Abstract
Metal tolerance proteins (MTP) as divalent cation transporters are essential for plant metal tolerance and homeostasis. However, the characterization and the definitive phylogeny of the MTP gene family in Fagopyrum tartaricum, and their roles in response to metal stress are still unknown. In the present study, MTP genes in Fagopyrum tartaricum were identified, and their phylogenetic relationships, structural characteristics, physicochemical parameters, as well as expression profiles under five metal stresses including Fe, Mn, Cu, Zn, and Cd were also investigated. Phylogenetic relationship analysis showed that 12 Fagopyrum tartaricum MTP genes were classified into three major clusters and seven groups. All FtMTPs had typical structural features of the MTP gene family and were predicted to be located in the cell vacuole. The upstream region of FtMTPs contained abundant cis-acting elements, implying their functions in development progress and stress response. Tissue-specific expression analysis results indicated the regulation of FtMTPs in the growth and development of Fagopyrum tataricum. Besides, the expression of most FtMTP genes could be induced by multiple metals and showed different expression patterns under at least two metal stresses. These findings provide useful information for the research of the metal tolerance mechanism and genetic improvement of Fagopyrum tataricum.
1. Introduction
Trace elements, including metallic elements such as iron (Fe), manganese (Mn), copper (Cu), and zinc (Zn), are important to maintain normal growth and development in plants [1,2]. However, the growth and production of plants could be inhibited if they absorb excessive amounts of these trace elements [3]. In addition, some toxic heavy metal ions, including lead (Pb) and cadmium (Cd), could also be absorbed by plants because these metal ions have similar chemical structures with trace elements [4]. Recently, due to the rapid development of global industrialization and urbanization, the average content of metallic elements, including heavy metal elements in soil, have gradually increased, causing heavy metal stresses and affecting crop production [5,6]. Precise homeostasis of metallic elements is essential for plant growth and development [7,8,9,10].
The cation diffusion facilitator (CDF) family, which has been widely found in all six kingdoms of living things, plays a vital role in metal tolerance and homeostasis of organisms [11,12]. There are three common features in the CDF family members: an N-terminal signature sequence, a cation efflux domain, and approximately six predicted transmembrane regions [13]. Meanwhile, the CDF family members were clustered into three categories, called Zn-CDF, Zn/Fe-CD,F and Mn-CDF, which had different substrate specificities [14]. In plants, CDF family members are considered metal tolerance proteins (MTP). There are 12 and 10 members in Arabidopsis thaliana and Oryza sativa, respectively [15]. These MTP proteins have been found to be involved in transporting various metallic elements, including Fe, Mn, Cu, Zn, Ni, Co and Cd [16]. For instance, AtMTP1 and AtMTP3, two members of Zn-CDF, play essential roles in Zn and Co tolerance by transporting Zn2+ and Co2+ to the vacuole [17,18,19,20], while two more genes (AtMTP5 and AtMTP12) of Zn-CDF forms a functional complex during Zn2+ transportation into Golgi [21]. MTP8 is the most thoroughly investigated Mn-CDF family members, which contributes to Mn detoxification in A. thaliana and O. sativa by sequestering Mn in vacuoles [22,23,24]. Up to now, many MTP gene family members have already been found in non-model plants species, including Sorghum bicolor [25], Vitis vinifera [26], and Camellia sinensis [27], etc. However, the functional assignment of these MTP proteins identified recently are still unknown.
Tartary buckwheat (Fagopyrum tataricum Gaertn) belongs to the genus Fagopyrum in the family Polygonaceae. As a traditional and irreplaceable crop [6], it is widely cultivated in many countries, including China, Japan, South Korea, India, and Russia [28]. Due to its vital nutritional and medicinal values, it has been processed into a variety of foods, including tea, noodles, and shakima, which are popular all over the world [29]. However, it was reported that Tartary buckwheat had the ability to uptake toxic metals (Pb, Cd, and Hg) from the growth medium and accumulate them in the edible parts [30,31,32,33], and our previous research also found excessively high levels of some heavy metals in Tartary buckwheat tea [34]. These results suggest that heavy metal stress is one of the non-negligible factors restricting Tartary buckwheat production and food safety. Although a few previous studies have shown that Tartary buckwheat seedlings increase the activities of some antioxidant enzymes in response to oxidative stress caused by heavy metals [33] and promote the compartmentalization of heavy metals in vacuoles to reduce heavy metal concentrations in protoplasts [31], the molecular mechanism of heavy metal tolerance and detoxification of Tartary buckwheat is still largely unknown.
Thus, the purposes of this study are to (1) identify MTP gene family members from the genome of Tartary buckwheat; (2) systematically analyze sequences and structural characteristics of Tartary buckwheat MTP gene family members; (3) perform expression analysis of Tartary buckwheat MTP genes in response to various metal stresses. Results of this study will not only provide useful information for subsequent studies on the mechanism involved in heavy metal tolerance of Tartary buckwheat but also contribute to the genetic improvement of Tartary buckwheat.
2. Results
2.1. Identification and Physicochemical Parameters of MTPs in the Tartary Buckwheat Genome
