A Novel Strategy to Reveal the Landscape of Crossovers in an F1 Hybrid Population of Populus deltoides and Populus simonii
Abstract
:1. Introduction
2. Results
2.1. SNP Genotype Data
2.2. Difference of CO Rates between Two Parents
2.3. Investigation of CO Interference in Both Species
3. Discussion
4. Materials and Methods
4.1. Plant Material and Sequencing Data
4.2. SNP Genotyping
4.3. Comparison of CO Rates between Two Parents
4.4. Analysis of CO Interference in Both Parents
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rice, W.R. Experimental tests of the adaptive significance of sexual recombination. Nat. Rev. Genet. 2002, 3, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Barrientos, D.; Engelstädter, J.; Rieseberg, L.H. Recombination rate evolution and the origin of species. Trends Ecol. Evol. 2016, 31, 226–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, D. Genetic Recombination. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 277–280. [Google Scholar]
- Keeney, S.; Giroux, C.N.; Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 1997, 88, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Mercier, R.; Mézard, C.; Jenczewski, E.; Macaisne, N.; Grelon, M. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 2015, 66, 297–327. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.; Cohen, P.E. Control of meiotic crossovers: From double-stand break formation to designation. Annu. Rev. Genet. 2016, 50, 175–210. [Google Scholar] [CrossRef] [Green Version]
- Jones, G.H.; Franklin, F.C. Meiotic crossing-over: Obligation and interference. Cell 2006, 126, 246–248. [Google Scholar] [CrossRef] [Green Version]
- Mézard, C.; Tagliaro Jahns, M.; Grelon, M. Where to cross? New insights into the location of meiotic crossovers. Trends Genet. 2015, 31, 393–401. [Google Scholar] [CrossRef]
- do Vale Martins, L.; Yu, F.; Zhao, H.; Dennison, T.; Lauter, N.; Wang, H.; Deng, Z.; Thompson, A.; Semrau, K.; Rouillard, J.-M.; et al. Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat. Commun. 2019, 10, 4604. [Google Scholar] [CrossRef]
- Cole, F.; Baudat, F.; Grey, C.; Keeney, S.; De Massy, B.; Jasin, M. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat. Genet. 2014, 46, 1072–1080. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Bitoun, E.; Altemose, N.; Davies, R.W.; Davies, B.; Myers, S.R. A high-resolution map of non-crossover events reveals impacts of genetic diversity on mammalian meiotic recombination. Nat. Commun. 2019, 10, 3900. [Google Scholar] [CrossRef] [Green Version]
