Bacteriophage Control of Pseudomonas savastanoi pv. glycinea in Soybean
Abstract
:1. Introduction
2. Results
2.1. Bacterial Strains
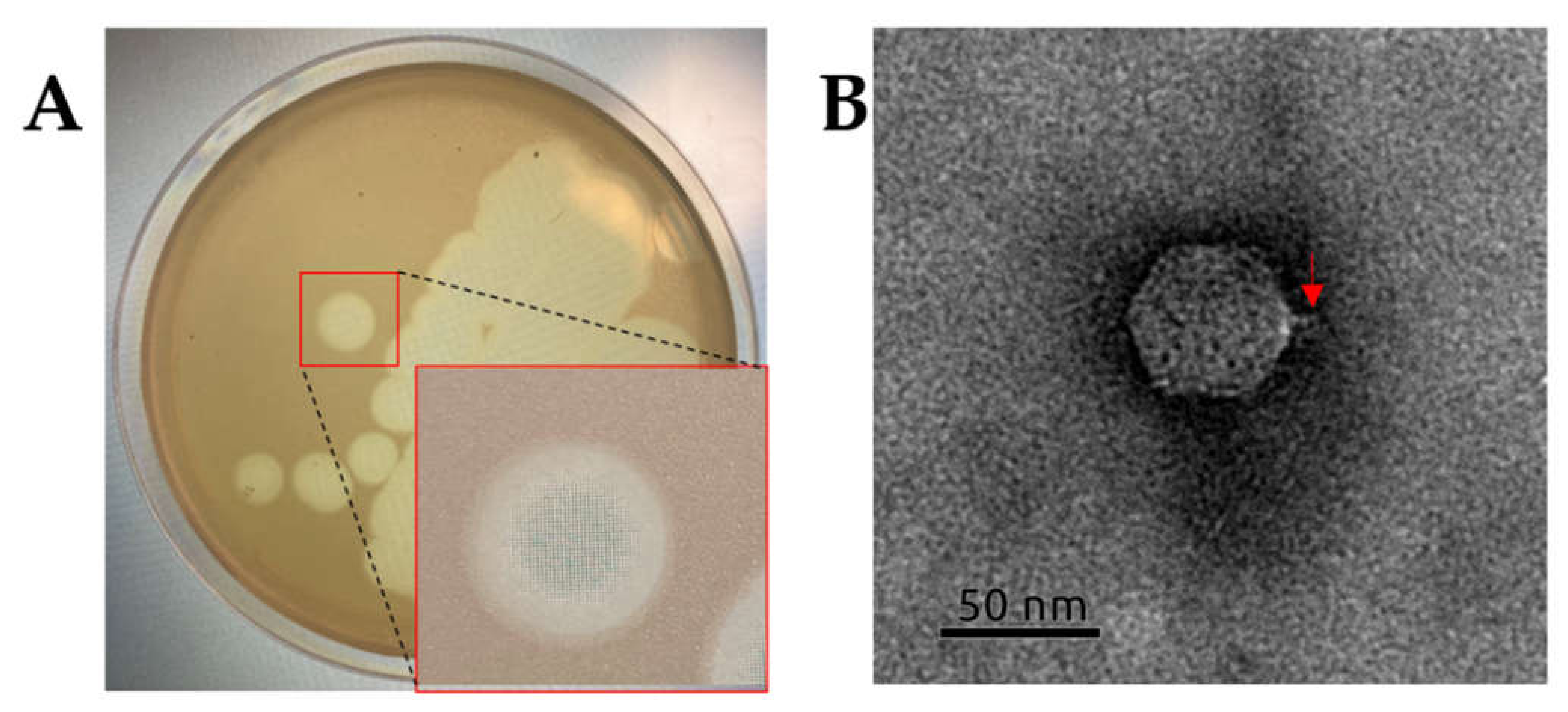
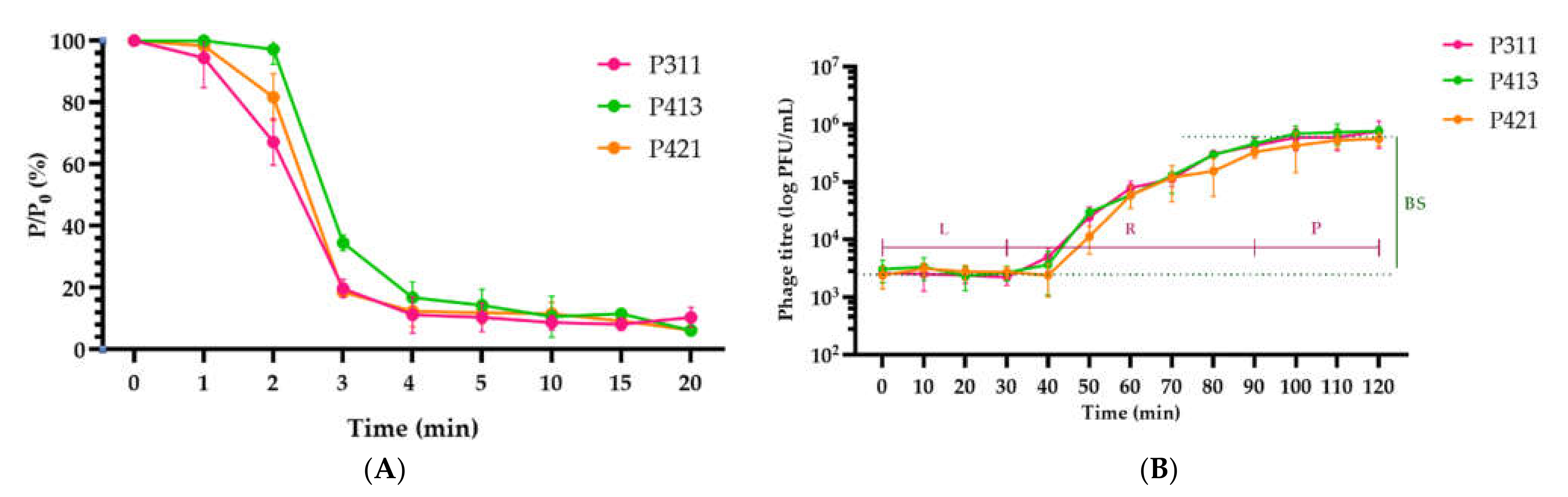
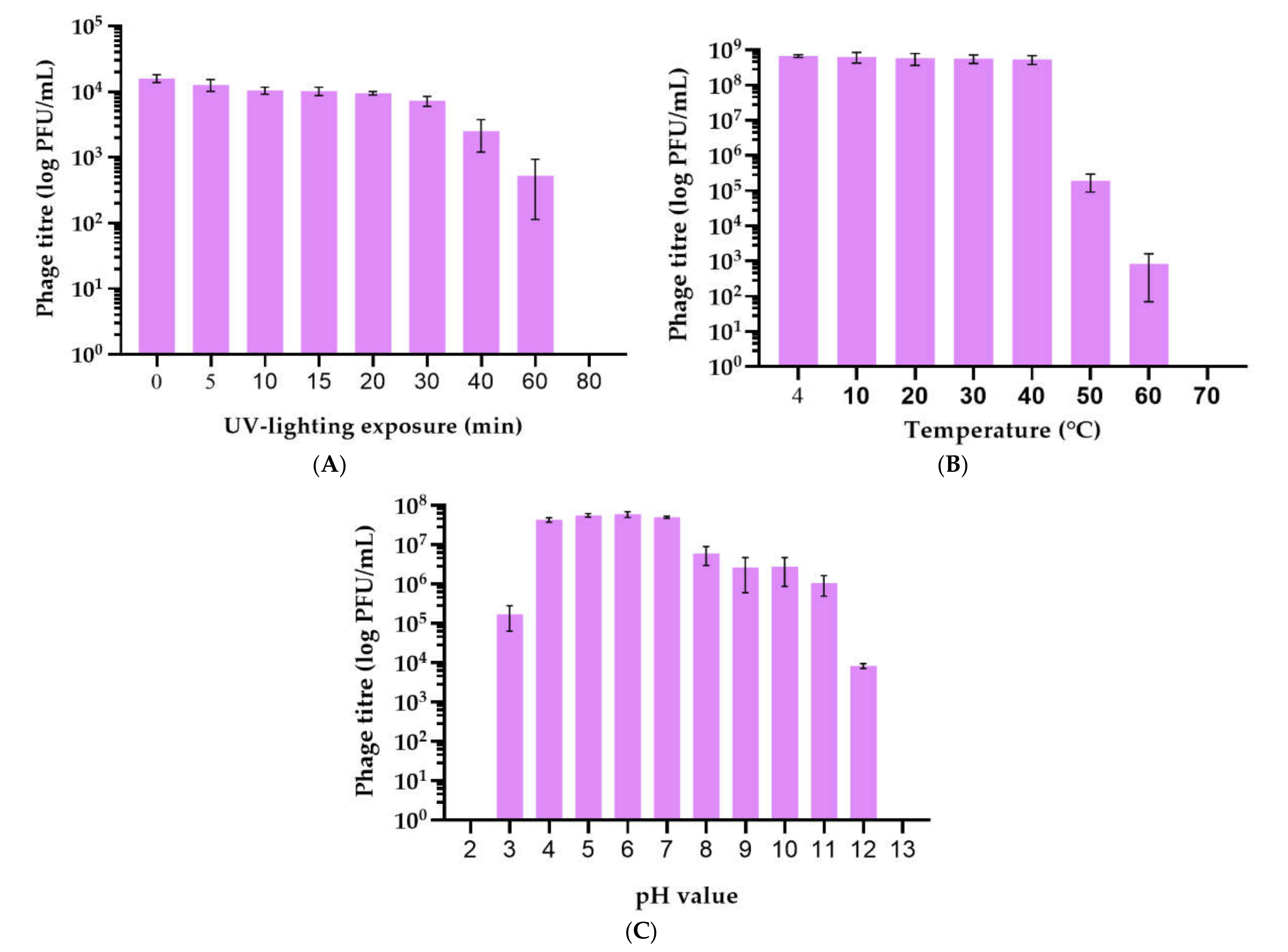
2.2. Isolation and General Properties of Pseudomonas savastanoi pv. glycinea Bacteriophages
2.3. Phage P421 Genomic and Phylogenetic Characterisation
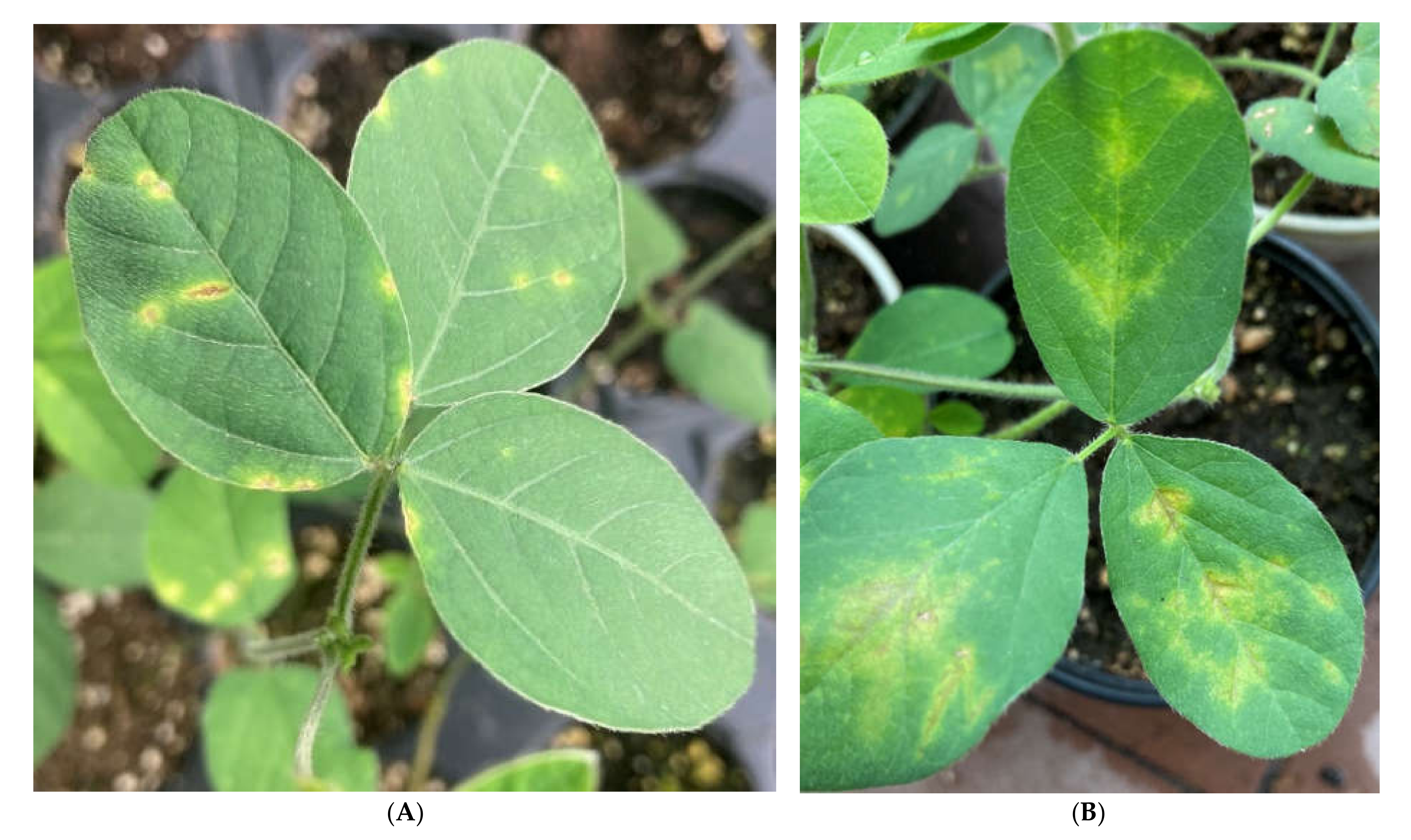
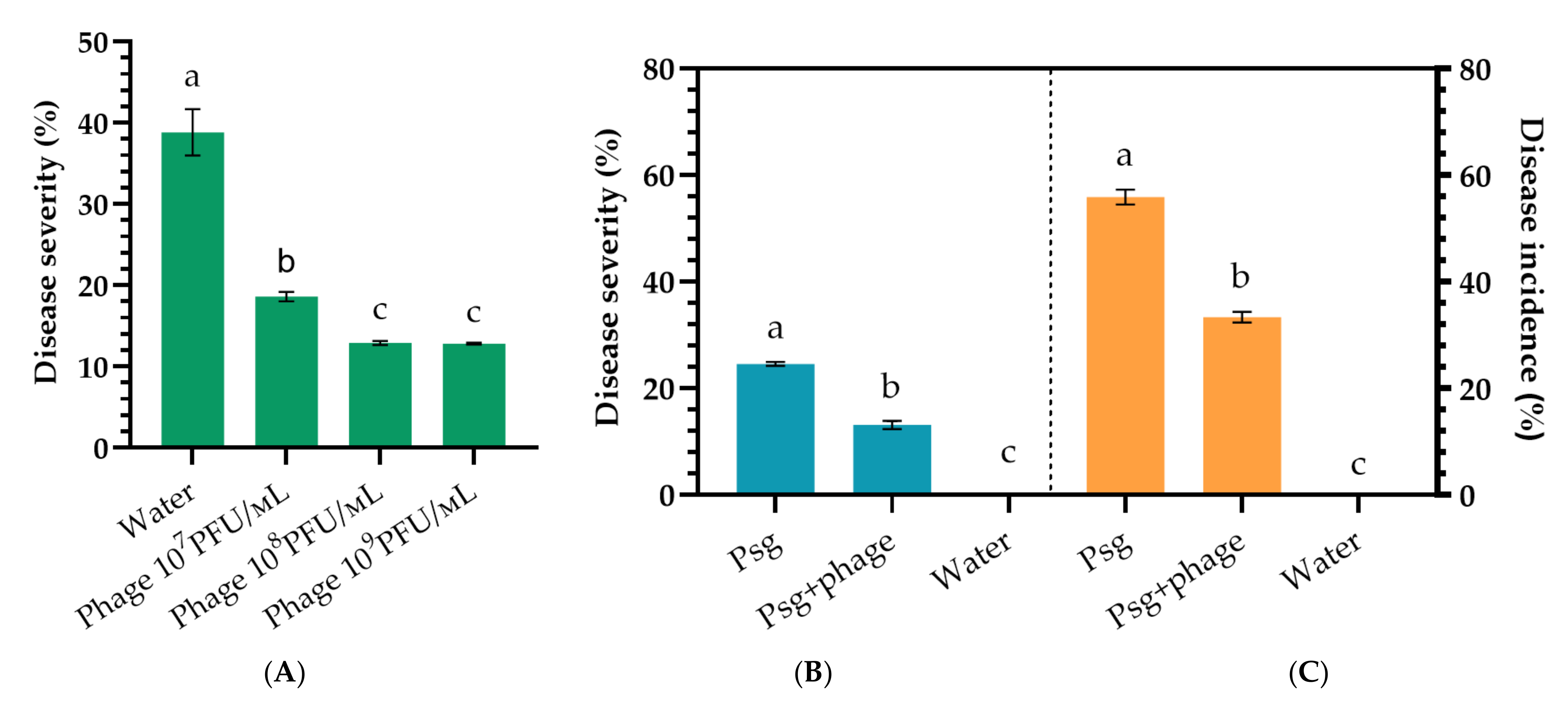
2.4. Phage Efficacy against Psg Leaf Infection
2.5. Phage Efficacy against Psg Seed Infection
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Bacteriophage Isolation, Propagation and Purification
4.3. Electron Microscopy
4.4. Determination of Phage Host Specificity
4.5. Phage Adsorption and One-Step Growth Experiments
4.6. Phage Stability under Different Conditions
4.7. Genome Sequencing and Annotation
4.8. Phylogeny and Taxonomy Studies
4.9. Phage Control of Bacterial Blight on Soybean
4.9.1. Artificial Latent Infection of Soybean Seeds
4.9.2. Phage Application on Seeds
4.9.3. Artificial Infection of Soybean Leaves
4.9.4. Phage Application on Leaves
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.; Wang, X.; Lu, Y.; Bhusal, S.J.; Song, Q.; Cregan, P.B.; Yen, Y.; Brown, M.; Jiang, G.L. Genome-Wide Scan for Seed Composition Provides Insights into Soybean Quality Improvement and the Impacts of Domestication and Breeding. Mol. Plant 2018, 11, 460–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagtap, G.P.; Dhopte, S.B.; Dey, U. Original Article Bio-Efficacy of Different Antibacterial Antibiotic, Plant Extracts and Bioagents against Bacterial Blight of Soybean Caused by Pseudomonas Syringae Pv. Glycinea. Sci. J. Microbiol. 2012, 1, 1–9. [Google Scholar]
- Young, J.M.; Saddler, G.S.; Takikawa, Y.; de Boer, S.H.; Vauterin, L.; Gardan, L.; Gvozdyak, R.I.; Stead, D.E. Names of Plant Pathogenic Bacteria 1864–1995. Rev. Plant Pathol. 1996, 75, 721–763. [Google Scholar]