According to the similarities with Arabidopsis MTP genes and MTP gene family features, 12 Fagopyrum tataricum MTP (FtMTP) genes were successfully identified from the Tartary buckwheat genome. Subsequently, based on their homology with the corresponding Arabidopsis MTP genes, these FtMTP genes were named FtMTP1, FtMTP3.1, FtMTP3.2, FtMTP5, FtMTP6, FtMTP7.1, FtMTP7.2, FtMTP8.1, FtMTP8.2, FtMTP9, FtMTP11, and FtMTP12, respectively. Moreover, sequence analysis was performed to investigate the characteristics of the FtMTP gene family. As shown in Table 1, the peptide length of FtMTP proteins ranged from 230 to 722 amino acids, with FtMTP12 and FtMTP5 having the largest and smallest length, respectively. The isoelectric point of FtMTP7.1 was predicted to be 9.23, whereas the isoelectric points of the remaining 11 FtMTP proteins were below 7.00. Besides, of the 12 FtMTP proteins, the predicted molecular weight of FtMTP proteins ranged from 25.41 to 80.51 KDa, and the numbers of predicted transmembrane helices ranged from 1 (FtMTP6) to 10 (FtMTP12). The grand average of hydropathicity (GRAVY) was an index reflecting the hydrophobicity of a protein; the GRAVY value greater than zero indicated that the protein was predicted to be hydrophobic protein, while less than zero indicated the protein was predicted to be hydrophilic protein. Regarding the GRAVY values of the FtMTP proteins, four were hydrophobic, and eight were hydrophilic. In addition, the predicted subcellular localization results showed that all FtMTP proteins were located on the vacuolar membrane, and FtMTP9 was also located on the cell membrane.

Table 1.
Sequences and characteristic analysis of the FtMTP proteins.
2.2. Phylogenetic Tree of 67 MTP Proteins from Six Species
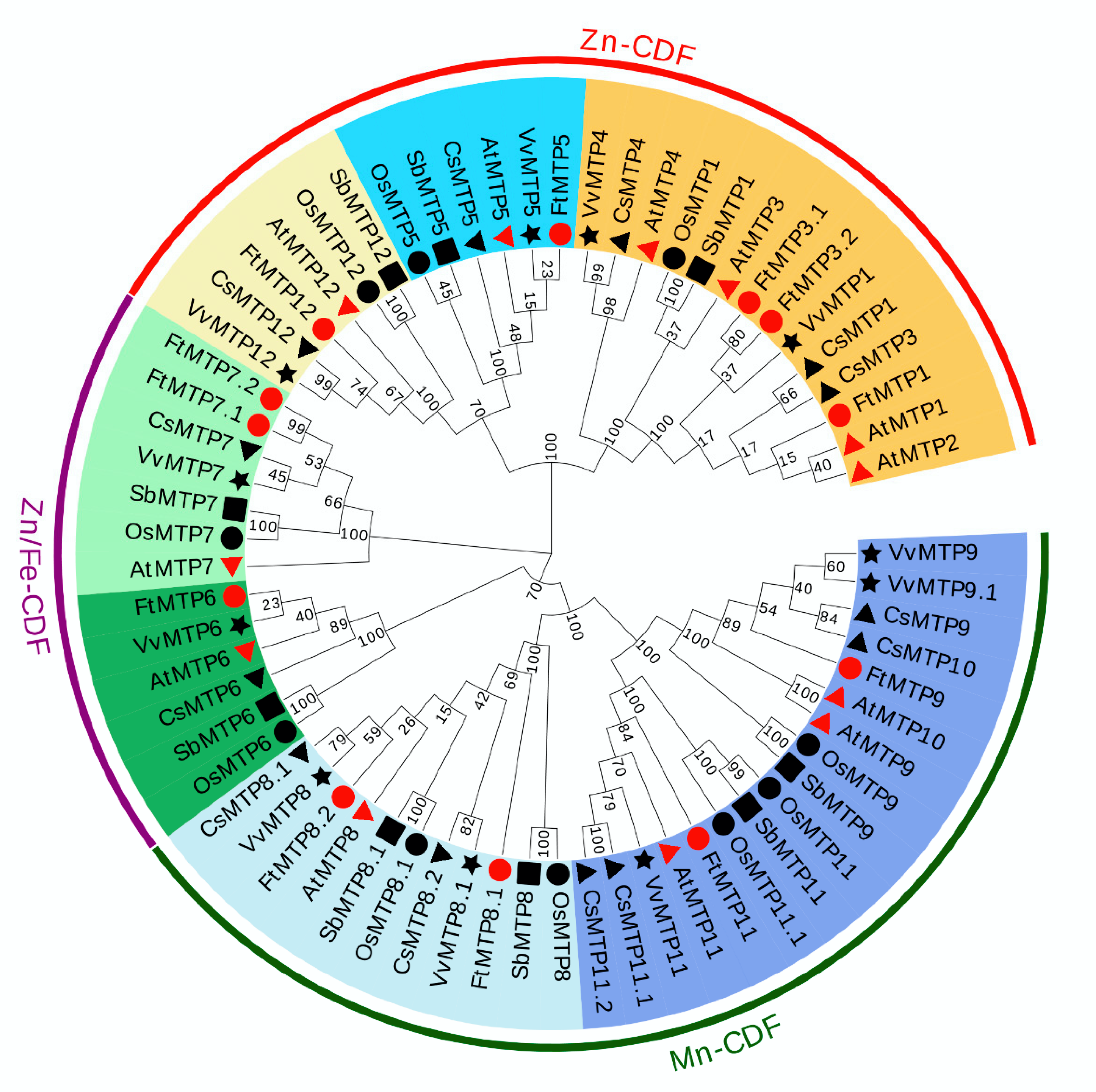
In order to investigate the definitive phylogenetic relationship among MTP gene family members, the phylogenetic tree constructed of 67 MTP proteins from six species, Tartary buckwheat (12), Arabidopsis (12), Rice (10), Sorghum (9), Tea plant (13), and Grape (11) was investigated. According to their similarities at protein level, these 67 MTP proteins were clustered into three major clusters, including Zn-CDFs, Zn/Fe-CDFs and Mn-CDFs, and seven groups, namely group 1, 5, 6, 7, 8, 9 and 12 (Figure 1). However, the numbers of MTP protein in each group, as well as the numbers of group in each cluster were diverse. Of the seven groups, group 1, 5 and 12 belonged to Zn-CDFs, group 6 and 7 were constructed Zn/Fe-CDFs, and group 8 and 9 made up Mn-CDFs. Furthermore, group 1 contained 14 MTP proteins, which were AtMTP1, AtMTP2, AtMTP3, AtMTP4, OsMTP1, SbMTP1, CsMTP1, CsMTP3, CsMTP4, VvMTP1, VvMTP4, FtMTP1, FtMTP3.1 and FtMTP3.2. Group 5 was composed of 6 MTP proteins, i.e., AtMTP5, OsMTP5, SbMTP5, CsMTP5, VvMTP5 and FtMTP5. Similarly, group 6 was composed of six MTP proteins, AtMTP6, OsMTP6, SbMTP6, CsMTP6, VvMTP6 and FtMTP6. Besides, seven MTP proteins, AtMTP7, OsMTP7, SbMTP7, CsMTP7, VvMTP7, FtMTP7.1 and FtMTP7.2 formed group 7. For group 8, there were 11 MTP proteins, namely AtMTP8, OsMTP8, OsMTP8.1, SbMTP8, CsMTP8.1, CsMTP8.2, VvMTP8, FtMTP8.1 and FtMTP8.2. The members of group 9 were seventeen, with AtMTP9, AtMTP10, AtMTP11, OsMTP9, OsMTP11, OsMTP11.1, SbMTP9, SbMTP11, CsMTP9, CsMTP10, CsMTP11.1, CsMTP11.2, VvMTP9, VvMTP9.1, VvMTP11, FtMTP9 and FtMTP11. Meanwhile, six MTP proteins including AtMTP12, OsMTP12, SbMTP12, CsMTP12, VvMTP12 and FtMTP12 belonged to group 12.
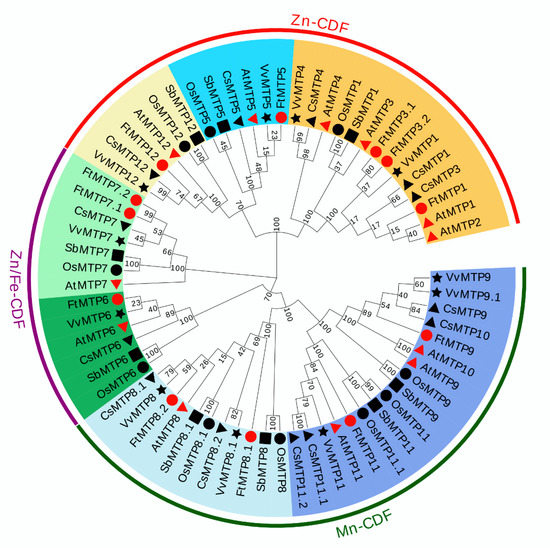
Figure 1.
A phylogenetic tree based on 67 MTP proteins: 12 Arabidopsis (marked by red triangle), 10 rice (black circle), 11 grape (black star), 9 sorghum (black square), 13 tea plant (black triangle), and 12 Tartary buckwheat (red circle). The phylogenetic tree was constructed using the neighbor-joining (NJ) method and MEGA 7.0 software at 1000 replications bootstrap. The diagrams were compiled, labeled, color-coded, and visualized using EvolView v3.
2.3. Phylogenetic Tree, Gene Structure and Motif Patterns of FtMTPs
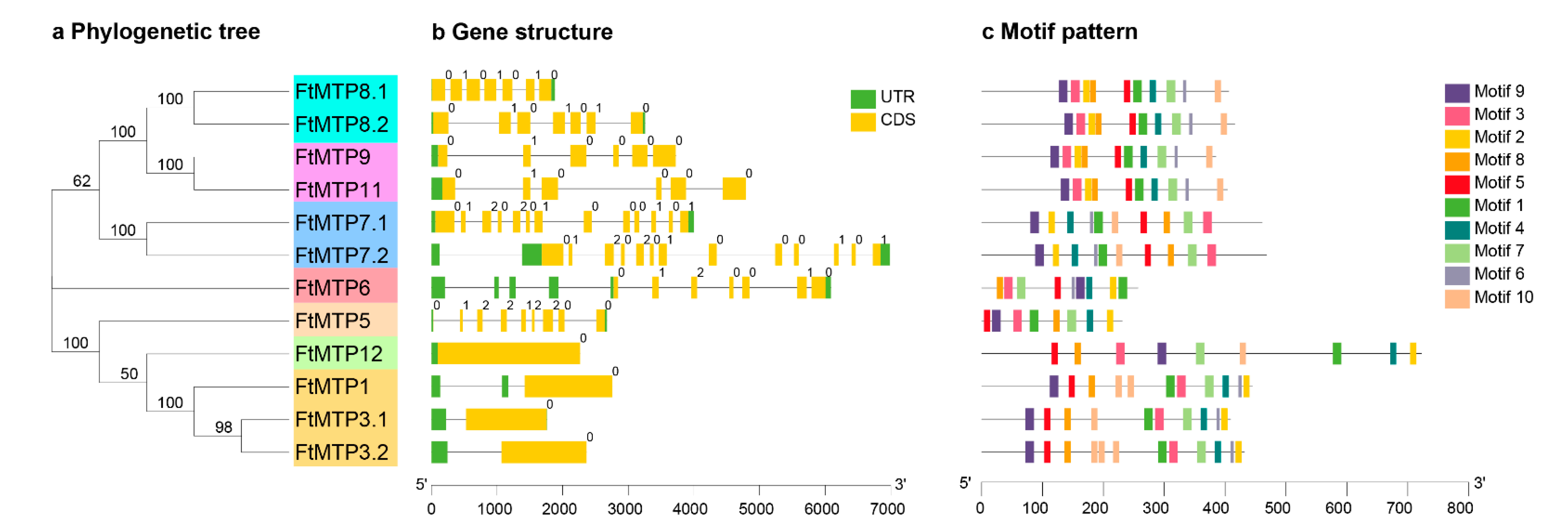
As shown in Figure 2a, Tartary buckwheat MTP gene family members were divided into three clusters (Zn-CDFs, Zn/Fe-CDFs, and Mn-CDFs) and seven groups (1, 5, 6, 7, 8, 9, and 12). Of the seven groups, groups 1, 5, and 12 belonged to Zn-CDFs, groups 6 and 7 were constructed Zn/Fe-CDFs, and groups 8 and 9 made up Mn-CDFs. Group 1 was the largest with three members, followed by groups 7, 8, and 9 (2 genes), whereas groups 5, 6, and 12 each had only one gene. All FtMTPs contained exon and intron; however, the number and length of exon or intron were diverse among different clusters (Figure 2b). Regarding exons, Zn/Fe-CDFs had 7 or 13 exons, Mn-CDFs contained 6 or 7 exons, and Zn-CDFs had only one exon expect for group 5 (9 exons). For introns, Zn/Fe-CDFs were the largest with 12–15 introns, followed by Mn-CDFs (6–8 introns), whereas Zn-CDFs had 1–2 introns except for Group 5 (10 introns). The conserved motifs of FtMTP proteins were predicted and analyzed using MEME depending on the amino acid sequences with ten motifs (Figure 2c). It was primarily observed that the number, type, or order of motifs was similar within the same group in addition to that among different clusters. These ten motifs appeared at least once in groups 1, 7, 8, and 9, whereas groups 6 and 12 lacked motifs 10 and 6, respectively, and Group 5 lost both motifs 6 and 10. In addition, FtMTP1 and FtMTP3.2, two members belonged to Group 1, containing two and three motif 10, respectively.
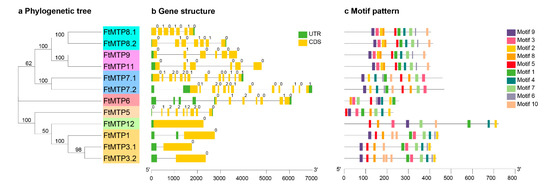
Figure 2.
Phylogenetic tree, gene structure, and motif pattern analysis of FtMTPs. (a) the neighbor-joining phylogenetic tree was constructed with MEGA 7.0 using FtMTP amino acid sequences with 1000 replications bootstrap; (b) exon-intron structure of FtMTP genes where yellow boxes represented the exons, the black lines represented the introns, and the green boxes represented the untranslated regions (UTRs), with size scales detailed at the bottom; (c) the motif patterns of FtMTP proteins using ten conserved motifs (Supplementary Figure S1) are represented by the unique color mentioned in the legend on the top lift, with size scales detailed at the bottom.