- Drouaud, J.; Mercier, R.; Chelysheva, L.; Berard, A.; Falque, M.; Martin, O.; Zanni, V.; Brunel, D.; Mezard, C. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 2007, 3, 1096–1107. [Google Scholar] [CrossRef] [PubMed]
- Sardell, J.M.; Cheng, C.D.; Dagilis, A.J.; Ishikawa, A.; Kitano, J.; Peichel, C.L.; Kirkpatrick, M. Sex differences in recombination in Sticklebacks. G3-Genes Genomes Genet. 2018, 8, 1971–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apuli, R.P.; Bernhardsson, C.; Schiffthaler, B.; Robinson, K.M.; Jansson, S.; Street, N.R.; Ingvarsson, P.K. Inferring the genomic landscape of recombination rate variation in European Aspen (Populus tremula). G3-Genes Genomes Genet. 2020, 10, 299–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachman, M.W. Variation in recombination rate across the genome: Evidence and implications. Curr. Opin. Genet. Dev. 2003, 12, 657–663. [Google Scholar] [CrossRef]
- Stapley, J.; Feulner, P.G.D.; Johnston, S.E.; Santure, A.W.; Smadja, C.M. Variation in recombination frequency and distribution across eukaryotes: Patterns and processes. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160455. [Google Scholar] [CrossRef]
- Wilfert, L.; Gadau, J.; Schmid-Hempel, P. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity 2007, 98, 189–197. [Google Scholar] [CrossRef]
- Dumont, B.L.; Payseur, B.A. Evolution of the genomic rate of recombination in mammals. Evolution 2008, 62, 276–294. [Google Scholar] [CrossRef]
- Jaramillo-Correa, J.P.; Verdú, M.; González-Martínez, S.C. The contribution of recombination to heterozygosity differs among plant evolutionary lineages and life-forms. BMC Evol. Biol. 2010, 10, 22. [Google Scholar] [CrossRef] [Green Version]
- Jensen-Seaman, M.I.; Furey, T.S.; Payseur, B.A.; Lu, Y.T.; Roskin, K.M.; Chen, C.F.; Thomas, M.A.; Haussler, D.; Jacob, H.J. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004, 14, 528–538. [Google Scholar] [CrossRef] [Green Version]
- Fang, L.; Ye, J.; Li, N.; Zhang, Y.; Li, S.; Wong, G.K.-S.; Wang, J. Positive correlation between recombination rate and nucleotide diversity is shown under domestication selection in the chicken genome. Chin. Sci. Bull. 2008, 53, 746–750. [Google Scholar] [CrossRef]
