Phenotype and Genotype Interaction Underlying Distributive Characteristic for Awn Development in Rice
Abstract
:1. Introduction
2. Results
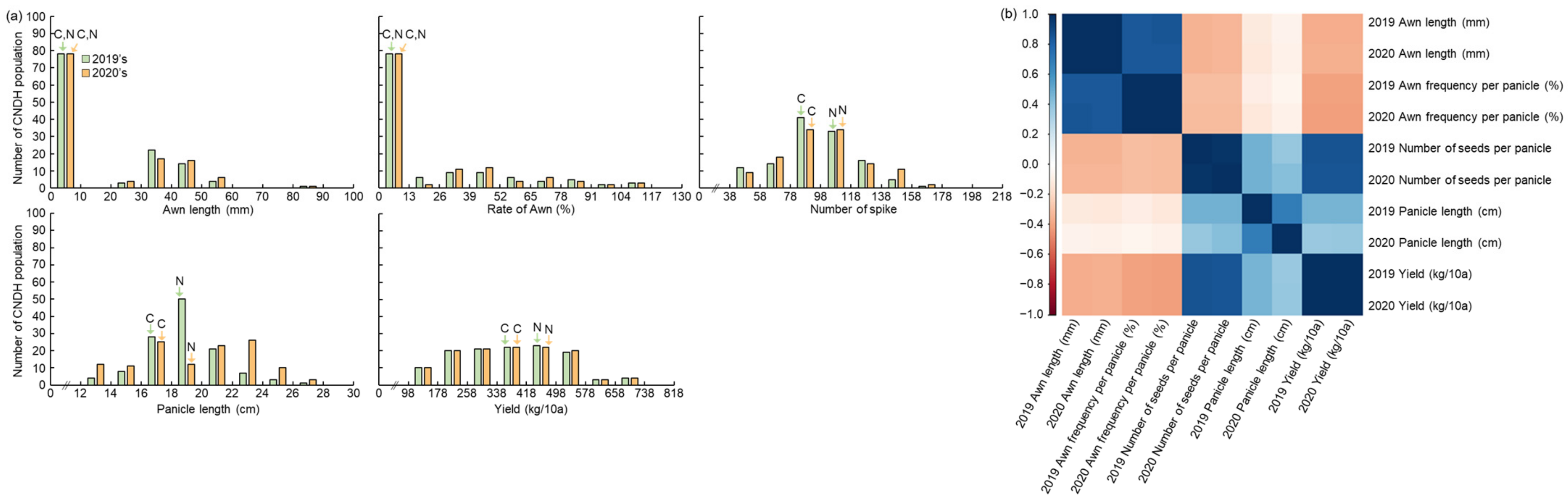
2.1. Phenotype Characterization Associated with Awn Elongation in 120 Cheongcheong/Nagdong Double Haploid (CNDH) Population
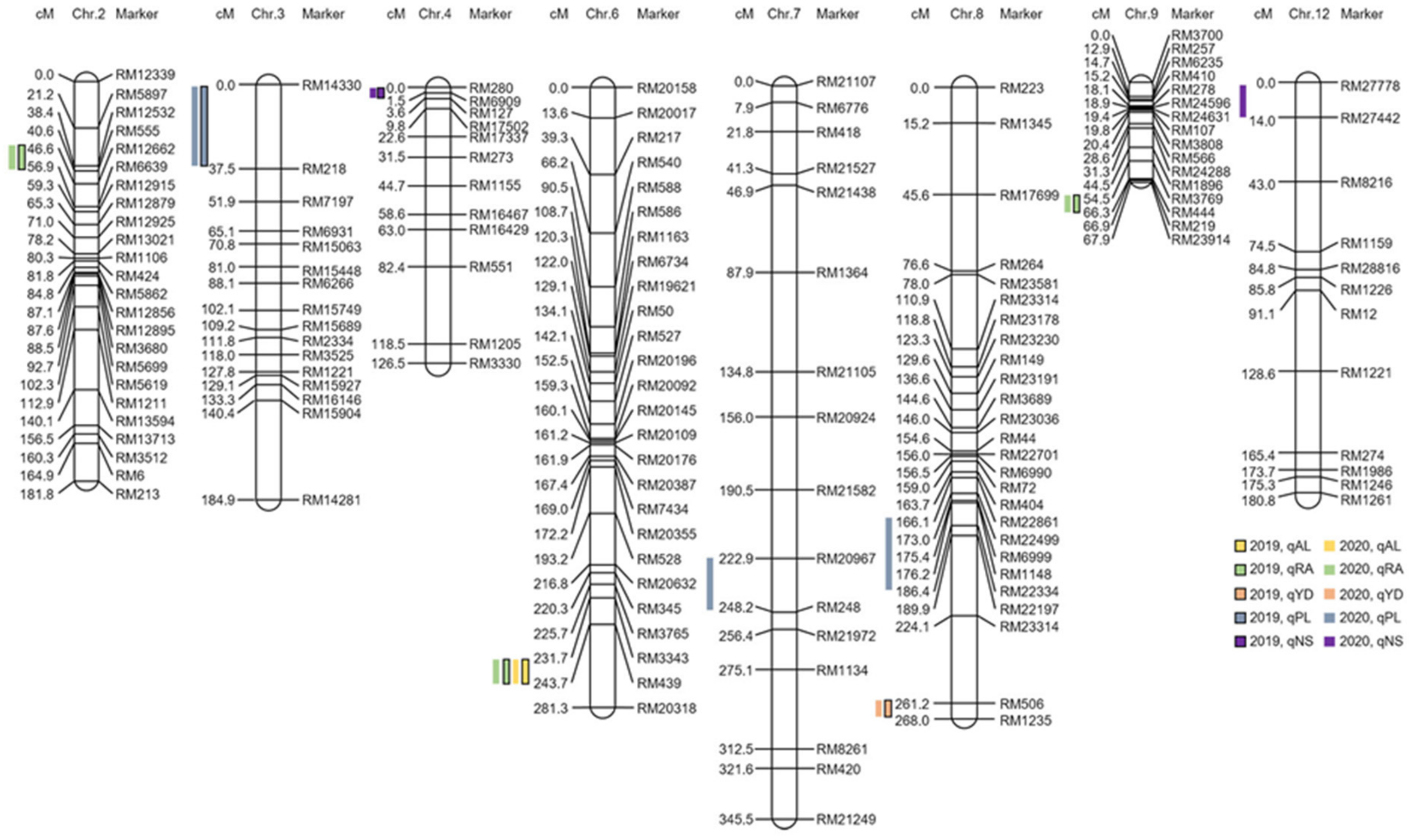
2.2. Awn Elongation and Yield Based on QTL Mapping and Physical Map Construct
2.3. Search for Associate Genes Associated with Awn Development Based on QTL Mapping
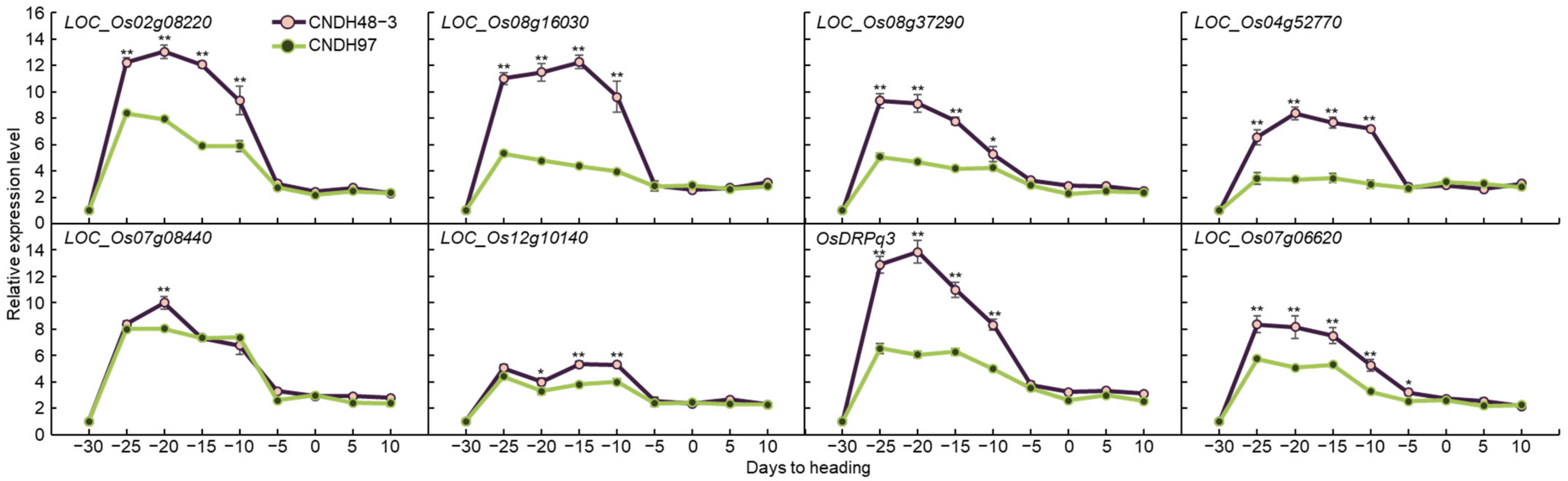
2.4. Associate Gene Expression Level at the Panicle Formation Stage
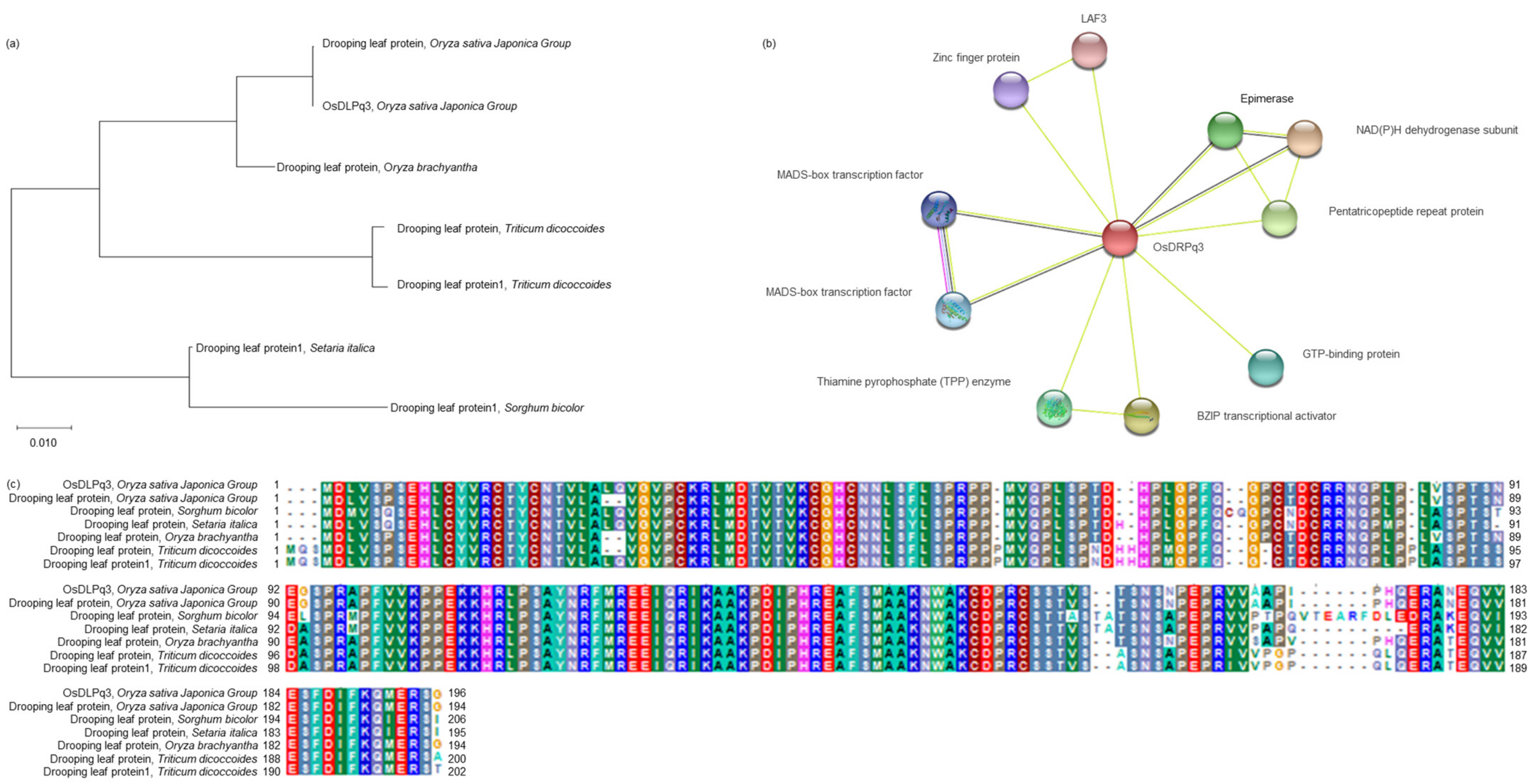
2.5. Analysis of the Phylogenetic Tree and Homology Sequence
3. Discussion
4. Materials and Methods
4.1. Plant Material and Field Design
4.2. Measurement of Awn Related Phenotype and Yield
4.3. Construction of Genetic Map and QTL Analysis Associated with Awn Elongation in Rice
4.4. Screening and Annotation of Associate Genes
4.5. Genomic DNA Extraction and PCR Protocol
4.6. RNA Extraction
4.7. Analysis of the Associate Gene Expression Level
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.T.; Marciano, A.P.; Loresto, G.C. Morpho-agronomic variousness and economic potentials of Oryza glaberrima and wild species in the genus Oryza. In Proceedings of the Meeting on African Rice Species, ORSTOM-IRAT, Paris, France, 25–26 January 1977; pp. 67–75. [Google Scholar]
- Kovach, M.J.; Sweeney, M.T.; McCouch, S.R. New insights into the history of rice domestication. Trends Genet. 2007, 23, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Grundbacher, F.J. THE physiological function of the cereal awn. Bot. Rev. 1963, 29, 366–381. [Google Scholar] [CrossRef]