2.4. Structure Variation of the FtMTP Proteins
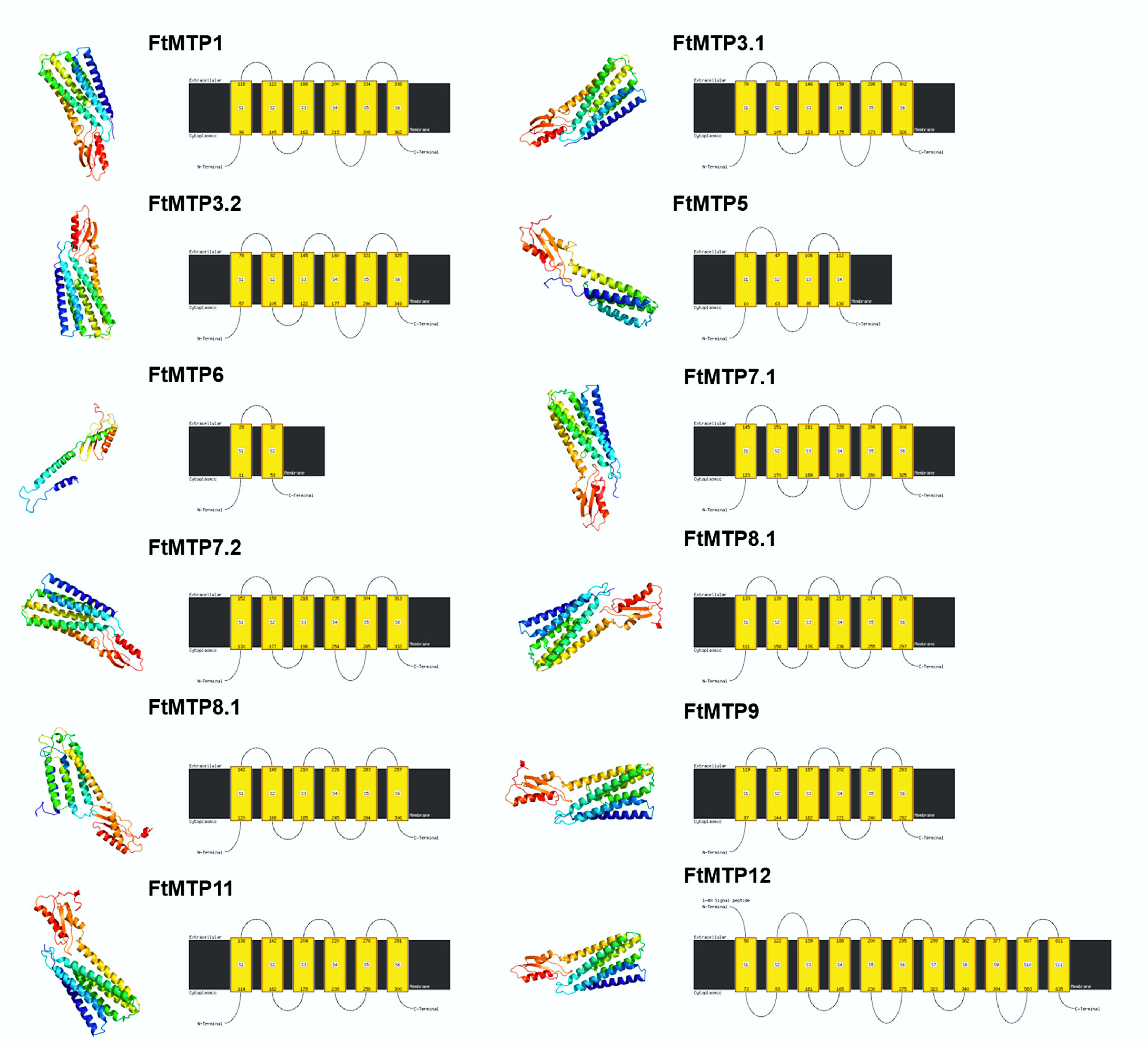
To further explore the structure variation, twelve FtMTP proteins were modeled using Phyre2 with normal mode, and the results were shown in Figure 3. It could be clearly seen that all FtMTP proteins contained α-helices and β-sheets. Expect for FtMTP7.2, which contained two β-sheets, the rest of the proteins contained three β-sheets, and except for FtMTP5 and FtMTP6, which contain six and four α-helices, respectively, the rest of the proteins contained eight α-helices. Besides that, three members, FtMTP1, FtMTP3.1, and FtMTP3.2, in Group 1 contained six transmembrane helices. Proteins belonging to groups 5 and 7 had four transmembrane helices. There was only two transmembrane helices in Group 6, which was the least of all seven groups. Group 8 had four (FtMTP8.2) or five (FtMTP8.1) transmembrane helices, whereas Group 9 had four (FtMTP11) or six (FtMTP9) transmembrane helices, and Group 12 contained the largest number (10) of transmembrane helices among seven groups. Based on these results, the numbers of transmembrane helices of the FtMTP proteins in the same group were similar, whereas variable among different groups, indicating that the diversity of the structure endowed the functional complexity of the FtMTP gene family.
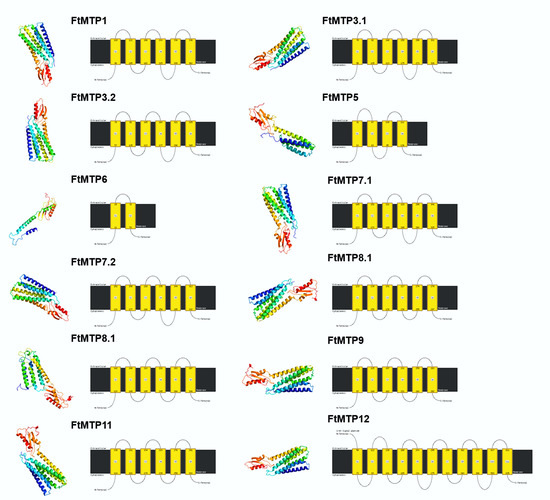
Figure 3.
Structure prediction analyses of FtMTP proteins. Models had been generated by using Phyre 2 server in normal mode and were visualized by rainbow color from N to C terminus and organized in FtMTP1, and FtMTP3.1 to FtMTP12. For the left pictures, the coils and the smooths represented alpha-helices and beta-sheets, respectively; For the right pictures, the black line represented protein sequences, and the yellow boxes represented transmembrane helices.
2.5. Cis-Regulatory Elements of the FtMTPs
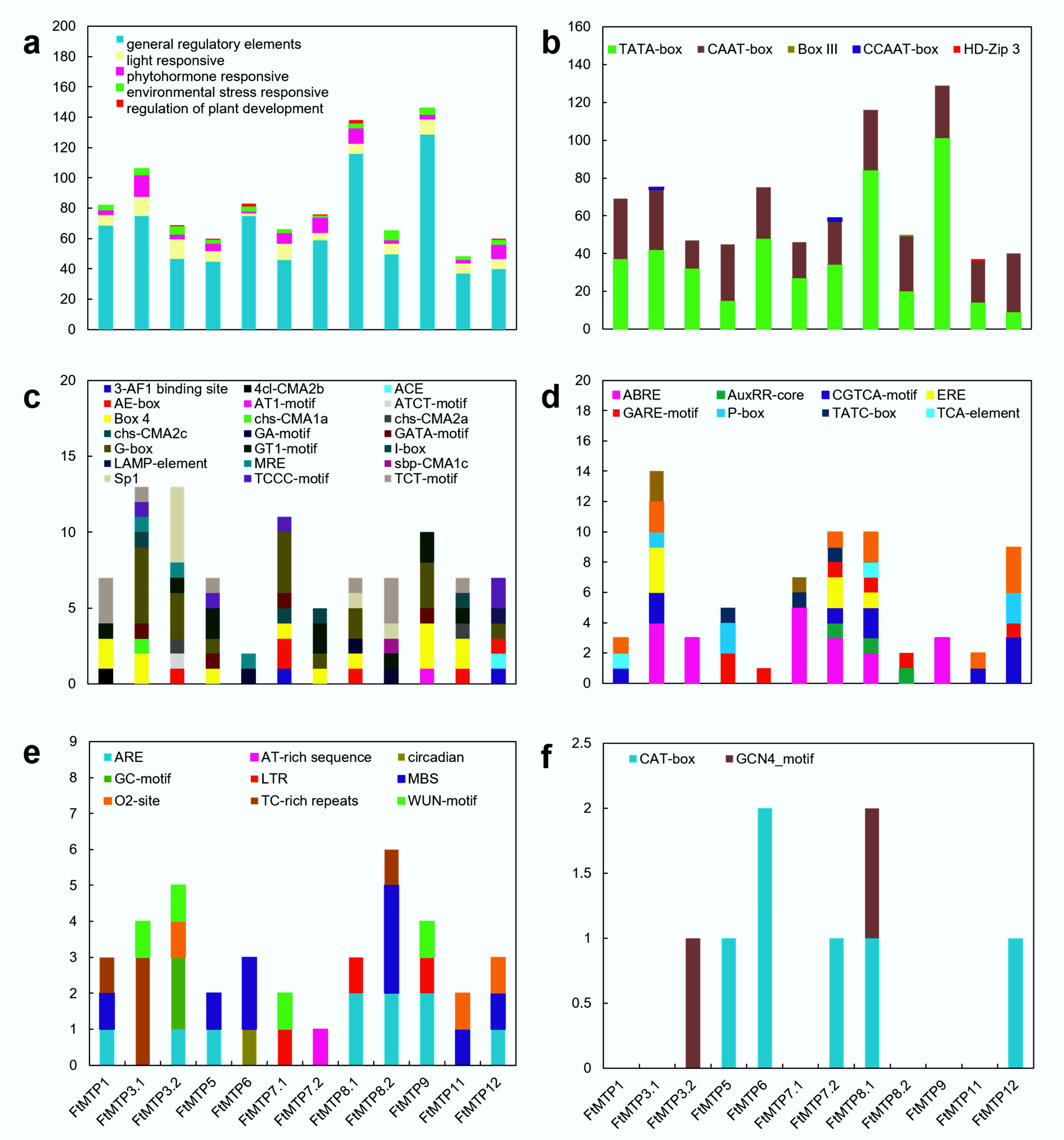
To better understand the transcription regulatory mechanism of FtMTP genes, cis-regulatory elements (CREs) in the 1500 bp upstream of these genes were analyzed. After removing unknown motifs, a total of 47 CREs were found in the promoter of FtMTP genes, and these CREs were further classified into five groups, general regulatory elements, light response elements, hormone response elements, environmental stress response elements, and regulation of plant development elements (Figure 4a). The overall frequency of these CREs in FtMTP genes and their frequency in the upstream region of each corresponding gene were very diverse. General regulatory elements, light response elements, hormone response elements, and environmental stress response elements were frequently found in all of the FtMTP genes, but regulation of plant development elements was specifically found in FtMTP3.2, FtMTP5, FtMTP6, FtMTP7.2, FtMTP8.1, and FtMTP12. Further analysis showed that five types of general regulatory elements, including TATA-box, CAAT-box, Box Ⅲ, CCAAT-box, and HD-ZIP 3, were identified in the upstream region of FtMTP genes. TATA- and CAAT-boxes were frequently found in all of the FtMTP genes, while Box Ⅲ was specifically found in FtMTP8.2, CCAAT-box was specifically present in FtMTP3.1, and HD-ZIP 3 specifically existed in FtMTP11 (Figure 4b). There were 21 types of light-responsive elements and ten types of hormone response elements involved in ethylene, abscisic acid, auxin, MeJA, salicylic acid, and gibberellin mediated responses were observed in all of the