- Tiley, G.P.; Burleigh, J.G. The relationship of recombination rate, genome structure, and patterns of molecular evolution across angiosperms. BMC Evol. Biol. 2015, 15, 194. [Google Scholar] [CrossRef] [Green Version]
- Jabbari, K.; Wirtz, J.; Rauscher, M.; Wiehe, T. A common genomic code for chromatin architecture and recombination landscape. PLoS ONE 2019, 14, e0213278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreissig, S.; Mascher, M.; Heckmann, S. Variation in recombination rate is shaped by domestication and environmental conditions in barley. Mol. Biol. Evol. 2019, 36, 2029–2039. [Google Scholar] [CrossRef]
- Kawakami, T.; Wallberg, A.; Olsson, A.; Wintermantel, D.; De Miranda, J.R.; Allsopp, M.; Rundlöf, M.; Webster, M.T. Substantial heritable variation in recombination rate on multiple scales in honeybees and bumblebees. Genetics 2019, 212, 1101–1119. [Google Scholar] [CrossRef] [PubMed]
- de Massy, B. Distribution of meiotic recombination sites. Trends Genet. 2003, 19, 514–522. [Google Scholar] [CrossRef]
- Mézard, C.; Vignard, J.; Drouaud, J.; Mercier, R. The road to crossovers: Plants have their say. Trends Genet. 2007, 23, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, J.; Sang, M.; Jiang, L.; Zhao, B.; Cheng, T.; Zhang, Q.; Wu, R. Landscaping crossover interference across a genome. Trends Plant Sci. 2017, 22, 894–907. [Google Scholar] [CrossRef]
- Sturtevant, A.H. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zool. 1913, 14, 43–59. [Google Scholar] [CrossRef]
- Auger, D.L.; Sheridan, W.F. Negative crossover interference in maize translocation heterozygotes. Genetics 2001, 159, 1717–1726. [Google Scholar] [CrossRef]
- Berchowitz, L.; Copenhaver, G. Genetic Interference: Dont Stand So Close to Me. Curr. Genom. 2010, 11, 91–102. [Google Scholar] [CrossRef] [Green Version]
- Strickland, W.N. An analysis of interference in Aspergillus nidulans. Proc. R. Soc. Lond. Ser. B-Biol. Sci. 1958, 149, 82–101. [Google Scholar] [CrossRef]
