A New Ilarvirus Found in French Hydrangea
Abstract
:1. Introduction
2. Results
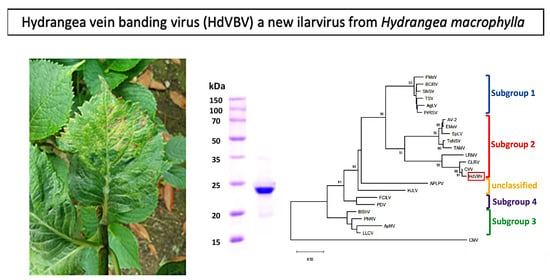
2.1. Symptomatology
2.2. Nucleotide Sequence and Genome Organization
2.3. Phylogenetic Relationships
2.4. Electron Microscopy, Coat Protein Mass Determination and Serological Detection
3. Discussion
4. Materials and Methods
4.1. Virus Source, Experimental Host Range and Graft Transmission
4.2. Virus Purification and Electron Microscopy Observation
4.3. Determination of Coat Protein Molecular Weight, Antibody Production and Serological Assays
4.4. Total RNA Extraction, Sequencing and Genome Assembly
4.5. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Govaerts, R.; Nic Lughadha, E.; Black, N.; Turner, R.; Paton, A. The World Checklist of Vascular Plants, a continuously updated resource for exploring global plant diversity. Sci. Data 2021, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Bi, G.; Zhao, X.; Harkess, R.L.; Scagel, C. Nitrogen fertilization container type and irrigation frequency affect mineral nutritient uptake of hydrangea. Water 2020, 12, 1987. [Google Scholar] [CrossRef]
- Bertaccini, A.; Paltrinieri, S.; Contaldo, N.; Cavicchi, L.; Mori, N.; Bellardi, M.G. Severe disease induced by viruses and phytoplasmas in Hydrangea in Italy. Acta Hortic. 2015, 1072, 105–112. [Google Scholar] [CrossRef]
- Machado Caballero, J.E.; Lockhart, B.E.; Mason, S.L.; Daughtrey, M. Identification and properties of a carlavirus causing chlorotic mottle of florists’ hydrangea (H. macrophylla) in the United States. Plant Dis. 2009, 93, 891–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bujarski, J.; Gallitelli, D.; García-Arenal, F.; Pallás, V.; Palukaitis, P.; Reddy, M.K.; Wang, A. ICTV Virus Taxonomy Profile: Bromoviridae. J. Gen. Virol. 2019, 100, 1206–1207. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V. Evolution and taxonomy of positive-strand RNA viruses: Implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 375–430. [Google Scholar] [CrossRef]
- Kasteel, D.T.; van der Wel, N.N.; Jansen, K.A.; Goldbach, R.W.; van Lent, J.W. Tubule-forming capacity of the movement proteins of alfalfa mosaic virus and brome mosaic virus. J. Gen. Virol. 1997, 78, 2089–2093. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Maiss, E.; Adam, G.; Casper, R. Prunus necrotic ringspot ilarvirus: Nucleotide sequence of RNA3 and the relationship to other ilarviruses based on coat protein comparison. J. Gen. Virol. 1995, 76, 1073–1079. [Google Scholar] [CrossRef]
- Bachman, E.J.; Scott, S.W.; Xin, G.; Vance, V.B. The complete nucleotide sequence of prune dwarf ilarvirus RNA 3: Implications for coat protein activation of genome replication in ilarviruses. Virology 1994, 201, 127–131. [Google Scholar] [CrossRef]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Sanchez-Navarro, J.A.; Scott, S.W. The molecular biology of Ilarviruses. Adv. Virus Res. 2013, 87, 139–181. [Google Scholar]
- Le, S.Q.; Gascuel, O. An improved general amino-acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, M.J.; Antoniw, J.F. DPVweb: A comprehensive database of plant and fungal virus genes and genomes. Nucleic Acids Res. 2006, 34, D382–D385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, D.D.; Ward, C.W.; Brunt, A.A. The Potyviridae; CAB International: Wallingford, CT, USA, 1994; p. 516. [Google Scholar]
- Aparicio, F.; Aparicio-Sanchis, R.; Gadea, J.; Sánchez-Navarro, J.Á.; Pallás, V.; Murguía, J.R. A Plant Virus Movement Protein Regulates the Gcn2p Kinase in Budding Yeast. PLoS ONE 2011, 6, e27409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, H. How to Identify and Treat Hydrangea Diseases. 2019. Available online: https://gardenerspath.com/how-to/disease-and-pests/hydrangea-diseases/ (accessed on 20 December 2021).