FtMTP genes (Figure 4c,d). These hormone response elements identified in FtMTP8.1 might participate in these six plant hormones mediated responses. However, only ABRE or GARE-motif involved in abscisic acid or gibberellin-mediated responses were resolved in FtMTP3.2 and FtMTP6, while FtMTP5 contained ABRE, GARE-motif, and TATC-box elements. Nine types of environmental stress response elements involved in anaerobic induction, stress and defense, drought, low temperature, and other abiotic stresses were frequently found in all of the FtMTP genes (Figure 4e). Most FtMTP genes shared ARE element, which was essential for the anaerobic stress responses, while MBS element (MYB binding site), which was involved in drought stress responses, were specifically found in FtMTP5, FtMTP6, FtMTP8.2, FtMTP11 and FtMTP12, and LTR element involved in low-temperature stress were specifically found in FtMTP7.1, FtMTP8.1, and FtMTP9. CREs involved in the regulation of plant development elements, i.e., CAT-box and GCN4_motif, which were essential for meristem or endosperm development, were also investigated (Figure 4f). It could be clearly found that GCN4_motif was specifically found in FtMTP3.2 and FtMTP8.1, yet CAT-box was specifically observed in FtMTP5, FtMTP6, FtMTP7.2, FtMTP8.1, and FtMTP12.
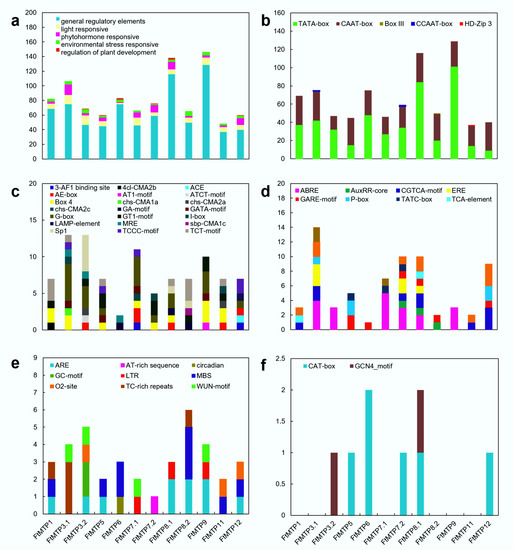
Figure 4.
Cis-regulatory elements (CREs) of FtMTP genes. (a) members and numbers of CREs of FtMTP genes; (b) members and numbers of general regulatory elements of FtMTP genes; (c) members and numbers of light-responsive elements of FtMTP genes; (d) members and numbers of phytohormone responsive elements of FtMTP genes; (e) members and numbers of environmental stress-responsive elements of FtMTP genes; (f) members and numbers of regulation of plant development elements of FtMTP genes. The value used in the vertical axis in these figures represented the number of corresponding cis-regulatory elements.
2.6. Tissues-Specific Expression Profile of FtMTPs
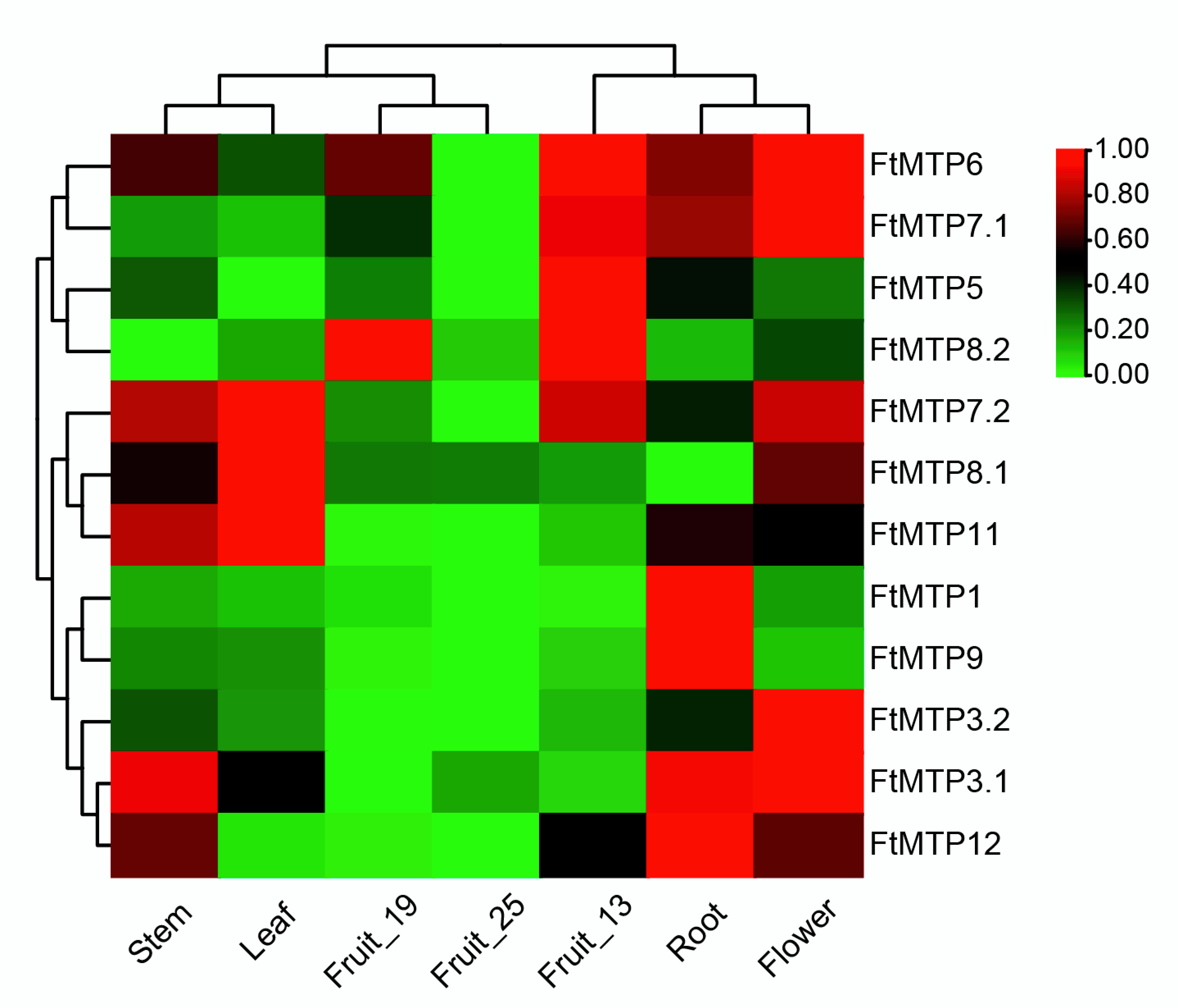
In Figure 5, it showed the expression of FtMTPs from different Tartary buckwheat tissues under different developmental stages, including root, stem, leaf, flower, fruit at 13 days of development (fruit_13), fruit at 19 days of development (fruit_19), fruit at 25 days of development (fruit_25). From the results, FtMTP3.1 and FtMTP3.2, which belonged to Group 1, expressed highest in flower, and lowest in fruit_19 or fruit_25, but the other Group 1 member, FtMTP1, had the highest and the lowest expression in root and fruit_13, respectively. A member of Group 5, FtMTP5, showed the highest and lowest expression levels in fruit_13 and leaf, respectively. FtMTP6, the gene from Group 6 displayed the highest expression in flower and fruit_13, and the lower expression in fruit_25. In addition, all members in groups 7 and 8 showed tissue-specific expression patterns. FtMTP7.1 was expressed highly in flowers, while FtMTP7.2 was strongly expressed in leaves. The expression of FtMTP8.1 in leaves was highest compared to other tissues; however, FtMTP8.2, the other member in Group 8, showed the highest expression in both fruit_13 and fruit_19. FtMTP9 was strongly expressed in roots and weakly expressed in fruit_19 and fruit_25, and FtMTP11 exhibited the highest and lowest expression in leaves and fruit_25, respectively. FtMTP12, a Group 12 member, expressed higher in roots, stems, flowers, and fruit_13, but lower in leaf, fruit_19 and fruit_25.