- Campbell, C.L.; Bhérer, C.; Morrow, B.E.; Boyko, A.R.; Auton, A. A pedigree-based map of recombination in the domestic dog genome. G3 Genesgenetics 2016, 6, 3517–3524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petkov, P.M.; Broman, K.W.; Szatkiewicz, J.P.; Paigen, K. Crossover interference underlies sex differences in recombination rates. Trends Genet. 2007, 23, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Torgasheva, A.A.; Borodin, P.M. Immunocytological analysis of meiotic recombination in the Gray Goose (Anser anser). Cytogenet. Genome Res. 2017, 151, 27–35. [Google Scholar] [CrossRef]
- Hou, Y.; Fan, W.; Yan, L.; Li, R.; Lian, Y.; Huang, J.; Li, J.; Xu, L.; Tang, F.; Xie, X.S.; et al. Genome analyses of single human oocytes. Cell 2013, 155, 1492–1506. [Google Scholar] [CrossRef] [Green Version]
- Campbell, C.L.; Furlotte, N.A.; Eriksson, N.; Hinds, D.; Auton, A. Escape from crossover interference increases with maternal age. Nat. Commun. 2015, 6, 6260. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; O’Connell, J.R.; Vanraden, P.M.; Shen, B.; Padhi, A.; Sun, C.; Bickhart, D.M.; Cole, J.B.; Null, D.J.; Liu, G.E. Cattle sex-specific recombination and genetic control from a large pedigree analysis. PLoS Genet. 2015, 11, e1005387. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Shen, B.; Jiang, J.; Li, J.; Ma, L. Effect of sex, age and genetics on crossover interference in cattle. Sci. Rep. 2016, 6, 37698. [Google Scholar] [CrossRef] [Green Version]
- Basu-Roy, S.; Gauthier, F.; Giraut, L.; Mezard, C.; Falque, M.; Martin, O.C. Hot regions of noninterfering crossovers coexist with a nonuniformly interfering pathway in Arabidopsis thaliana. Genetics 2013, 195, 769–779. [Google Scholar] [CrossRef] [Green Version]
- Cronk, Q.C. Plant eco-devo: The potential of poplar as a model organism. New Phytol. 2005, 166, 39–48. [Google Scholar] [CrossRef]
- Gaudet, M.; Jorge, V.; Paolucci, I.; Beritognolo, I.; Mugnozza, G.S.; Sabatti, M. Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genet. Genomes 2008, 4, 25–36. [Google Scholar] [CrossRef]
- Mousavi, M.; Tong, C.; Liu, F.; Tao, S.; Shi, J. De novo SNP discovery and genetic linkage mapping in Poplar using restriction site associated DNA and whole-genome sequencing technologies. BMC Genom. 2016, 17, 656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pakull, B.; Groppe, K.; Meyer, M.; Markussen, T.; Fladung, M. Genetic linkage mapping in aspen (Populus tremula L. and Populus tremuloides Michx.). Tree Genet. Genomes 2009, 5, 505–515. [Google Scholar] [CrossRef]