- Prom, L.K.; Venette, J.R. Races of Pseudomonas Syringae Pv. Glycinea on Commercial Soybean in Eastern North Dakota. Plant Dis. 1997, 81, 541–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abo-Moch, F.; Mavridis, A.; Rudolph, K. Determination of Races of Pseudomonas Syringae Pv. Glycinea Occurring in Europe. J. Phytopathol. 1995, 143, 1–5. [Google Scholar] [CrossRef]
- Ignjatov, M.; Milošević, M.; Nikolić, Z.; Vujaković, M.; Petrović, D. Characterization of Pseudomonas Savastanoi Pv. Glycinea Isolates from Vojvodina. Phytopathol. Pol. 2007, 45, 43–54. [Google Scholar]
- Shepherd, L.M.; Block, C.C. Chapter 13: Detection of Pseudomonas savastanoi pv. glycinea in Soybean Seeds. In Detection of Plant-Pathogenic Bacteria in Seed and Other Planting Material, 2nd ed.; The American Phytopathological Society: St. Paul, MN, USA, 2017; ISBN 978-0-89054-539-3. [Google Scholar]
- Monteil, C.L.; Yahara, K.; Studholme, D.J.; Mageiros, L.; Méric, G.; Swingle, B.; Morris, C.E.; Vinatzer, B.A.; Sheppard, S.K. Population-Genomic Insights into Emergence, Crop Adaptation and Dissemination of Pseudomonas Syringae Pathogens. Microb. Genom. 2016, 2, e000089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giesler, L.J.; Weissling, T.J. Bacterial Diseases of Soybean; The Board of Regents of the University of Nebraska on behalf of the University of Nebraska–Lincoln Extension: Lincoln, NE, USA, 2011; p. G2058. [Google Scholar]
- Keen, N.T.; Buzzell, R.I. New Disease Resistance Genes in Soybean against Pseudomonas Syringae Pv Glycinea: Evidence That One of Them Interacts with a Bacterial Elicitor. Theor. Appl. Genet. 1991, 81, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Sotelo, J.P.; Oddino, C.; Giordano, D.F.; Carezzano, M.E.; Oliva, M.D.L.M. Effect of Thymus Vulgaris Essential Oil on Soybeans Seeds Infected with Pseudomonas Syringae. Physiol. Mol. Plant Pathol. 2021, 116, 101735. [Google Scholar] [CrossRef]