- Elbaum, R.; Zaltzman, L.; Burgert, I.; Fratzl, P. The role of wheat awns in the seed dispersal unit. Science 2007, 316, 884–886. [Google Scholar] [CrossRef]
- Amarasinghe, Y.P.J.; Kuwata, R.; Nishimura, A.; Phan, P.D.T.; Ishikawa, R.; Ishii, T. Evaluation of Domestication Loci Associated with Awnlessness in Cultivated Rice, Oryza sativa. Rice 2020, 13, 26. [Google Scholar] [CrossRef]
- Shuaib, M.; Bahadur, S.; Hussain, F. Enumeration of genetic diversity of wild rice through phenotypic trait analysis. Gene Rep. 2020, 21, 100797. [Google Scholar] [CrossRef]
- Abebe, T.; Melmaiee, K.; Berg, V.; Wise, R.P. Drought response in the spikes of barley: Gene expression in the lemma, palea, awn, and seed. Funct. Integr. Genom. 2010, 10, 191–205. [Google Scholar] [CrossRef]
- Toriba, T.; Suzaki, T.; Yamaguchi, T.; Ohmori, Y.; Tsukaya, H.; Hirano, H.Y. Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 2010, 22, 1452–1462. [Google Scholar] [CrossRef] [Green Version]
- Tsudamori, M. Seishoku-kikan to shiteno ine noge no kachi (Values of the rice awn as a reproductive organ). Nissakuki 1933, 5, 380–390. [Google Scholar]
- Magwa, R.A.; Zhao, H.; Yao, W.; Xie, W.; Yang, L.; Xing, Y.; Bai, X. Genomewide association analysis for awn length linked to the seed shattering gene qSH1 in rice. J. Genet. 2016, 95, 639–646. [Google Scholar] [CrossRef]
- Liller, C.B.; Walla, A.; Boer, M.P.; Hedley, P.; Macaulay, M.; Effgen, S.; von Korff, M.; van Esse, G.W.; Koornneef, M. Fine mapping of a major QTL for awn length in barley using a multiparent mapping population. Theor. Appl. Genet. 2017, 130, 269–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Q.; Chen, Y.; Fu, L.; Zhou, S.; Chen, J.; Zhao, X.; Zhang, D.; Ouyang, S.; Wang, Z.; Li, D.; et al. QTL mapping of flag leaf traits in common wheat using an integrated high-density SSR and SNP genetic linkage map. Euphytica 2016, 208, 337–351. [Google Scholar] [CrossRef]