- Paolucci, I.; Gaudet, M.; Jorge, V.; Beritognolo, I.; Terzol, S.; Kuzminsky, E.; Muleo, R.; Mugnozza, G.S.; Sabatti, M. Genetic linkage maps of Populus alba L. and comparative mapping analysis of sex determination across Populus species. Tree Genet. Genomes 2010, 6, 863–875. [Google Scholar] [CrossRef]
- Tong, C.; Li, H.; Wang, Y.; Li, X.; Shi, J. Construction of high-density linkage maps of Populus deltoides × P. simonii using restriction-site associated DNA sequencing. PLoS ONE 2016, 11, e0150692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Street, N.R.; Scofield, D.G.; Ingvarsson, P.K. Natural selection and recombination rate variation shape nucleotide polymorphism across the genomes of three related Populus species. Genetics 2016, 202, 1185–1200. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.N.; Yao, D.; Chen, Y.H.; Yang, W.G.; Zhao, W.; Gao, H.; Tong, C.F. De novo genome assembly of Populus simonii further supports that Populus simonii and Populus trichocarpa belong to different sections. G3-Genes Genomes Genet. 2020, 10, 455–466. [Google Scholar] [CrossRef] [Green Version]
- Fierst, J.L. Using linkage maps to correct and scaffold de novo genome assemblies: Methods, challenges, and computational tools. Front. Genet. 2015, 6, 220. [Google Scholar] [CrossRef]
- Tong, C.; Yao, D.; Wu, H.; Chen, Y.; Yang, W.; Zhao, W. High-quality SNP linkage maps improved QTL mapping and genome assembly in Populus. J. Hered. 2020, 111, 515–530. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Yao, D.; Wu, H.; Chen, Y.; Yang, W.; Gao, H.; Tong, C. gmRAD: An integrated SNP calling pipeline for genetic mapping with RADseq across a hybrid population. Brief. Bioinform. 2020, 21, 329–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broman, K.W.; Weber, J.L. Characterization of human crossover interference. Am. J. Hum. Genet. 2000, 66, 1911–1926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraut, L.; Falque, M.; Drouaud, J.; Pereira, L.; Martin, O.C.; Mézard, C. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011, 7, e1002354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagercrantz, U.; Lydiate, D.J. RFLP mapping in Brassica nigra indicates differing recombination rates in male and female meioses. Genome 1995, 38, 255–264. [Google Scholar] [CrossRef]