- Balogh, B.; Jones, J.B.; Iriarte, F.B.; Momol, M.T. Phage Therapy for Plant Disease Control. Curr. Pharm. Biotechnol. 2010, 11, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Vu, N.T.; Oh, C.S. Bacteriophage Usage for Bacterial Disease Management and Diagnosis in Plants. Plant Pathol. J. 2020, 36, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Lukianova, A.A.; Shneider, M.M.; Evseev, P.V.; Shpirt, A.M.; Bugaeva, E.N.; Kabanova, A.P.; Obraztsova, E.A.; Miroshnikov, K.K.; Senchenkova, S.N.; Shashkov, A.S.; et al. Morphologically Different Pectobacterium Brasiliense Bacteriophages PP99 and PP101: Deacetylation of O-Polysaccharide by the Tail Spike Protein of Phage PP99 Accompanies the Infection. Front. Microbiol. 2019, 10, 3147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Addy, H.S.; Wahyuni, W.S. Nucleic Acid and Protein Profile of Bacteriophages That Infect Pseudomonas Syringae Pv. Glycinea, Bacterial Blight on Soybean. Agric. Agric. Sci. Procedia 2016, 9, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Susianto, G.; Farid, M.M.; Dhany, N.R.; Addy, H.S. Host Range for Bacteriophages That Infect Bacterial Blight Pathogen on Soybean. Procedia Environ. Sci. 2014, 20, 760–766. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.D. Bacteriophages from Sewage Specific for Fluorescent Phytopathogenic Pseudomonads. Phytopathology 1983, 73, 403. [Google Scholar] [CrossRef]
- Bull, C.T.; Boer, S.H.D.; Denny, T.P.; Firrao, G.; Saux, M.F.-L.; Saddler, G.S.; Scortichini, M.; Stead, D.E.; Takikawa, Y. Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J. Plant Pathol. 2010, 92, 551–592. [Google Scholar] [CrossRef]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-Encoded Virion-Associated Enzymes to Overcome the Carbohydrate Barriers during the Infection Process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [Green Version]
- Pethybridge, S.J.; Nelson, S.C. Leaf Doctor: A New Portable Application for Quantifying Plant Disease Severity. Plant Dis. 2015, 99, 1310–1316. [Google Scholar] [CrossRef] [Green Version]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and Bacterial Plant Diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Svircev, A.; Roach, D.; Castle, A. Framing the Future with Bacteriophages in Agriculture. Viruses 2018, 10, 218. [Google Scholar] [CrossRef] [Green Version]