- Li, Y.; Mmbaga, M.T.; Zhou, B.; Joshua, J.; Rotich, E.; Parikh, L. Diseases of Hydrangea. In Handbook of Florists’ Crops Diseases; McGovern, R., Elmer, W., Eds.; Springer: Cham, Switzerland, 2018; pp. 1–19. [Google Scholar]
- Marrou, J. Amélioration des méthodes de transmission mécanique des virus par adsorption des inhibiteurs d’infection sur charbon végétal. C. R. Acad. Agric. Fr. 1967, 53, 972–981. [Google Scholar]
- Lot, H.J.; Marrou, J.; Quiot, B.; Esvan, C. Contribution à l’étude du virus de la mosaïque du concombre (CMV). II. Méthode de purification rapide du virus. Ann. Phytopath. 1972, 4, 25–38. [Google Scholar]
- Milne, R.G.; Lesemann, D.E. Immunosorbent electron microscopy in plant virus studies. Methods Virol. 1984, 8, 85–101. [Google Scholar]
- PM 7/126 (1) Electron microscopy in diagnosis of plant viruses. EPPO Bull. 2015, 45, 450–453. [CrossRef]
- Wang, Z.; Fang, S.G.; Zhang, Z.; Han, C.; Li, D.; Yu, J. Development of an ID-ELISA for the detection of rice black-streaked dwarf virus in plants. J. Virol. Methods 2006, 134, 61–65. [Google Scholar] [CrossRef]
- Laemmli, U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Ramasso, E.; Roggero, P.; Milne, R.G.; Boccardo, G.; Lisa, V.; Bozzano, G.; Crotti, A.; Bassetti, G. A tomato disease induced by spinach latent ilarvirus. Petria 1995, 5 (Suppl. S1), 42–43. [Google Scholar]
- Untiveros, M.; Perez-Egusquiza, Z.; Clover, G. PCR assays for the detection of members of the genus Ilarvirus and family Bromoviridae. J. Virol. Methods 2010, 165, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Eiras, M.R.; Resende, R.O.; Missiaggia, A.A.; De Avila, A.C. RT-PCR and Dot Blot hybridization methods for a universal detection of tospoviruses. Fitopat. Bras. 2001, 26, 170–175. [Google Scholar] [CrossRef]
- Li, Y.Y.; Zhang, R.N.; Xiang, H.Y.; Abouelnasr, H.; Li, D.W.; Yu, J.L.; McBeath, J.H.; Han, C.-G. Discovery and characterization of a novel carlavirus infecting potatoes in China. PLoS ONE 2013, 8, e69255. [Google Scholar] [CrossRef] [Green Version]
- Maliogka, V.; Dovas, C.; Efthimiou, K.; Katis, N. Detection and differentiation of Comoviridae species using a semi-nested RT-PCR and a phylogenetic analysis based on the polymerase protein. J. Phytopath. 2004, 152, 404–409. [Google Scholar] [CrossRef]
- Tian, T.; Klaassen, V.A.; Soong, J.; Wisler, G.; Duffus, J.E.; Falk, B.W. Generation of cDNAs specific to lettuce Infectious yellows closterovirus and other whitefly-transmitted viruses by RT–PCR and degenerate oligonucleotide primers to the closterovirus gene encoding the HSP70 homologue. Phytopathology 1996, 86, 1167–1173. [Google Scholar] [CrossRef]
- Dovas, C.I.; Katis, N.I. A spot nested RT-PCR method for the simultaneous detection of member of Vitivirus and Foveavirus genera in grapevine. J. Virol. Methods 2003, 107, 99–106. [Google Scholar] [CrossRef]
- Sabanadzovic, S.; Abou Ghanem-Sabanadzovic, N.; Gorbalenya, A.E. Permutation of the active site of putative RNA-dependent RNA polymerase in a newly identified species of plant alpha-like virus. Virology 2009, 394, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, A.; Mackenzie, A. A primer pair for amplifying part of the genome of all potyvirids by RT-PCR. J. Virol. Methods 1997, 63, 9–16. [Google Scholar] [CrossRef]
- Parrella, G.; Sorrentino, D. Identification af a Cucumber mosaic virus isolate from Passiflora edulis in southern Italy and validation of subgroup identification by in silico restriction fragment length polymorphism. J. Phytopath. 2009, 157, 762–767. [Google Scholar] [CrossRef]
- Parrella, G.; Lanave, C.; Marchoux, G.; Finetti-Sialer, M.M.; Di Franco, A.; Gallitelli, D. Evidence for two distinct subgroups of Alfalfa mosaic virus (AMV) from France and Italy and their relationships with other AMV strains. Arch. Virol. 2000, 145, 2659–2667. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [Green Version]






| CVV | EMoV | AV-2 | SpLV | TAMV | ToNSV | CLRV | LRMV | |
|---|---|---|---|---|---|---|---|---|
| Nucleotides | ||||||||
| RNA 1 | 85 | 86 | 85 | 79 | 69 | 70 | 70 | 67 b |
| RNA 2 | 83 | 80 | 88 | 77 | 75 | 74 | 75 | 69 |
| RNA 3 | 81 | 79 | 85 | 71 | 65 | 67 | 68 | 66 |
| Amino acids | ||||||||
| p1 | 91/95 c | 91/95 | 90/95 | 87/92 | 69/80 | 71/84 | 72/84 | na |
| p2a | 87/91 | 82/88 | 90/93 | 74/81 | 70/78 | 63/72 | 67/74 | 60/72 |
| p2b | 79/85 | 68/76 | 85/90 | 67/77 | 72/83 | 69/84 | 66/75 | 54/70 |
| p3a | 91/95 | 35/51 | 34/51 | 33/48 | 30/48 | 35/51 | 75/83 | 48/63 |
| p3b | 76/86 | 75/86 | 81/88 | 66/79 | 58/71 | 63/75 | 60/74 | 37/49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parrella, G.; Troiano, E. A New Ilarvirus Found in French Hydrangea. Plants 2022, 11, 944. https://doi.org/10.3390/plants11070944
Parrella G, Troiano E. A New Ilarvirus Found in French Hydrangea. Plants. 2022; 11(7):944. https://doi.org/10.3390/plants11070944
Chicago/Turabian StyleParrella, Giuseppe, and Elisa Troiano. 2022. "A New Ilarvirus Found in French Hydrangea" Plants 11, no. 7: 944. https://doi.org/10.3390/plants11070944
APA StyleParrella, G., & Troiano, E. (2022). A New Ilarvirus Found in French Hydrangea. Plants, 11(7), 944. https://doi.org/10.3390/plants11070944