- Kianian, P.M.A.; Wang, M.H.; Simons, K.; Ghavami, F.; He, Y.; Dukowic-Schulze, S.; Sundararajan, A.; Sun, Q.; Pillardy, J.; Mudge, J.; et al. High-resolution crossover mapping reveals similarities and differences of male and female recombination in Maize. Nat. Commun. 2018, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Grattapaglia, D.; Sederoff, R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: Mapping strategy and RAPD markers. Genetics 1994, 137, 1121–1137. [Google Scholar] [CrossRef]
- Maliepaard, C.; Jansen, J.; Van Ooijen, J.W. Linkage analysis in a full-sib family of an outbreeding plant species: Overview and consequences for applications. Genet. Res. 1997, 70, 237–250. [Google Scholar] [CrossRef] [Green Version]
- Tong, C.; Zhang, B.; Shi, J. A hidden Markov model approach to multilocus linkage analysis in a full-sib family. Tree Genet. Genomes 2010, 6, 651–662. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef]
- Lenormand, T.; Dutheil, J. Recombination difference between sexes: A role for haploid selection. PLoS Biol. 2005, 3, 396–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Jiang, L.; Ye, M.; Zhu, X.; Wu, R. The genomic landscape of crossover interference in the desert tree Populus euphratica. Front. Genet. 2019, 10, 440. [Google Scholar] [CrossRef] [PubMed]
- Housworth, E.A.; Stahl, F.W. Crossover interference in humans. Am. J. Hum. Genet. 2003, 73, 188–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Ooijen, J.W. JoinMap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma B. V.: Wageningen, The Netherlands, 2006. [Google Scholar]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newburg, L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]
- Lander, E.S.; Botstein, D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 1989, 121, 185–199. [Google Scholar] [CrossRef]
- Zeng, Z.-B. Precision mapping of quantitative trait loci. Genetics 1994, 136, 1457–1468. [Google Scholar] [CrossRef]
- Bartholomé, J.; Mandrou, E.; Mabiala, A.; Jenkins, J.; Nabihoudine, I.; Klopp, C.; Schmutz, J.; Plomion, C.; Gion, J.M. High-resolution genetic maps of Eucalyptus improve Eucalyptus grandis genome assembly. New Phytol. 2015, 206, 1283–1296. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]

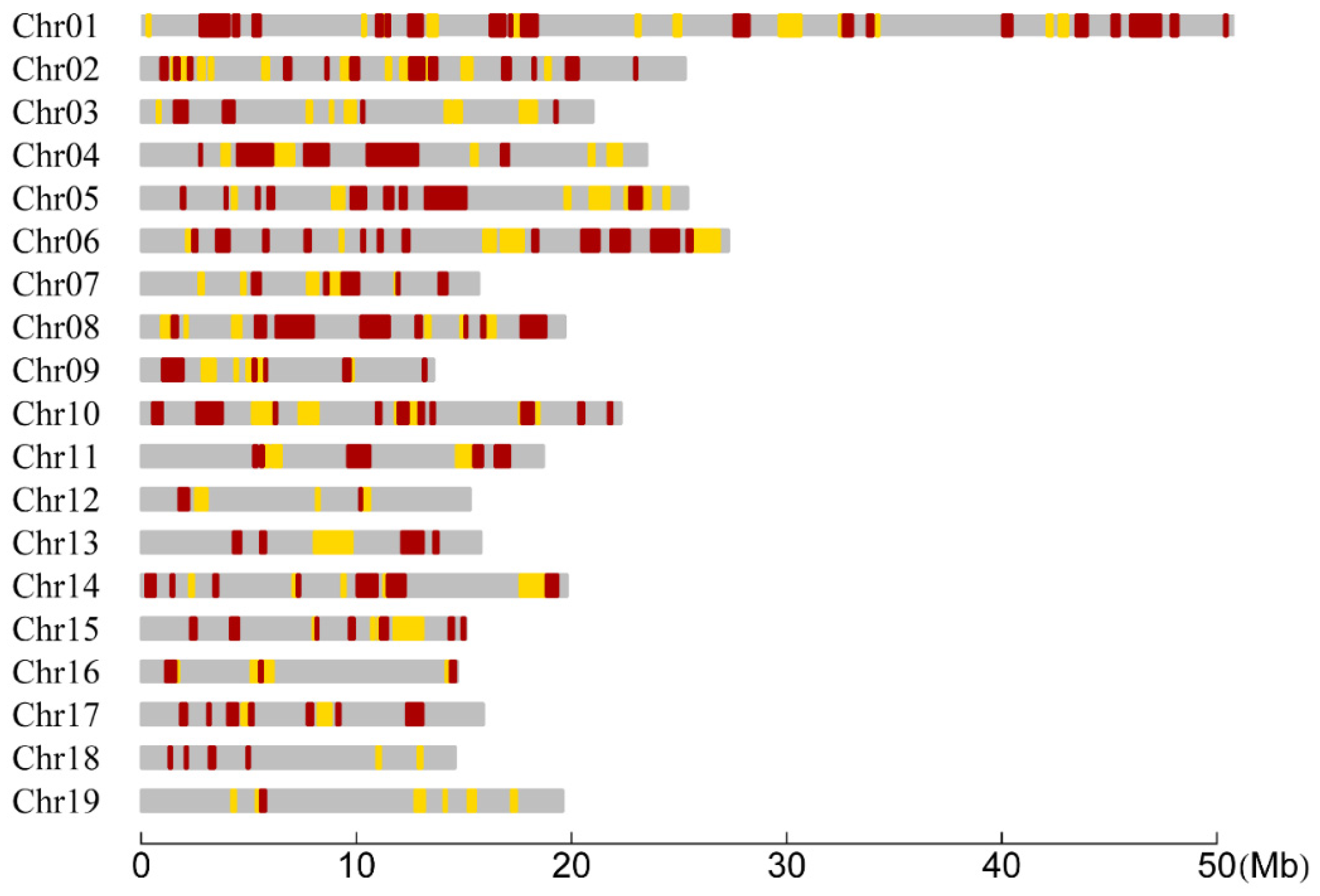