- Holtappels, D.; Fortuna, K.; Lavigne, R.; Wagemans, J. The Future of Phage Biocontrol in Integrated Plant Protection for Sustainable Crop Production. Curr. Opin. Biotechnol. 2021, 68, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.-C.; Moon, J.-K.; Park, S.-K.; Kim, M.-S.; Lee, K.; Lee, S.R.; Jeong, N.; Choi, M.S.; Kim, N.; Kang, S.-T.; et al. Genetic Diversity Patterns and Domestication Origin of Soybean. Theor. Appl. Genet. 2019, 132, 1179–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatain-Ly, M.H. The Factors Affecting Effectiveness of Treatment in Phages Therapy. Front. Microbiol. 2014, 5, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gašić, K.; Kuzmanović, N.; Ivanović, M.; Prokić, A.; Šević, M.; Obradović, A. Complete Genome of the Xanthomonas Euves catoria Specific Bacteriophage KΦ1, Its Survival and Potential in Control of Pepper Bacterial Spot. Front. Microbiol. 2018, 9, 2021. [Google Scholar] [CrossRef] [PubMed]
- Flores, O.; Retamales, J.; Núñez, M.; León, M.; Salinas, P.; Besoain, X.; Yañez, C.; Bastías, R. Characterization of Bacteriophages against Pseudomonas Syringae Pv. Actinidiae with Potential Use as Natural Antimicrobials in Kiwifruit Plants. Microorganisms 2020, 8, 974. [Google Scholar] [CrossRef]
- Quiñones-Aguilar, E.E.; Reyes-Tena, A.; Hernández-Montiel, L.G.; Rincón-Enríquez, G. Bacteriophages in the Biological Control of Pseudomonas Syringae Pv. Phaseolicola, Causal Agent of Halo Blight in Bean. Ecosist. Recur. Agropecu. 2018, 5, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Hassan, E.O. Biocontrol of Halo Blight of Bean Caused by Pseudomonas Phaseolicola. Int. J. Virol. 2014, 10, 235–242. [Google Scholar] [CrossRef]
- Pinheiro, L.A.M.; Pereira, C.; Frazão, C.; Balcão, V.M.; Almeida, A. Efficiency of Phage Φ6 for Biocontrol of Pseudomonas Syringae Pv. Syringae: An in Vitro Preliminary Study. Microorganisms 2019, 7, 286. [Google Scholar] [CrossRef] [Green Version]