- Xiong, L.Z.; Liu, K.D.; Dai, X.K.; Xu, C.G.; Zhang, Q. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 1999, 98, 243–251. [Google Scholar] [CrossRef]
- Bai, X.; Luo, L.; Yan, W.; Kovi, M.R.; Zhan, W.; Xing, Y. Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 2010, 11, 16. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Liu, H.; Zhou, T.; Gu, B.; Huang, X.; Shangguan, Y.; Zhu, J.; Li, Y.; Zhao, Y.; Wang, Y.; et al. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 2013, 25, 3360–3376. [Google Scholar] [CrossRef] [Green Version]
- Hua, L.; Wang, D.R.; Tan, L.; Fu, Y.; Liu, F.; Xiao, L.; Zhu, Z.; Fu, Q.; Sun, X.; Gu, P.; et al. LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 2015, 27, 1875–1888. [Google Scholar] [CrossRef] [Green Version]
- Bessho-Uehara, K.; Wang, D.R.; Furuta, T.; Minami, A.; Nagai, K.; Gamuyao, R.; Asano, K.; Angeles-Shim, R.B.; Shimizu, Y.; Ayano, M.; et al. Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc. Natl. Acad. Sci. USA 2016, 113, 8969–8974. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Singh, N.; Mishra, S.; Tripathi, K.; Singh, B.P.; Rai, V.; Singh, A.K.; Singh, N.K. Morphological and molecular data reveal three distinct populations of indian wild rice Oryza rufipogon griff. Species complex. Front. Plant Sci. 2018, 9, 123. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, Z.; Sun, X.; Zhu, X.; Li, B.; Li, J.; Guo, H.; Chen, C.; Pan, Y.; Liang, Y.; et al. Natural alleles of GLA for grain length and awn development were differently domesticated in rice subspecies japonica and indica. Plant Biotechnol. J. 2019, 17, 1547–1559. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.-j.; Du, C.-q.; Dai, B.-c.; Chen, D.-l.; Chen, J.-g.; Cai, D.-t. Studies on the Growth Habits and Characteristics of Two Polyploid Indica-Japonica Hybrid Rice with Powerful Heterosis. Agric. Sci. China 2007, 6, 265–274. [Google Scholar] [CrossRef]