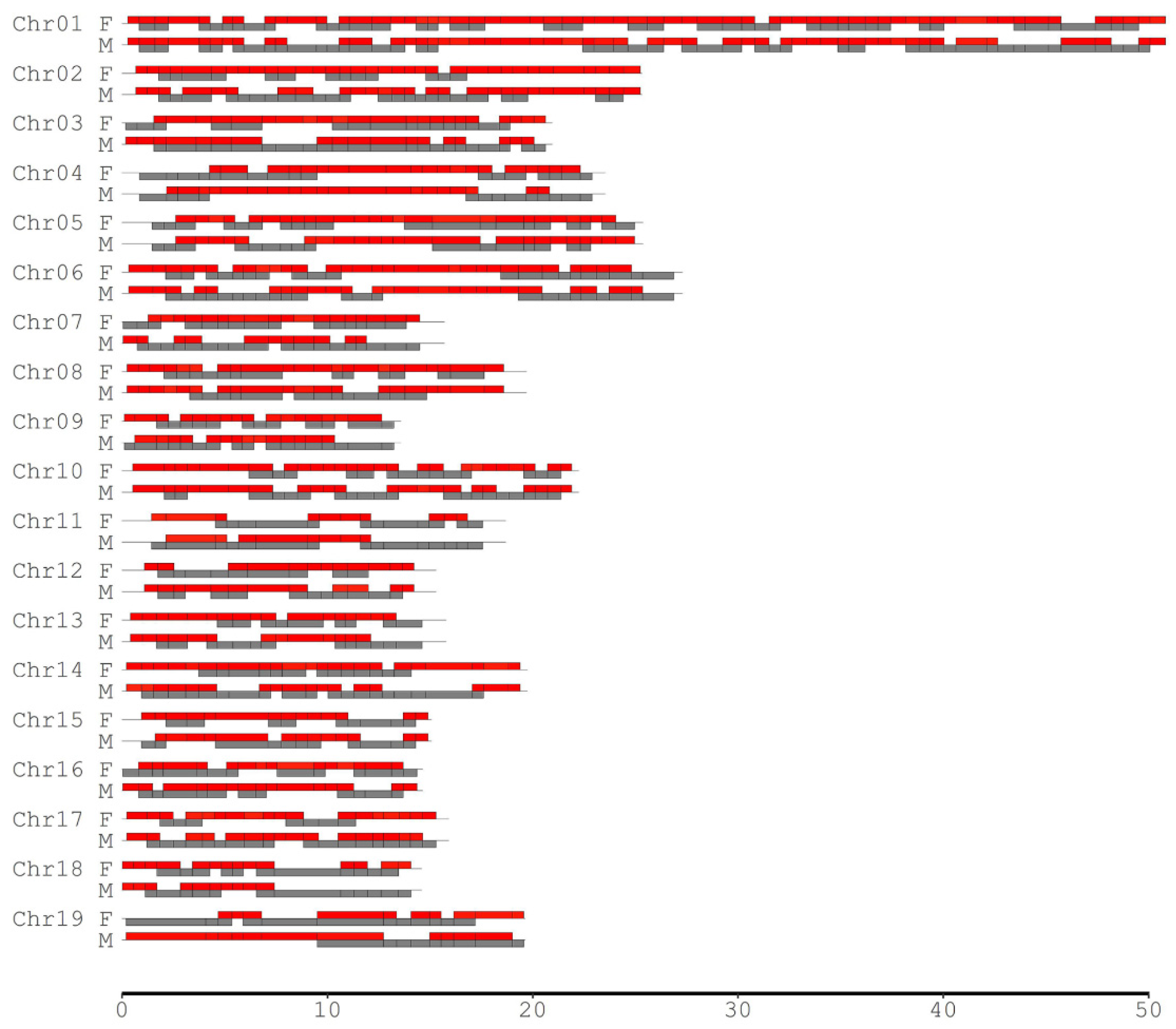
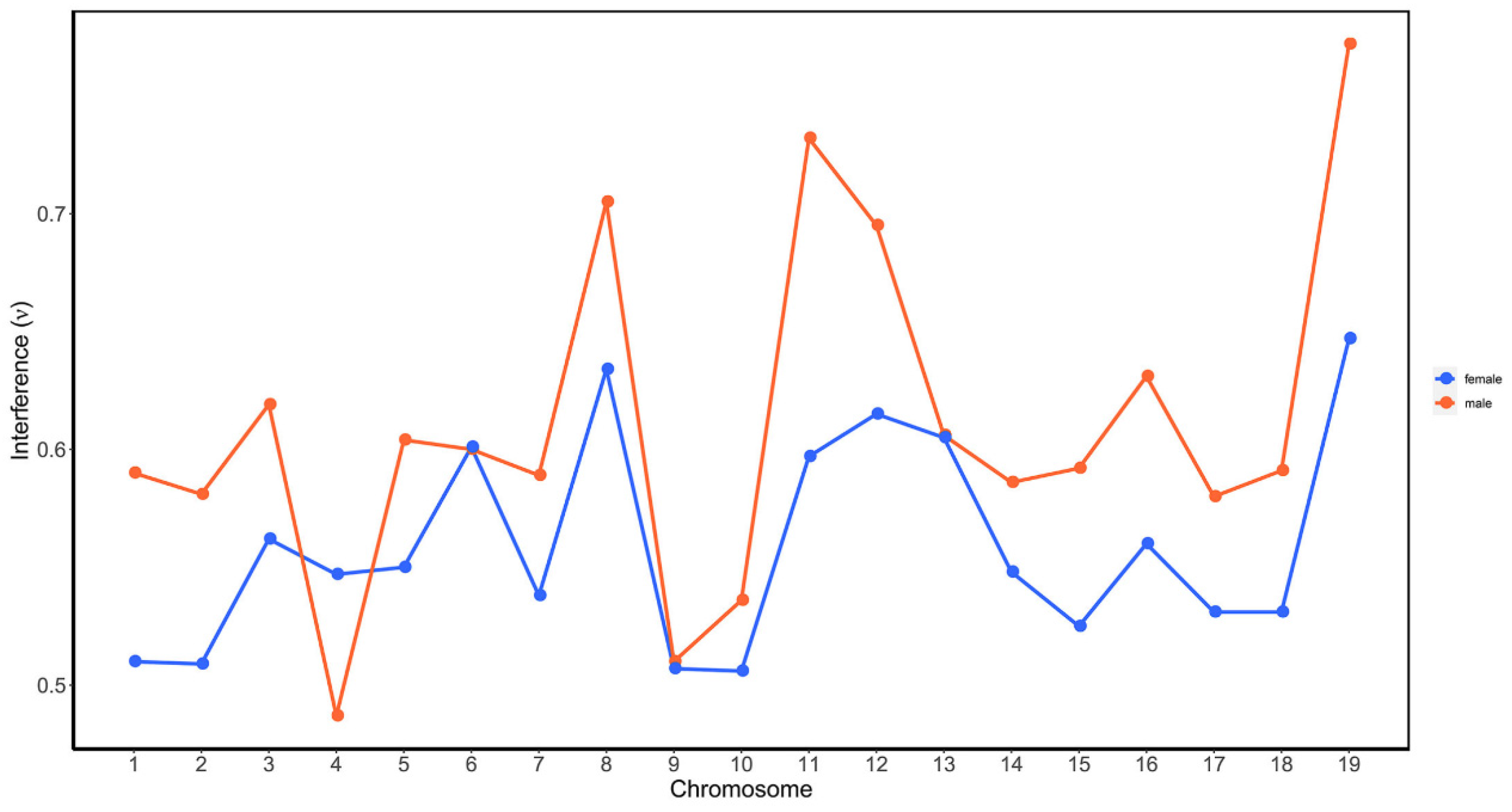

| Chrom. | RAD Tag Number | Length Range (Kb) | Length Mean (Kb) | Female CO Rate Range | Female CO Rate Mean | Male CO Rate Range | Male CO Rate Mean |
|---|---|---|---|---|---|---|---|
| Chr01 | 151 | 101~1492 | 337 | 0.005~0.150 | 0.053 | 0.004~0.166 | 0.040 |
| Chr02 | 88 | 102~1191 | 282 | 0.008~0.111 | 0.047 | 0.005~0.118 | 0.043 |
| Chr03 | 67 | 100~1502 | 313 | 0.005~0.113 | 0.046 | 0.004~0.117 | 0.044 |
| Chr04 | 67 | 103~1370 | 338 | 0.004~0.137 | 0.044 | 0.008~0.111 | 0.040 |
| Chr05 | 71 | 105~2195 | 336 | 0.008~0.202 | 0.060 | 0.004~0.100 | 0.040 |
| Chr06 | 83 | 100~1490 | 324 | 0.008~0.192 | 0.057 | 0.005~0.202 | 0.046 |
| Chr07 | 49 | 102~1175 | 301 | 0.004~0.170 | 0.052 | 0.009~0.102 | 0.042 |
| Chr08 | 51 | 110~1566 | 374 | 0.004~0.165 | 0.056 | 0.014~0.225 | 0.066 |
| Chr09 | 54 | 101~1310 | 248 | 0.008~0.147 | 0.043 | 0.008~0.095 | 0.038 |
| Chr10 | 82 | 100~1056 | 265 | 0.005~0.164 | 0.050 | 0.004~0.123 | 0.039 |
| Chr11 | 41 | 108~2405 | 403 | 0.004~0.127 | 0.060 | 0.009~0.123 | 0.039 |
| Chr12 | 37 | 103~2025 | 365 | 0.009~0.145 | 0.050 | 0.008~0.159 | 0.052 |
| Chr13 | 42 | 100~1755 | 346 | 0.008~0.152 | 0.048 | 0.004~0.089 | 0.039 |
| Chr14 | 60 | 106~1990 | 325 | 0.004~0.203 | 0.045 | 0.005~0.147 | 0.045 |
| Chr15 | 46 | 102~2311 | 314 | 0.012~0.110 | 0.044 | 0.004~0.136 | 0.040 |
| Chr16 | 40 | 109~1659 | 374 | 0.005~0.167 | 0.052 | 0.005~0.158 | 0.043 |
| Chr17 | 49 | 102~869 | 314 | 0.008~0.148 | 0.053 | 0.009~0.101 | 0.042 |
| Chr18 | 47 | 100~2837 | 306 | 0.004~0.201 | 0.047 | 0.004~0.204 | 0.044 |
| Chr19 | 32 | 104~3687 | 625 | 0.008~0.120 | 0.056 | 0.012~0.158 | 0.054 |
| Chrom. | Interval | Sig. Interval | Sig. Region | 100–500 kb | 500 Kb–1 Mb | ≥1 Mb | Cover. (Mb) a | Percent (%) b | Chrom. Size (Mb) c |
|---|---|---|---|---|---|---|---|---|---|
| Chr01 | 150 | 41 | 11 | 15 | 6 | 4 | 13.06 | 25.69 | 50.84 |
| Chr02 | 87 | 32 | 8 | 13 | 3 | 1 | 6.86 | 27.09 | 25.34 |
| Chr03 | 66 | 13 | 2 | 8 | 3 | 0 | 3.70 | 17.63 | 20.97 |
| Chr04 | 66 | 16 | 4 | 5 | 2 | 3 | 7.92 | 33.65 | 23.53 |
| Chr05 | 70 | 23 | 5 | 9 | 3 | 2 | 7.32 | 28.83 | 25.39 |
| Chr06 | 82 | 25 | 8 | 9 | 4 | 3 | 8.46 | 30.98 | 27.31 |
| Chr07 | 48 | 10 | 2 | 6 | 0 | 1 | 3.43 | 21.82 | 15.71 |