- Rabiey, M.; Roy, S.R.; Holtappels, D.; Franceschetti, L.; Quilty, B.J.; Creeth, R.; Sundin, G.W.; Wagemans, J.; Lavigne, R.; Jackson, R.W. Phage Biocontrol to Combat Pseudomonas Syringae Pathogens Causing Disease in Cherry. Microb. Biotechnol. 2020, 13, 1428–1445. [Google Scholar] [CrossRef]
- Jørgensen, J.B.; Djurhuus, A.M.; Carstens, A.B.; Kot, W.; Neve, H.; Morris, C.E.; Hansen, L.H. Presentation of Three Novel Tailed Phages Targeting Multiple Strains of Pseudomonas Syringae. Phage 2020, 1, 245–250. [Google Scholar] [CrossRef]
- Girard, L.; Lood, C.; Höfte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas Putida Group. Microorganisms 2021, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Saati-Santamaría, Z.; Peral-Aranega, E.; Velázquez, E.; Rivas, R.; García-Fraile, P. Phylogenomic Analyses of the Genus Pseudomonas Lead to the Rearrangement of Several Species and the Definition of New Genera. Biology 2021, 10, 782. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Cruz, G.; Smith, C.M.; Wiebe, K.F.; Villanueva, S.M.; Klonowski, A.R.; Cassone, B.J. Applications of Next-Generation Sequencing for Large-Scale Pathogen Diagnoses in Soybean. Plant Dis. 2019, 103, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Wang, D.; Bradley, C.A.; Zhao, Y. Genome Sequence Analyses of Pseudomonas Savastanoi Pv. Glycinea and Subtractive Hybridization-Based Comparative Genomics with Nine Pseudomonads. PLoS ONE 2011, 6, e16451. [Google Scholar] [CrossRef]
- Kovalyova, I.V.; Kropinski, A.M. The Complete Genomic Sequence of Lytic Bacteriophage Gh-1 Infecting Pseudomonas Putida—Evidence for Close Relationship to the T7 Group. Virology 2003, 311, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Frampton, R.; Acedo, E.; Young, V.; Chen, D.; Tong, B.; Taylor, C.; Easingwood, R.; Pitman, A.; Kleffmann, T.; Bostina, M.; et al. Genome, Proteome and Structure of a T7-Like Bacteriophage of the Kiwifruit Canker Phytopathogen Pseudomonas Syringae Pv. Actinidiae. Viruses 2015, 7, 3361–3379. