Schwenkert, S. The ACT Domain in Chloroplast Precursor–Phosphorylating STY Kinases Binds Metabolites and Allosterically Regulates Kinase Activity. J. Biol. Chem. 2019, 294, 17278–17288. [Google Scholar] [CrossRef]
- Lamberti, G.; Drurey, C.; Soll, J.; Schwenkert, S. The Phosphorylation State of Chloroplast Transit Peptides Regulates Preprotein Import. Plant Signal Behav. 2011, 6, 1918–1920. [Google Scholar] [CrossRef][Green Version]
- Oreb, M.; Zoryan, M.; Vojta, A.; Maier, U.G.; Eichacker, L.A.; Schleiff, E. Phospho-Mimicry Mutant of AtToc33 Affects Early Development of Arabidopsis Thaliana. FEBS Lett. 2007, 581, 5945–5951. [Google Scholar] [CrossRef]
- Pottosin, I.; Shabala, S. Transport Across Chloroplast Membranes: Optimizing Photosynthesis for Adverse Environmental Conditions. Mol. Plant 2016, 9, 356–370. [Google Scholar] [CrossRef]
- Bose, J.; Munns, R.; Shabala, S.; Gilliham, M.; Pogson, B.; Tyerman, S.D. Chloroplast Function and Ion Regulation in Plants Growing on Saline Soils: Lessons from Halophytes. J. Exp. Bot. 2017, 68, 3129–3143. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.B.; Eisenhut, M.; Schneider, A. Chloroplast Transition Metal Regulation for Efficient Photosynthesis. Trends Plant Sci. 2020, 25, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Vogel, M.O.; Moore, M.; König, K.; Pecher, P.; Alsharafa, K.; Lee, J.; Dietz, K.-J. Fast Retrograde Signaling in Response to High Light Involves Metabolite Export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF Transcription Factors in Arabidopsis. Plant Cell 2014, 26, 1151–1165. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, F.; Dissanayake, I.M.; Lacombe, B.; Barbier, F. Sugar and Nitrate Sensing: A Multi-Billion-Year Story. Trends Plant Sci. 2021, 26, 352–374. [Google Scholar] [CrossRef]
- Liu, Y.; Duan, X.; Zhao, X.; Ding, W.; Wang, Y.; Xiong, Y. Diverse Nitrogen Signals Activate Convergent ROP2-TOR Signaling in Arabidopsis. Dev. Cell 2021, 56, 1283–1295. [Google Scholar] [CrossRef]
- Dong, Y.; Silbermann, M.; Speiser, A.; Forieri, I.; Linster, E.; Poschet, G.; Allboje Samami, A.; Wanatabe, M.; Sticht, C.; Teleman, A.A.; et al. Sulfur Availability Regulates Plant Growth via Glucose-TOR Signaling. Nat. Commun. 2017, 8, 1174. [Google Scholar] [CrossRef]
- Schepetilnikov, M.; Makarian, J.; Srour, O.; Geldreich, A.; Yang, Z.; Chicher, J.; Hammann, P.; Ryabova, L.A. GTPase ROP2 Binds and Promotes Activation of Target of Rapamycin, TOR, in Response to Auxin. EMBO J. 2017, 36, 886–903. [Google Scholar] [CrossRef]
- Margalha, L.; Valerio, C.; Baena-González, E. Plant SnRK1 Kinases: Structure, Regulation, and Function. Exp. Suppl. 2016, 107, 403–438. [Google Scholar] [CrossRef]
- Nukarinen, E.; Nägele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Börnke, F.; Hanson, J.; Teige, M.; Baena-Gonzalez, E.; et al. Quantitative Phosphoproteomics Reveals the Role of the AMPK Plant Ortholog SnRK1 as a Metabolic Master Regulator under Energy Deprivation. Sci. Rep. 2016, 6, 31697. [Google Scholar] [CrossRef]
- Nietzsche, M.; Schießl, I.; Börnke, F. The Complex Becomes More Complex: Protein-Protein Interactions of SnRK1 with DUF581 Family Proteins Provide a Framework for Cell- and Stimulus Type-Specific SnRK1 Signaling in Plants. Front. Plant Sci. 2014, 5, 54. [Google Scholar] [CrossRef]
- Soto-Burgos, J.; Bassham, D.C. SnRK1 Activates Autophagy via the TOR Signaling Pathway in Arabidopsis Thaliana. PLoS ONE 2017, 12, e0182591. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.-C.; Liu, X.; Fu, L.; Hou, Y.-J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 2018, 69, 100–112. [Google Scholar] [CrossRef]
- Belda-Palazón, B.; Adamo, M.; Valerio, C.; Ferreira, L.J.; Confraria, A.; Reis-Barata, D.; Rodrigues, A.; Meyer, C.; Rodriguez, P.L.; Baena-González, E. A Dual Function of SnRK2 Kinases in the Regulation of SnRK1 and Plant Growth. Nat. Plants 2020, 6, 1345–1353. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Chory, J. The Arabidopsis Transcriptome Responds Specifically and Dynamically to High Light Stress. Cell Rep. 2019, 29, 4186–4199. [Google Scholar] [CrossRef]