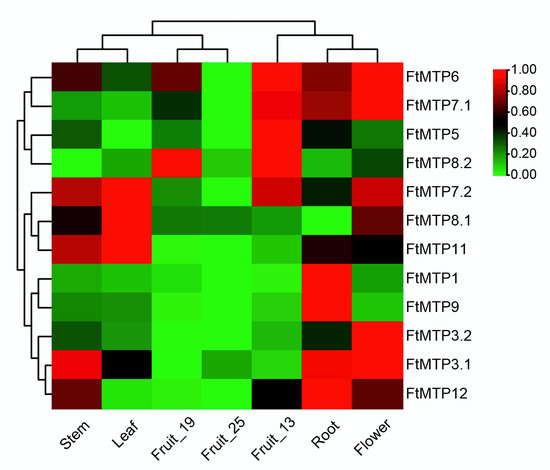
Figure 5.
Heatmap analysis of the expressions of FtMTPs in different Tartary buckwheat tissues at different developmental stages. Gene expression (FPKM) was normalized and visualized using the TBtools software. The Stem, Leaf, Fruit_19, Fruit_25, Fruit_13, Root, and Flower marked in the figure represented the stem, leaf, fruit at 19 days of development, fruit at 25 days of development, fruit at 13 days of development, root and flower of Tartary buckwheat, respectively.
2.7. Expression of FtMTPs under Multiple Metal Stress
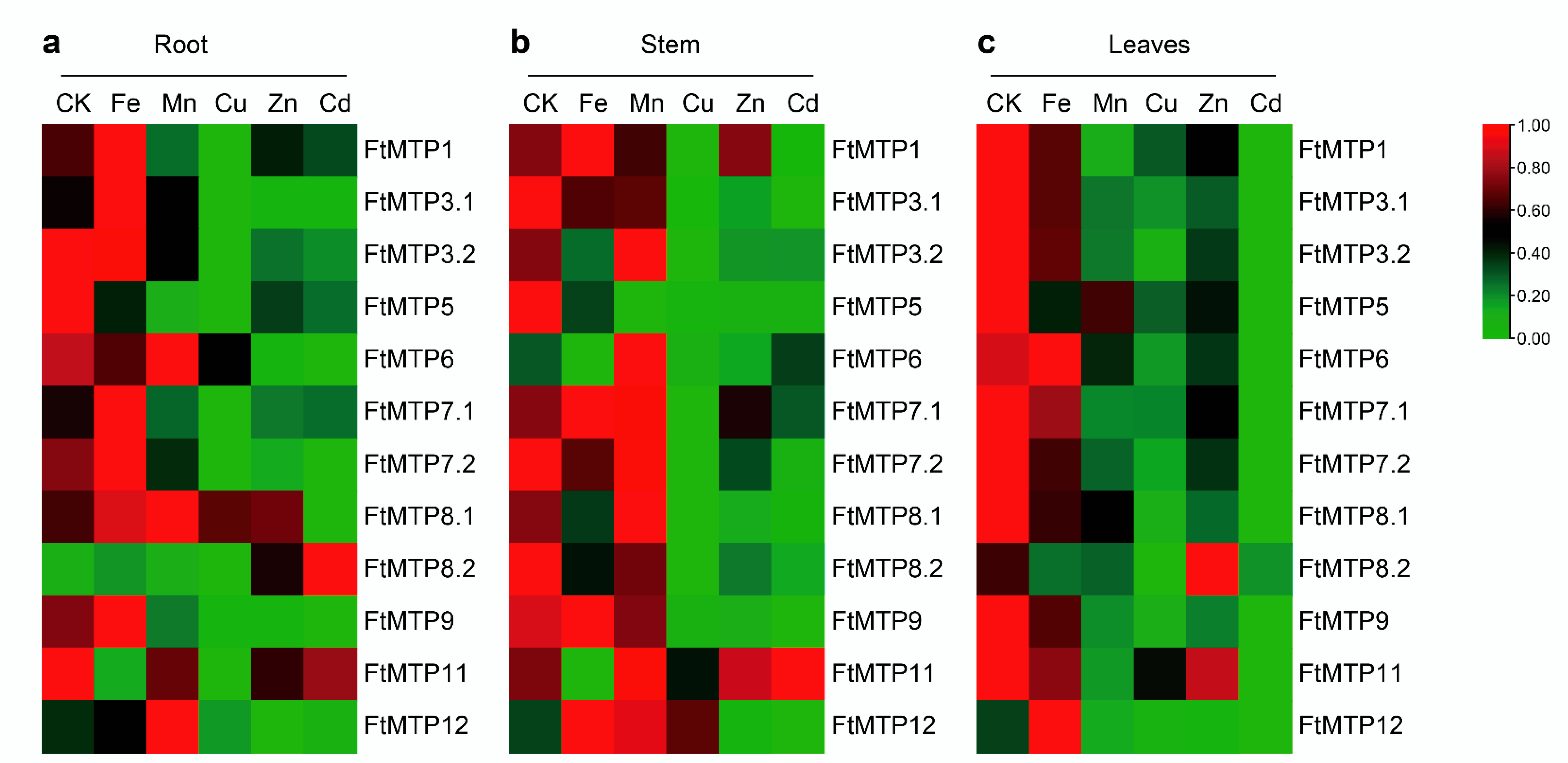
The expression of FtMTPs in Tartary buckwheat roots under normal conditions and five metal stresses were investigated by qRT-PCR and are shown in Figure 6a. Compared with the expression of FtMTPs in roots under normal condition (CK), Fe3+ treatment upregulated the expression of FtMTP1, FtMTP3.1, FtMTP7.1, FtMTP8.1, and FtMTP8.2, whereas downregulated the expression levels of FtMTP5 and FtMTP11. Similarly, Mn2+ treatment upregulated the expression of FtMTP8.1 and FtMTP12, while downregulated the expression of FtMTP1, FtMTP3.1, FtMTP5, FtMTP7.1, FtMTP7.2, and FtMTP9. Except for FtMTP8.1, the expression of all FtMTPs was reduced under Cu2+ treatment. Zn2+ treatment resulted in the expression of FtMTP8.2 upregulation, while the expression of FtMTP1, FtMTP3.1, FtMTP3.2, FtMTP5, FtMTP6, FtMTP7.1, FtMTP7.2, FtMTP9, and FtMTP12 downregulation. Only the expression of FtMTP8.2 was increased in response to Cd2+ treatment, and the expression levels of the remaining FtMTPs except for FtMTP11 were reduced.
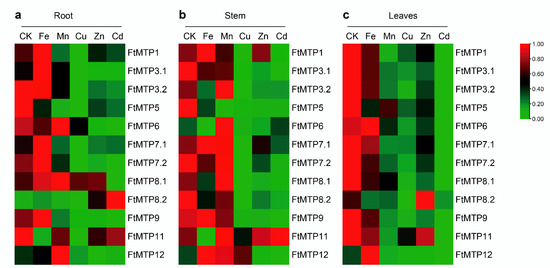
Figure 6.
Heatmap expressions of the abundance of FtMTPs in different Tartary buckwheat tissues under multiple metal stress. (a) heatmap analysis of the abundance of FtMTPs in Tartary buckwheat root, gene expression was normalized using zero to one method; (b) heatmap analysis of the abundance of FtMTPs in Tartary buckwheat stem, gene expression was normalized using zero to one method; (c) heatmap analysis of the abundance of FtMTPs in Tartary buckwheat leaves. Gene expression was normalized and visualized by the TBtools software. CK, Fe, Mn, Cu, and Zn, appearing in Figure 6a–c, represent the root, stem, and leaves of Tartary buckwheat under normal conditions, Fe3+, Mn2+, Cu2+, and Zn2+ treatments, respectively.
The expression of FtMTPs in Tartary buckwheat stems under normal condition and five metal stresses were examined and showed in Figure 6b. Compared with the expression of FtMTPs in Tartary buckwheat stems under normal condition (CK), Fe3+ treatment induced the expression of FtMTP12, whereas reduced the expression of FtMTP3.1, FtMTP3.2, FtMTP5, FtMTP6, FtMTP7.2, FtMTP8.1, FtMTP8.2, and FtMTP11. Similarly, Mn2+ treatment resulted in the expression of FtMTP3.2, FtMTP6, FtMTP8.1, and FtMTP12 upregulation, while the expression of FtMTP3.1, FtMTP5, FtMTP8.2, and FtMTP9 downregulation. Moreover, the expression of FtMTP6, FtMTP8.1, and FtMTP12 were upregulated in response to Cu2+ treatment, but the expression of FtMTP1, FtMTP3.1, FtMTP3.2, FtMTP5, FtMTP8.2, and FtMTP9 were downregulated. Expect for FtMTP1, FtMTP6, FtMTP7.1, and FtMTP11, the expression of the remaining eight FtMTPs were reduced under Zn2+ treatment. Besides, the expression of FtMTP1, FtMTP3.1, FtMTP3.2, FtMTP5, FtMTP7.1, FtMTP7.2, FtMTP8.1, FtMTP8.2, FtMTP9, and FtMTP12 were downregulated under Cd2+ treatment.
The expression of FtMTPs in Tartary buckwheat leaves under normal condition and five metal stresses were tested by qRT-PCR. As shown in Figure 6c, when compared to the expression of FtMTPs in Tartary buckwheat leaves under normal condition (CK), the expression of FtMTP12 upregulated in response to Fe3+ treatment, whereas the expression of most FtMTPs, including FtMTP1, FtMTP3.1, FtMTP3.2, FtMTP5, FtMTP7.2, FtMTP8.1, FtMTP8.2, and FtMTP9 downregulated. Mn2+, Cu2+, and Cd2+ treatments resulted in the expression of all FtMTPs reduced. Besides, except that the expression of FtMTP8.2 was upregulated under Zn2+ treatment, the expression of the remaining FtMTPs under Zn2+ treatment was reduced.