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, G.; Walkowiak-Nowicka, K.; Zemleduch-Barylska, A.; Mleczko, A.; Frąckowiak, P.; Nowaczyk, N.; Kozdrowska, E.; Barylski, J. Complete Genome Sequences of Two Novel Autographiviruses Infecting a Bacterium from the Pseudomonas Fluorescens Group. Arch. Virol. 2017, 162, 2907–2911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sillankorva, S.; Kluskens, L.D.; Lingohr, E.J.; Kropinski, A.M.; Neubauer, P.; Azeredo, J. Complete Genome Sequence of the Lytic Pseudomonas Fluorescens Phage ΦIBB-PF7A. Virol. J. 2011, 8, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahiwale, S.; Prakash, D.; Gajbhiye, M.; Jagdale, S.; Patil, N.; Kapadnis, B. BVPaP-3, a T7-Like Lytic Phage of Pseudomonas Aeruginosa: Its Isolation and Characterisation. Curr. Microbiol. 2012, 64, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Orynbayev, A.T.; Dzhalilov, F.S.U.; Ignatov, A.N. Improved Efficacy of Formulated Bacteriophage in Control of Black Rot Caused by Xanthomonas Campestris Pv. Campestris on Cabbage Seedlings. Arch. Phytopathol. Plant Prot. 2020, 53, 379–394. [Google Scholar] [CrossRef]
- Jones, J.; Jackson, L.; Balogh, B.; Obradovic, A.; Iriarte, F.; Momol, M. Bacteriophages for Plant Disease Control. Annu. Rev. Phytopathol. 2007, 45, 245–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, E. New Assays for Detection of Pseudomonas Syringae pv. glycinea in Soybean Seed. Plant Dis. 1995, 79, 12. [Google Scholar] [CrossRef]
- Mohan, S.K. An Improved Agar Plating Assay for Detecting Pseudomonas Syringae Pv. Syringae and P. s. Pv. Phaseolicola in Contaminated Bean Seed. Phytopathology 1987, 77, 1390. [Google Scholar] [CrossRef]
- Wensing, A.; Braun, S.D.; Büttner, P.; Expert, D.; Völksch, B.; Ullrich, M.S.; Weingart, H. Impact of Siderophore Production by Pseudomonas Syringae Pv. Syringae 22d/93 on Epiphytic Fitness and Biocontrol Activity against Pseudomonas Syringae Pv. Glycinea 1a/96. Appl. Environ. Microbiol. 2010, 76, 2704–2711. [Google Scholar] [CrossRef] [Green Version]
- Lelliott, R.A.; Billing, E.; Hayward, A.C. A Determinative Scheme for the Fluorescent Plant Pathogenic Pseudomonads. J. Appl. Bacteriol. 1966, 29, 470–489. [Google Scholar] [CrossRef] [PubMed]