- Svizzero, S.; Ray, A.; Chakraborty, D. Awn Reduction and the Domestication of Asian Rice: A Syndrome or Crop Improvement Trait? Econ. Bot. 2019, 73, 477–488. [Google Scholar] [CrossRef]
- Xu, S. Mapping quantitative trait loci by controlling polygenic background effects. Genetics 2013, 195, 1209–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, L.; Bruce, W. Genetics and Analysis of Quantitative Traits; Sinauer: Sunderland, MA, USA, 1998; Volume 1, pp. 535–557. [Google Scholar]
- Wang, T.; Zou, T.; He, Z.; Yuan, G.; Luo, T.; Zhu, J.; Liang, Y.; Deng, Q.; Wang, S.; Zheng, A.; et al. GRAIN LENGTH AND AWN 1 negatively regulates grain size in rice. J. Integr. Plant Biol. 2019, 61, 1036–1042. [Google Scholar] [CrossRef]
- Sang, T.; Ge, S. Understanding rice domestication and implications for cultivar improvement. Curr. Opin. Plant Biol. 2013, 16, 139–146. [Google Scholar] [CrossRef]
- Ikeda, M.; Miura, K.; Aya, K.; Kitano, H.; Matsuoka, M. Genes offering the potential for designing yield-related traits in rice. Curr. Opin. Plant Biol. 2013, 16, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Toriba, T.; Hirano, H.Y. The DROOPING LEAF and OsETTIN2 genes promote awn development in rice. Plant J. 2014, 77, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Hrtyan, M.; Šliková, E.; Hejátko, J.; Růžička, K. RNA processing in auxin and cytokinin pathways. J. Exp. Bot. 2015, 66, 4897–4912. [Google Scholar] [CrossRef] [Green Version]
- Gu, B.; Zhou, T.; Luo, J.; Liu, H.; Wang, Y.; Shangguan, Y.; Zhu, J.; Li, Y.; Sang, T.; Wang, Z.; et al. An-2 Encodes a Cytokinin Synthesis Enzyme that Regulates Awn Length and Grain Production in Rice. Mol. Plant 2015, 8, 1635–1650. [Google Scholar] [CrossRef]