3. Discussion
Soil heavy metal pollution caused by artificial activities such as mineral smelting, sewage irrigation, and large-scale application of pesticides and fertilizers has become one of the abiotic factors restricting crop growth and production, thus food safety [5,6]. Metal tolerance proteins (MTPs) as divalent cation transporters involve in metal uptake and transportation and play vital roles in metal tolerance and detoxification of crops. In the present study, a total of 12 FtMTPs were identified in F. tartaricum, which is similar to that in A. thaliana [15], but higher than that in O. sativa [15], V. vinifera [26], and S. bicolor [25] and less than that in C. sinensis [27], P. trichocarpa [35], and N. tabacum [36]. However, homologs of AtMTP2, AtMTP4, and AtMTP10 could not be detected in the F. tartaricum, whereas two AtMTP3 orthologs, FtMTP3.1 and FtMTP3.2, two AtMTP7 orthologs, FtMTP7.1 and FtMTP7.2, and two AtMTP8, FtMTP8.1 and FtMTP8.2, were identified (Table 1 and Figure 1). These results indicate that gene loss and duplication might have occurred in the evolution of the MTP gene family in F. tartaricum. Meanwhile, the characteristic analysis of FtMTP proteins was processed, and all FtMTP proteins were predicted to be located in the vacuolar membrane (Table 1), suggesting that FtMTPs might mainly participate in the transportation of heavy metals to vacuoles. Interestingly, the FtMTP9 was also predicted to be located in the cell membrane. This result indicated that FtMTP9 might play an essential role in heavy metal transportation between Tartary buckwheat cells. The function of FtMTP9 for heavy metal tolerance in Tartary buckwheat needs to be investigated in future studies.
There were three structural features of MTP proteins, an N-terminal signature sequence, a cation efflux domain, and approximately six predicted transmembrane regions [13]. In this study, the cation efflux domain and the modified signature sequence were found in all FtMTP proteins, although some other motifs were not present or more than one in certain FtMTP members, as shown in Figure 2. Besides, FtMTP6 had the smallest peptide length (256 amino acid), MW (28.54 KDa), and TMHs (1 TMHs), while FtMTP12 had the largest peptide length (722 amino acid), MW (80.51 KDa), and TMHs (10 TMHs) (Table 1 and Figure 3). These results were consistent with the characteristics of the MTP gene family in other plant species, including C. sinensis [27], P. trichocarpa [35], and N. tabacum [36], which indicated that the structure and function conservation of the MTP gene family in plant evolutionary events.
In general, phylogenetic analysis is one of the strategies to predict the function of uncharacterized genes [37]. For instance, the reported gene families in F. tartaricum, including NAC [38], AP2/ERF [39], GRAS [40], ARF [41], bZIP [42], Bhlh [43], MADS [44], SPL [45], BBX [46], andNF-Y [47], showed similar gene functions between the phylogenetic orthologs. This study showed FtMTPs and AtMTPs were extremely similar at the protein level, and the results of the phylogenetic analysis showed that their orthologs were closely clustered together (Figure 1), thus indicating FtMTPs might have similar functions with their corresponding AtMTP orthologs.
Gene expression regulation is involved in different stages of plant growth and development. Normally, transcriptional regulation and post-transcriptional regulation are two aspects in which gene expression regulation happens [35]. Transcriptional regulation is an important way to regulate gene expression level, while cis-acting regulatory elements involving in this process through interactions with RNA polymerase and specific transcription factors [35]. Here, different cis-acting regulatory elements were processed in the upstream region of each FtMTP (Figure 4), which revealed regulating events at the initiation of transcription. All FtMTP genes contained light-responsive cis-acting elements, phytohormone responsive cis-acting elements, environmental stress-response cis-acting elements, and also general regulatory elements, as shown in Figure 4, indicating that the expression of FtMTPs genes could be affected by photoperiod, plant hormones, and environmental stress. This result also implied that all MTPs genes of Tartary buckwheat could be transcriptionally regulated by multiple stimulates. Additionally, only FtMTP3.2, FtMTP5, FtMTP6, FtMTP7.2, FtMTP8.1, and FtMTP12 contained cis-acting elements related to plant development regulation, indicating that these FtMTPs might also be related to the growth and development of Tartary buckwheat. In particular, FtMTP9 was detected to have the largest number of regularity elements, suggesting that it might be involved in many regulation pathways. Overall, abundant cis-acting regulatory elements located upstream of these FtMTPs imply that they might play an important role not only in heavy metal stress responses but also in multiple stress responses and plant development.
In addition, analysis of gene expression patterns could also provide insights into gene functions [37]. Comprehensive analysis of expression patterns of FtMTPs in three tissues under five metal stresses (Figure 5 and Figure 6), this study formed a relatively comprehensive function prediction of FtMTPs. Interestingly, previous studies reported that Zn-CDFs such as AtMTP1, AtMTP3, AtMTP5, AtMTP12, and their orthologs, PtdMTP1, CsMTP1, CitMTP1, CisMTP3, CitMTP5, and CitMTP12 were mainly involved in Zn and Cd detoxification [17,20,37,48,49]; however, in this study, only the member of Zn-CDFs, FtMTP1 were induced in the stems after treatment with excessive amounts of Zn, and no Zn-CDFs in F. tartaricum were induced in the roots, stems or leaves after treatment with excessive amounts of Cd. This may be attributed to the Zn and Cd concentrations used in our experiment, which means that the response of these genes may require appropriate concentrations of Zn and Cd treatment. Besides, this study also found that the Zn-CDF in F. tartaricum except for FtMTP5 induced in the roots, stem, or leaves after exposure to high levels of Fe, suggesting the potential roles of these genes in Fe homeostasis, which are consistent with previous research. Mn-CDF, such as the members of MTP8, MTP9, MTP10, and MTP11, play vital roles in Mn transport and detoxification [22,24,50,51,52]. In the present study, FtMTP8.1 in roots and stems were highly expressed after the Mn treatment; in addition, the expression of FtMTP8.1 in roots was also induced by Fe or Zn treatment. For the other three Mn-CDFs in F. tartaricum, the expression of FtMTP8.2 in roots and leaves were upregulated by Cd and Zn treatment, respectively, FtMTP9 in roots and stems were induced after Fe treatment, and FtMTP11 in stems was upregulated by Mn, Zn, and Cd treatment. This finding supports those of previous studies. For Zn/Fe-CDFs, including MTP6 and MTP7, it is mainly involved in the uptake, transport, and homeostasis of Fe or Zn. This study found that FtMTP7.1 and FtMTP7.2 in roots, FtMTP7.1 in stems were highly expressed after excessive Fe treatment, and FtMTP6 in roots and stems and FtMTP7.1 in stems were upregulated by Mn treatment, which has been proven by previous studies. Although most of the MTP genes have been fully characterized in many plant species, the role of MTP genes in Cu tolerance and detoxification in plants remains unknown. From the result of this study, the expression of FtMTP8.1 in roots and the expression of FtMTP12 in stems were significantly upregulated after exposure to excessive amounts of Cu. Similar results have been found in C. sinensis [37]. Therefore, more in-depth investigations on Cu detoxification of these MTP genes need to be carried out in the future.
4. Materials and Methods
4.1. Identification, Physicochemical Parameters of MTPs in Tartary Buckwheat
Tartary buckwheat genome was downloaded from MBKBASE (http://mbkbase.org (accessed on 20 October 2021)). Protein sequences of 55 MTPs in Arabidopsis thaliana, Oryza sativa, Sorghum bicolor, Vitis vinifera and Camellia sinensis (Supplementary Table S1) were downloaded from the National Center for Biotechnology (NCBI) (https://www.ncbi.nlm.nih.gov/ (accessed on 20 October 2021)), the Rice Genome Annotation Project [53] (http://rice.uga.edu/ (accessed on 20 October 2021)) and Phytozome [54] (https://phytozome-next.jgi.doe.gov/ (accessed on 20 October 2021)) respectively. In order to identify the candidate MTP genes in Tartary buckwheat genome, all known Arabidopsis MTP genes were used as query sequences to search against the Tartary buckwheat genome by a BLASTp search at a score value of ≥ 100 and e-value ≤ 1 × 10−5 [55]. After that, the MTP gene sequences were retrieved from the Tartary buckwheat genome. Then, domains and functional sites in each Tartary buckwheat candidate MTP gene were examined with Pfam tool [56] (http://pfam.xfam.org/ (accessed on 23 October 2021)) and Smart program [57] (http://smart.embl.de/smart/batch.pl (accessed on 23 October 2021)), and all candidate Tartary buckwheat MTP genes containing cation efflux family (CDF) domain (PF01545) were verified. After removing redundant sequences, 12 MTP genes (Supplementary Table S2) in the Tartary buckwheat genome were successfully identified. Subsequently, the ProtParam tool of ExPaSy [58] (https://www.expasy.org/ (accessed on 5 November 2021)) was used to calculate the molecular weight (MW), the theoretical isoelectric point (pI), and the grand average of hydropathicity (GRAVY) of proteins. Putative transmembrane regions in proteins were predicted using the TMHMM Server V2.0 [59] (http://www.cbs.dtu.dk/services/TMHMM/ (accessed on 5 November 2021)). Subcellular localization (SL) of FtMTP proteins were predicting with the Plant-mPLoc version 2.0 [60,61,62,63] (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/ (accessed on 5 November 2021)).
4.2. Phylogenetic Analysis
ClustalW [64] was used for MTP gene sequences alignment among Fagopyrum tartaricum, Arabidopsis thaliana, Oryza sativa, Sorghum bicolor, Vitis vinifera, and Camellia sinensis. Phylogenetic trees were constructed using MEGA 7.0 software [65] with neighbor-joining method [66] and 1000 bootstrap test replicates [67], then drawn with EvolView V3 [68] (https://www.evolgenius.info/evolview/ (accessed on 10 December 2021)).