- Bereswill, S.; Bugert, P.; Völksch, B.; Ullrich, M.; Bender, C.L.; Geider, K. Identification and Relatedness of Coronatine-Producing Pseudomonas Syringae Pathovars by PCR Analysis and Sequence Determination of the Amplification Products. Appl. Environ. Microbiol. 1994, 60, 2924–2930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, S.F.; Guttman, D.S. Evolution of the Core Genome of Pseudomonas Syringae, a Highly Clonal, Endemic Plant Pathogen. Appl. Environ. Microbiol. 2004, 70, 1999–2012. [Google Scholar] [CrossRef] [Green Version]
- Rzhetsky, A.; Nei, M. A Simple Method for Estimating and Testing Minimum-Evolution Trees. Mol. Biol. Evol. 1992, 9, 945. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; ISBN 0-19-513584-9. [Google Scholar]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Van Twest, R.; Kropinski, A.M.; Twest, R.; Kropinski, A.M. Bacteriophage Enrichment from Water and Soil. In Bacteriophages; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 15–21. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Basic phage electron microscopy. In Bacteriophages: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2009; Volume 1, pp. 113–126. ISBN 978-1-58829-682-5. [Google Scholar]
- Kutter, E. Phage Host Range and Efficiency of Plating. In Bacteriophages Methods and Protocols: Isolation, Characterization, and Interactions; Humana Press: New York, NY, USA, 2009; Volume 501, ISBN 978-1-58829-682-5. [Google Scholar]
- Tong, Z.; Sadowsky, M.J. A Selective Medium for the Isolation and Quantification of Bradyrhizobium Japonicum and Bradyrhizobium Elkanii Strains from Soils and Inoculants. Appl. Environ. Microbiol. 1994, 60, 581–586. [Google Scholar] [CrossRef] [Green Version]
- Adams, M. Bacteriophages; StatPearls Publishing: Treasure Island, FL, USA, 1959; p. 620. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying Bacterial Genes and Endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Gabler, F.; Nam, S.-Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Geneious|Bioinformatics Software for Sequence Data Analysis. Available online: https://www.geneious.com/ (accessed on 11 November 2021).