- Moreno, J.C.; Mi, J.; Alagoz, Y.; Al-Babili, S. Plant Apocarotenoids: From Retrograde Signaling to Interspecific Communication. Plant J. 2021, 105, 351–375. [Google Scholar] [CrossRef]
- Mallén-Ponce, M.J.; Pérez-Pérez, M.E.; Crespo, J.L. Photosynthetic Assimilation of CO2 Regulates TOR Activity. Proc. Natl. Acad. Sci. USA 2022, 119, e2115261119. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, F.; Que, Y.; Wang, K.; Yu, L.; Li, Z.; Maozhi, R. Expression Profiling and Functional Analysis Reveals That TOR Is a Key Player in Regulating Photosynthesis and Phytohormone Signaling Pathways in Arabidopsis. Front. Plant Sci. 2015, 6, 677. [Google Scholar] [CrossRef]
- Upadhyaya, S.; Rao, B.J. Reciprocal Regulation of Photosynthesis and Mitochondrial Respiration by TOR Kinase in Chlamydomonas Reinhardtii. Plant Direct 2019, 3, e00184. [Google Scholar] [CrossRef]
- Liu, Y.; Bassham, D.C. TOR Is a Negative Regulator of Autophagy in Arabidopsis Thaliana. PLoS ONE 2010, 5, e11883. [Google Scholar] [CrossRef]
- Kajikawa, M.; Fukuzawa, H. Algal Autophagy Is Necessary for the Regulation of Carbon Metabolism Under Nutrient Deficiency. Front Plant Sci. 2020, 11, 36. [Google Scholar] [CrossRef]
- Van Leene, J.; Han, C.; Gadeyne, A.; Eeckhout, D.; Matthijs, C.; Cannoot, B.; De Winne, N.; Persiau, G.; Van De Slijke, E.; Van de Cotte, B.; et al. Capturing the Phosphorylation and Protein Interaction Landscape of the Plant TOR Kinase. Nat. Plants 2019, 5, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Izumi, M. Chlorophagy Is ATG Gene-Dependent Microautophagy Process. Plant Signal. Behav. 2019, 14, 1554469. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.-Y.; Bassham, D.C. Combating Stress: The Interplay between Hormone Signaling and Autophagy in Plants. J. Exp. Bot. 2020, 71, 1723–1733. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Izumi, M.; Wada, S.; Makino, A. Roles of Autophagy in Chloroplast Recycling. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1837, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose–TOR Signalling Reprograms the Transcriptome and Activates Meristems. Nature 2013, 496, 181–186. [Google Scholar] [CrossRef]
- Scarpin, M.R.; Leiboff, S.; Brunkard, J.O. Parallel Global Profiling of Plant TOR Dynamics Reveals a Conserved Role for LARP1 in Translation. eLife 2020, 9, e58795. [Google Scholar] [CrossRef]
- Dobrenel, T.; Mancera-Martínez, E.; Forzani, C.; Azzopardi, M.; Davanture, M.; Moreau, M.; Schepetilnikov, M.; Chicher, J.; Langella, O.; Zivy, M.; et al. The Arabidopsis TOR Kinase Specifically Regulates the Expression of Nuclear Genes Coding for Plastidic Ribosomal Proteins and the Phosphorylation of the Cytosolic Ribosomal Protein S6. Front Plant Sci. 2016, 7, 1611. [Google Scholar] [CrossRef]
- Chen, G.-H.; Liu, M.-J.; Xiong, Y.; Sheen, J.; Wu, S.-H. TOR and RPS6 Transmit Light Signals to Enhance Protein Translation in Deetiolating Arabidopsis Seedlings. Proc. Natl. Acad. Sci. USA 2018, 115, 12823–12828. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Alessandro, S. Coordination of Chloroplast Activity with Plant Growth: Clues Point to TOR. Plants 2022, 11, 803. https://doi.org/10.3390/plants11060803
D’Alessandro S. Coordination of Chloroplast Activity with Plant Growth: Clues Point to TOR. Plants. 2022; 11(6):803. https://doi.org/10.3390/plants11060803
Chicago/Turabian StyleD’Alessandro, Stefano. 2022. "Coordination of Chloroplast Activity with Plant Growth: Clues Point to TOR" Plants 11, no. 6: 803. https://doi.org/10.3390/plants11060803
APA StyleD’Alessandro, S. (2022). Coordination of Chloroplast Activity with Plant Growth: Clues Point to TOR. Plants, 11(6), 803. https://doi.org/10.3390/plants11060803