4.3. Gene Structure and Motif Analysis
Intron-exon structures of FtMTP genes were analyzed using the TBtools software [69]. Pfam tool [56] (http://pfam.xfam.org/ (accessed on 12 December 2021)) and MEME program [70] (https://meme-suite.org/meme/ (accessed on 12 December 2021)) were used to analyze conserved domains and motifs in FtMTP gene sequences, respectively. Subsequently, domain and motif diagrams were drawn with the TBtools [69].
4.4. Protein Modeling and Prediction
All FtMTP amino acid sequences were submitted to Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2/ (accessed on 20 December 2021)) and 3D protein structure models for FtMTP genes were Predicted with homology modeling under the normal mode [71].
4.5. Cis-Regulatory Elements Prediction
The promoter sequence for each FtMTP gene was defined as 1500 bp upstream from the start codon and extracted with TBtools software [69]. All the cis-regulatory elements (CREs) in promoter sequence were analyzed and identified by the PlantCARE database [72] (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 30 December 2021)).
4.6. Transcriptome Data Analysis
The expression data of Tartary buckwheat MTP genes in seven tissues, including root, stem, leaf, flower, and fruit under three development stages, were obtained from the Tartary buckwheat Database [73] (http://tbd.sicau.edu.cn/index/gev/index.html (accessed on 10 January 2022)), and the HeatMap Illustrator of TBtools software [69], were used for visualization of the expression data.
4.7. Plant Cultivation and Stress Treatments
Tartary buckwheat seeds were sown in soil pots under natural conditions. After growth for one week, buckwheat seedlings were transplanted into uniform pots (d = 9.4 cm, h = 8 cm) with uniform soil (one seedling in a pot) under greenhouse conditions (22 to 25 °C, 12 h light and 12 h darkness, 50% to 60% relative humidity). Three days later, plants were irrigated with five metal ion solutions respectively. Sources and concentrations of metal ions are shown in Table 2. Plants were irrigated with 100 mL water (controls) or solution (treatments), and the pots were placed in culture dishes. After treatment for three days, roots, stems, and leaves from the same plant of each treatment were harvested separately, washed with water three times, frozen by liquid nitrogen, ground to powder, and then stored at −80 °C for RNA isolation. Three biological replicates were used for each treatment.

Table 2.
Concentrations and sources of different metal ions used in this study.
4.8. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNAs were isolated using the Eastep™ Super Total RNA Extraction Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The purity, integrity, and concentration of RNA were determined with a NanoDrop1000 (NanoDrop Technologies, Inc., Wilmington, DE, USA). Furthermore, 3 µg of RNA was reverse-transcribed to cDNA using a GoScript TM Reverse Transcription System (Promega, Madison, WI, USA). The optimal forward and reverse primers (Supplementary Table S3) designed by an online tool Primer-BLAST10 was used for qRT-PCR analysis. Then, qRT-PCR was conducted in triplicate using cDNAs prepared from different tissues with various treatments with a FastStart Universal SYBR Green Master (Rox, Roche, Indianapolis, IN, USA) and the ABI 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Thermal cycling parameters were as follows: 95 °C for 10 min, followed by 40 cycles of 94 °C for 5 s, 60 °C for 15 s, and 72 °C for 34 s. FtH3 gene was amplified as an internal control. Relative gene expression levels were obtained by dividing extrapolated transcript levels of target genes by levels of FtH3 (the internal control) from the same sample.
4.9. Statistical Analysis
All statistical analyses were performed using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA), and results were expressed as means ± SD. The gene expression was normalized using the TBtools software.
5. Conclusions
In this study, 12 MTP protein-encoding gene members were identified from the Tartary buckwheat genome, and they were clustered into seven groups based on their phylogenetic relationships. The physicochemical parameters and three-dimensional structures of proteins belonging to the same group were similar but varied among different groups. The analysis of cis-acting elements showed that the expression of the FtMTP gene family might be regulated by various environmental signals such as plant growth, development, light response, and stress responses. Besides, the expression of the FtMTP gene family was specific to different tissues or different developmental stages of plants, and the expression of most FtMTP genes could be induced by specific metal ions. These findings will provide basic and useful information for future research on the metal tolerance mechanisms and the genetic improvement of Fagopyrum tataricum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11070850/s1, Figure S1: The detailed information about ten motifs of FtMTP proteins. Table S1: Protein sequences of 55 MTPs used in this study, Table S2: Sequences of FtMTPs, Table S3: Optimal forward and reverse primers for qRT-PCR analysis.
Author Contributions
Conceptualization, Z.L., C.W., J.S., M.Z., and X.Z.; methodology, Z.L., H.C., and C.W.; investigation, Z.L., C.W., J.Z., and K.W.; writing—original draft preparation, Z.L.; writing—review and editing, C.W., J.S., H.C., M.Z., and X.Z.; supervision, M.Z. and X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by National Key R&D Program of China (2019YFD1000702-6, 2018YFD1000706-3).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wyszkowski, M.; Brodowska, M.S. Content of Trace Elements in Soil Fertilized with Potassium and Nitrogen. Agriculture 2020, 10, 398. [Google Scholar] [CrossRef]
- Liu, J.; Gao, Y.; Tang, Y.; Wang, D.; Chen, X.; Yao, Y.; Guo, Y. Genome-Wide Identification, Comprehensive Gene Feature, Evolution, and Expression Analysis of Plant Metal Tolerance Proteins in Tobacco Under Heavy Metal Toxicity. Front. Genet. 2019, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Eid, E.M.; Alrumman, S.A.; Galal, T.M.; El-Bebany, A.F. Regression models for monitoring trace metal accumulations by Faba sativa Bernh. plants grown in soils amended with different rates of sewage sludge. Sci. Rep. 2019, 9, 5443. [Google Scholar] [CrossRef] [PubMed]
- Nishida, S.; Tanikawa, R.; Ishida, S.; Yoshida, J.; Mizuno, T.; Nakanishi, H.; Furuta, N. Elevated Expression of Vacuolar Nickel Transporter Gene IREG2 Is Associated With Reduced Root-to-Shoot Nickel Translocation in Noccaea japonica. Front. Plant Sci. 2020, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Sabra, M.; Aboulnasr, A.; Franken, P.; Perreca, E.; Wright, L.P.; Camehl, I. Beneficial Root Endophytic Fungi Increase Growth and Quality Parameters of Sweet Basil in Heavy Metal Contaminated Soil. Front. Plant Sci. 2018, 9, 1726. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-J.; Zaidul, I.S.M.; Suzuki, T.; Mukasa, Y.; Hashimoto, N.; Takigawa, S.; Noda, T.; Matsuura-Endo, C.; Yamauchi, H. Comparison of phenolic compositions between common and tartary buckwheat (Fagopyrum) sprouts. Food Chem. 2008, 110, 814–820. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Sun, W.; Wang, T. The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis. Int. J. Mol. Sci. 2019, 20, 2424. [Google Scholar] [CrossRef] [Green Version]
- Nomura, T.; Itouga, M.; Kojima, M.; Kato, Y.; Sakakibara, H.; Hasezawa, S. Copper mediates auxin signalling to control cell differentiation in the copper moss Scopelophila cataractae. J. Exp. Bot. 2015, 66, 1205–1213. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.; Zhou, X.; Ma, Y.; Xu, E.; Yu, Y.; Liu, Y.; Chen, X.; Zhang, W. Expression of a Brassica napus metal transport protein (BnMTP3) in Arabidopsis thaliana confers tolerance to Zn and Mn. Plant Sci. 2021, 304, 110754. [Google Scholar] [CrossRef]
- Barabasz, A.; Klimecka, M.; Kendziorek, M.; Weremczuk, A.; Ruszczynska, A.; Bulska, E.; Antosiewicz, D.M. The ratio of Zn to Cd supply as a determinant of metal-homeostasis gene expression in tobacco and its modulation by overexpressing the metal exporter AtHMA4. J. Exp. Bot. 2016, 67, 6201–6214. [Google Scholar] [CrossRef] [Green Version]
- Tsunemitsu, Y.; Genga, M.; Okada, T.; Yamaji, N.; Ma, J.F.; Miyazaki, A.; Kato, S.I.; Iwasaki, K.; Ueno, D. A member of cation diffusion facilitator family, MTP11, is required for manganese tolerance and high fertility in rice. Planta 2018, 248, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Eroglu, S.; Giehl, R.F.H.; Meier, B.; Takahashi, M.; Terada, Y.; Ignatyev, K.; Andresen, E.; Kupper, H.; Peiter, E.; von Wiren, N. Metal Tolerance Protein 8 Mediates Manganese Homeostasis and Iron Reallocation during Seed Development and Germination. Plant Physiol. 2017, 174, 1633–1647. [Google Scholar] [CrossRef] [Green Version]
- Ueno, D.; Sasaki, A.; Yamaji, N.; Miyaji, T.; Fujii, Y.; Takemoto, Y.; Moriyama, S.; Che, J.; Moriyama, Y.; Iwasaki, K.; et al. A polarly localized transporter for efficient manganese uptake in rice. Nat. Plants 2015, 1, 15170. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Gu, D.; Li, Y.; Li, J.; Li, J.; Chen, X.; Zhang, W. Phylogenetic and Expression Analysis of Mn-CDF Transporters in Pear (Pyrus bretschneideri Rehd.). Plant Mol. Biol. Rep. 2019, 37, 98–110. [Google Scholar] [CrossRef]
- Li, J.; Zheng, L.; Fan, Y.; Wang, Y.; Ma, Y.; Gu, D.; Lu, Y.; Zhang, S.; Chen, X.; Zhang, W. Pear metal transport protein PbMTP8.1 confers manganese tolerance when expressed in yeast and Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2021, 208, 111687. [Google Scholar] [CrossRef] [PubMed]