- Takatsuji, H. Zinc-finger proteins: The classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 1999, 39, 1073–1078. [Google Scholar] [CrossRef]
- Yu, C.; Liu, Y.; Zhang, A.; Su, S.; Yan, A.; Huang, L.; Ali, I.; Liu, Y.; Forde, B.G.; Gan, Y. MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PLoS ONE 2015, 10, e0135196. [Google Scholar] [CrossRef]
- Takahisa, Y.; Yuko, Y.; Hiroyuki, K.; Takashi, M.; Udda, L.; Kazuhiro, S.; Masahiki, I.; Jobling, S.A.; Shin, T. A SHORT INTERNODES (SHI) family transcription factor gene regulates awn elongation and pistil morphology in barley. J. Exp. Bot. 2012, 63, 5223–5232. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, T.; Nagasawa, N.; Kawasaki, S.; Matsuoka, M.; Nagato, Y.; Hirano, H.Y. The Yabby Gene Drooping Leaf Regulates Carpel Specification and Midrib Development in Oryza sativa. Plant Cell 2004, 16, 500–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masoudi, B.; Mardi, M.; Hervan, E.M.; Bihamta, M.R.; Naghavi, M.R.; Nakhoda, B.; Bakhshi, B.; Ahmadi, M.; Tabatabaei, M.T.; Firouzabadi, M.H.D. Study of QTLs linked to awn length and their relationships with chloroplasts under control and saline environments in bread wheat. Genes Genom. 2019, 41, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Assmann, S.M. Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 2002, 14, 355–374. [Google Scholar] [CrossRef]
- Zhang, C.; Jian, J.; Feng, J.; Cui, Z.; Wu, X.; Sun, D. QTL identification for awn length based on 90K array mapping in wheat. Sci. Agric. Sin. 2018, 15, 17–25. [Google Scholar]
- Xiao, J.; Li, J.; Yuan, L.; Tanksley, S.D. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor. Appl. Genet. 1996, 92, 230–244. [Google Scholar] [CrossRef]