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Rooney, W.; Laird, J.; Chowdhury, M.; MacIntosh, C.; Deng, X.; McBride, P.; Milner, J. Pseudomonas Syringae Seed Infections. Available online: https://www.protocols.io/view/pseudomonas-syringae-seed-infections-ewov18beygr2/v1 (accessed on 23 February 2021).
- Ullrich, M.S.; Schergaut, M.; Boch, J.; Ullrich, B. Temperature-Responsive Genetic Loci in the Plant Pathogen Pseudomonas Syringae Pv. Glycinea. Microbiology 2000, 146, 2457–2468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shine, M.B.; Fu, D.-Q.; Kachroo, A. Airbrush Infiltration Method for Pseudomonas Syringae Infection Assays in Soybean. Bio-protocol 2015, 5, e1427. [Google Scholar] [CrossRef]
- Sibiya, M.; Sumbwanyambe, M. An Algorithm for Severity Estimation of Plant Leaf Diseases by the Use of Colour Threshold Image Segmentation and Fuzzy Logic Inference: A Proposed Algorithm to Update a “Leaf Doctor” Application. AgriEngineering 2019, 1, 15. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarakanov, R.I.; Lukianova, A.A.; Evseev, P.V.; Toshchakov, S.V.; Kulikov, E.E.; Ignatov, A.N.; Miroshnikov, K.A.; Dzhalilov, F.S.-U. Bacteriophage Control of Pseudomonas savastanoi pv. glycinea in Soybean. Plants 2022, 11, 938. https://doi.org/10.3390/plants11070938
Tarakanov RI, Lukianova AA, Evseev PV, Toshchakov SV, Kulikov EE, Ignatov AN, Miroshnikov KA, Dzhalilov FS-U. Bacteriophage Control of Pseudomonas savastanoi pv. glycinea in Soybean. Plants. 2022; 11(7):938. https://doi.org/10.3390/plants11070938
Chicago/Turabian StyleTarakanov, Rashit I., Anna A. Lukianova, Peter V. Evseev, Stepan V. Toshchakov, Eugene E. Kulikov, Alexander N. Ignatov, Konstantin A. Miroshnikov, and Fevzi S.-U. Dzhalilov. 2022. "Bacteriophage Control of Pseudomonas savastanoi pv. glycinea in Soybean" Plants 11, no. 7: 938. https://doi.org/10.3390/plants11070938