- Das, U.; Haque, A.F.M.M.; Bari, M.A.; Mandal, A.; Kabir, A.H. Computational characterization and expression profile of MTP1 gene associated with zinc homeostasis across dicot plant species. Gene Rep. 2021, 23, 101073. [Google Scholar] [CrossRef]
- Arrivault, S.; Senger, T.; Krämer, U. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 2006, 46, 861–879. [Google Scholar] [CrossRef]
- Kawachi, M.; Kobae, Y.; Mimura, T.; Maeshima, M. Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn2+/H+ antiporter of Arabidopsis thaliana, stimulates the transport activity. J. Biol. Chem. 2008, 283, 8374–8383. [Google Scholar] [CrossRef] [Green Version]
- Dräger, D.B.; Desbrosses-Fonrouge, A.G.; Krach, C.; Chardonnens, A.N.; Meyer, R.C.; Saumitou-Laprade, P.; Krämer, U. Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J. 2004, 39, 425–439. [Google Scholar] [CrossRef]
- Kobae, Y.; Uemura, T.; Sato, M.H.; Ohnishi, M.; Mimura, T.; Nakagawa, T.; Maeshima, M. Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 2004, 45, 1749–1758. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, T.; Kawachi, M.; Sato, Y.; Mori, H.; Kutsuna, N.; Hasezawa, S.; Maeshima, M. A high molecular mass zinc transporter MTP 12 forms a functional heteromeric complex with MTP 5 in the Golgi in Arabidopsis thaliana. FEBS J. 2015, 282, 1965–1979. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fujii, Y.; Yamaji, N.; Masuda, S.; Takemoto, Y.; Kamiya, T.; Yusuyin, Y.; Iwasaki, K.; Kato, S.-i.; Maeshima, M. Mn tolerance in rice is mediated by MTP8. 1, a member of the cation diffusion facilitator family. J. Exp. Bot. 2013, 64, 4375–4387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takemoto, Y.; Tsunemitsu, Y.; Fujii-Kashino, M.; Mitani-Ueno, N.; Yamaji, N.; Ma, J.F.; Kato, S.-I.; Iwasaki, K.; Ueno, D. The tonoplast-localized transporter MTP8. 2 contributes to manganese detoxification in the shoots and roots of Oryza sativa L. Plant Cell Physiol. 2017, 58, 1573–1582. [Google Scholar] [CrossRef]
- Eroglu, S.; Meier, B.; von Wirén, N.; Peiter, E. The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in Arabidopsis. Plant Physiol. 2016, 170, 1030–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricachenevsky, F.K.; Menguer, P.K.; Sperotto, R.A.; Williams, L.E.; Fett, J.P. Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front. Plant Sci. 2013, 4, 144. [Google Scholar] [CrossRef] [Green Version]
- Shirazi, Z.; Abedi, A.; Kordrostami, M.; Burritt, D.J.; Hossain, M.A. Genome-wide identification and characterization of the metal tolerance protein (MTP) family in grape (Vitis vinifera L.). 3 Biotech 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Q.; Xu, W.; Zhao, H.; Guo, F.; Wang, P.; Wang, Y.; Ni, D.; Wang, M.; Wei, C. Identification of MTP gene family in tea plant (Camellia sinensis L.) and characterization of CsMTP8. 2 in manganese toxicity. Ecotoxicol. Environ. Saf. 2020, 202, 110904. [Google Scholar] [CrossRef]
- Bonafaccia, G.; Marocchini, M.; Kreft, I. Composition and technological properties of the flour and bran from common and tartary buckwheat. Food Chem. 2003, 80, 9–15. [Google Scholar] [CrossRef]
- Peng, L.X.; Zou, L.; Wang, J.B.; Zhao, J.L.; Xiang, D.B.; Zhao, G. Flavonoids, Antioxidant Activity and Aroma Compounds Analysis from Different Kinds of Tartary Buckwheat Tea. Indian J. Pharm. Sci. 2015, 77, 661–667. [Google Scholar]
- Pirzadah, T.B.; Malik, B.; Tahir, I.; Irfan, Q.M.; Rehman, R.U. Characterization of mercury-induced stress biomarkers in Fagopyrum tataricum plants. Int. J. Phytoremediation 2018, 20, 225–236. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, Q.-f.; Li, J.; Xiong, J.; Zhou, L.-n.; He, S.-l.; Zhang, J.-q.; Chen, Z.-a.; He, S.-g.; Liu, H. Effects of exogenous sulfur on alleviating cadmium stress in tartary buckwheat. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirzadah, T.B.; Malik, B.; Tahir, I.; Hakeem, K.R.; Alharby, H.F.; Rehman, R.U. Lead toxicity alters the antioxidant defense machinery and modulate the biomarkers in Tartary buckwheat plants. Int. Biodeterior. Biodegrad. 2020, 151, 104992. [Google Scholar] [CrossRef]
- Wang, L.; Zheng, B.; Yuan, Y.; Xu, Q.; Chen, P. Transcriptome profiling of Fagopyrum tataricum leaves in response to lead stress. BMC Plant Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Z.; Huang, Y.; Jiang, Y.; Liu, Y.; Wen, W.; Li, H.; Shao, J.; Wang, C.; Zhu, X. Antioxidant capacity, metal contents, and their health risk assessment of tartary buckwheat teas. ACS Omega 2020, 5, 9724–9732. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, F.; Liu, J.; Xie, W.; Zhang, L.; Chen, Z.; Peng, Z.; Ou, Y.; Yao, Y. Genome-Wide Identification of Metal Tolerance Protein Genes in Populus trichocarpa and Their Roles in Response to Various Heavy Metal Stresses. Int. J. Mol. Sci. 2020, 21, 1680. [Google Scholar] [CrossRef] [Green Version]
- Shingu, Y.; Kudo, T.; Ohsato, S.; Kimura, M.; Ono, Y.; Yamaguchi, I.; Hamamoto, H. Characterization of genes encoding metal tolerance proteins isolated from Nicotiana glauca and Nicotiana tabacum. Biochem. Biophys. Res. Commun. 2005, 331, 675–680. [Google Scholar] [CrossRef]
- Fu, X.-Z.; Tong, Y.-H.; Zhou, X.; Ling, L.-L.; Chun, C.-P.; Cao, L.; Zeng, M.; Peng, L.-Z. Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene 2017, 629, 1–8. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 113. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Sun, W.; Ma, Z.; Zheng, T.; Huang, L.; Wu, Q.; Zhao, G.; Tang, Z.; Bu, T.; Li, C. Genome-wide investigation of the AP2/ERF gene family in tartary buckwheat (Fagopyum Tataricum). BMC Plant Biol. 2019, 19, 84. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Huang, L.; Ma, Z.; Sun, W.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, expression analysis and functional study of the GRAS gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 2019, 19, 342. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Ma, Z.; Wang, A.; Zheng, T.; Huang, L.; Sun, W.; Zhang, Y.; Jin, W.; Zhan, J.; Cai, Y. Genome-wide investigation of the auxin response factor gene family in Tartary buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2018, 19, 3526. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Wen, Y.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartary buckwheat. BMC Genom. 2019, 20, 483. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Jin, X.; Ma, Z.; Chen, H.; Liu, M. Basic helix–loop–helix (bHLH) gene family in Tartary buckwheat (Fagopyrum tataricum): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses. Int. J. Biol. Macromol. 2020, 155, 1478–1490. [Google Scholar] [CrossRef]
- Liu, M.; Fu, Q.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide investigation of the MADS gene family and dehulling genes in tartary buckwheat (Fagopyrum tataricum). Planta 2019, 249, 1301–1318. [Google Scholar] [CrossRef]
- Liu, M.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification of the SPL gene family in Tartary Buckwheat (Fagopyrum tataricum) and expression analysis during fruit development stages. BMC Plant Biol. 2019, 19, 299. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Li, H.; Huang, J.; Shi, T.; Meng, Z.; Chen, Q.; Deng, J. Genome-wide analysis of BBX gene family in Tartary buckwheat (Fagopyrum tataricum). PeerJ 2021, 9, e11939. [Google Scholar] [CrossRef]
- Yan, H.; Liu, C.; Zhao, J.; Ye, X.; Wu, Q.; Yao, T.; Peng, L.; Zou, L.; Zhao, G. Genome-wide analysis of the NF-Y gene family and their roles in relation to fruit development in Tartary buckwheat (Fagopyrum tataricum). Int. J. Biol. Macromol. 2021, 190, 487–498. [Google Scholar] [CrossRef]
- Desbrosses-Fonrouge, A.-G.; Voigt, K.; Schröder, A.; Arrivault, S.; Thomine, S.; Krämer, U. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 2005, 579, 4165–4174. [Google Scholar] [CrossRef]
- Blaudez, D.; Kohler, A.; Martin, F.; Sanders, D.; Chalot, M. Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 2003, 15, 2911–2928. [Google Scholar] [CrossRef] [Green Version]
- Peiter, E.; Montanini, B.; Gobert, A.; Pedas, P.; Husted, S.; Maathuis, F.J.; Blaudez, D.; Chalot, M.; Sanders, D. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 8532–8537. [Google Scholar] [CrossRef] [Green Version]
- Pedas, P.; Schiller Stokholm, M.; Hegelund, J.N.; Ladegård, A.H.; Schjoerring, J.K.; Husted, S. Golgi localized barley MTP8 proteins facilitate Mn transport. PLoS ONE 2014, 9, e113759. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Gruber, B.D.; Pittman, J.K.; White, R.G.; Leung, H.; Miao, Y.; Jiang, L.; Ryan, P.R.; Richardson, A.E. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 2007, 51, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Vatansever, R.; Filiz, E.; Eroglu, S. Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): Insights into metal homeostasis and biofortification. Biometals 2017, 30, 217–235. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. Proteom. Protoc. Handb. 2005, 571–607. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Large-scale plant protein subcellular location prediction. J. Cell. Biochem. 2007, 100, 665–678. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, K.-C. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 2005, 21, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 7, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, W.; Ma, Z.; Hu, Y.; Chen, H. Tartary buckwheat database (TBD): An integrative platform for gene analysis of and biological information on Tartary buckwheat. J. Zhejiang Univ.-Sci. B 2021, 22, 954–958. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).