- Lee, G.H.; Yun, B.W.; Kim, K.M. Analysis of QTLs associated with the rice quality related gene by double haploid populations. Int. J. Genom. 2014, 2014, 781832. [Google Scholar] [CrossRef] [Green Version]
- McCouch, S.R. Gene nomenclature system for rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef] [Green Version]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.C.; Iwamoto, M.; Abe, T.; et al. Rice annotation project database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro Version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, 1206–1213. [Google Scholar] [CrossRef] [Green Version]
- Sugiura, N.; Tang, D.; Kurokochi, H.; Saito, Y.; Ide, Y. Genetic structure of Quercus gilva Blume in Japan as revealed by chloroplast DNA sequences. Botany 2015, 93, 873–880. [Google Scholar] [CrossRef]
- Letunic, I.; Copley, R.R.; Pils, B.; Pinkert, S.; Schultz, J.; Bork, P. SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 2006, 34, 257–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.-R.; Son, J.H.; Kim, E.-G.; Jang, Y.-H.; Yun, B.-J.; Kim, K.-M. Phenotype and Genotype Interaction Underlying Distributive Characteristic for Awn Development in Rice. Plants 2022, 11, 851. https://doi.org/10.3390/plants11070851
Park J-R, Son JH, Kim E-G, Jang Y-H, Yun B-J, Kim K-M. Phenotype and Genotype Interaction Underlying Distributive Characteristic for Awn Development in Rice. Plants. 2022; 11(7):851. https://doi.org/10.3390/plants11070851
Chicago/Turabian StylePark, Jae-Ryoung, Ju Hyeong Son, Eun-Gyeong Kim, Yoon-Hee Jang, Byoung-Ju Yun, and Kyung-Min Kim. 2022. "Phenotype and Genotype Interaction Underlying Distributive Characteristic for Awn Development in Rice" Plants 11, no. 7: 851. https://doi.org/10.3390/plants11070851