| Chr08 | 50 | 20 | 7 | 5 | 3 | 3 | 7.43 | 37.72 | 19.71 |
| Chr09 | 53 | 11 | 3 | 3 | 3 | 0 | 3.28 | 24.14 | 13.60 |
| Chr10 | 81 | 24 | 4 | 9 | 1 | 3 | 6.35 | 28.53 | 22.25 |
| Chr11 | 40 | 15 | 4 | 3 | 1 | 2 | 3.94 | 21.04 | 18.70 |
| Chr12 | 36 | 7 | 3 | 3 | 1 | 0 | 1.65 | 10.75 | 15.30 |
| Chr13 | 41 | 7 | 2 | 3 | 0 | 2 | 3.58 | 22.65 | 15.80 |
| Chr14 | 59 | 17 | 5 | 6 | 2 | 1 | 5.21 | 26.35 | 19.76 |
| Chr15 | 45 | 12 | 3 | 8 | 0 | 1 | 3.43 | 22.75 | 15.07 |
| Chr16 | 39 | 8 | 3 | 1 | 1 | 1 | 2.13 | 14.52 | 14.66 |
| Chr17 | 48 | 13 | 2 | 5 | 3 | 0 | 3.23 | 20.29 | 15.92 |
| Chr18 | 46 | 6 | 0 | 6 | 0 | 0 | 1.01 | 6.91 | 14.60 |
| Chr19 | 31 | 7 | 1 | 6 | 0 | 0 | 1.81 | 9.20 | 19.64 |
| Total | 1138 | 307 | 77 | 123 | 36 | 27 | 93.78 | 23.80 | 394.11 |
| Parent | CoC | 0.5 Mb | 1.0 Mb | 2.0 Mb | 3.0 Mb | 4.0 Mb | 5.0 Mb |
|---|---|---|---|---|---|---|---|
| F | >1 | 214 | 64 | 13 | 9 | 3 | 0 |
| <1 | 76 | 34 | 29 | 25 | 25 | 14 | |
| Total (Coverage) | 290 (79.86%) | 98 (57.05%) | 42 (54.79%) | 34 (46.17%) | 28 (55.35%) | 14 (35.10%) | |
| M | >1 | 138 | 37 | 8 | 2 | 1 | 0 |
| <1 | 95 | 40 | 31 | 34 | 18 | 9 | |
| Total (Coverage) | 233 (69.77%) | 77 (46.35%) | 39 (52.44%) | 36 (55.50%) | 19 (38.28%) | 9 (22.78%) | |
| Both F and M | >1 | 81 | 18 | 1 | 1 | 0 | 0 |
| <1 | 17 | 3 | 12 | 14 | 11 | 4 | |
| Total (Coverage) | 168 (52.61%) | 37 (24.21%) | 16 (23.81%) | 17 (27.46%) | 12 (24.26%) | 4 (9.20%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Zhao, W.; Zhang, J.; Pan, Z.; Bai, S.; Tong, C. A Novel Strategy to Reveal the Landscape of Crossovers in an F1 Hybrid Population of Populus deltoides and Populus simonii. Plants 2022, 11, 1046. https://doi.org/10.3390/plants11081046
Li Z, Zhao W, Zhang J, Pan Z, Bai S, Tong C. A Novel Strategy to Reveal the Landscape of Crossovers in an F1 Hybrid Population of Populus deltoides and Populus simonii. Plants. 2022; 11(8):1046. https://doi.org/10.3390/plants11081046
Chicago/Turabian StyleLi, Zhiting, Wei Zhao, Jinpeng Zhang, Zhiliang Pan, Shengjun Bai, and Chunfa Tong. 2022. "A Novel Strategy to Reveal the Landscape of Crossovers in an F1 Hybrid Population of Populus deltoides and Populus simonii" Plants 11, no. 8: 1046. https://doi.org/10.3390/plants11081046
APA StyleLi, Z., Zhao, W., Zhang, J., Pan, Z., Bai, S., & Tong, C. (2022). A Novel Strategy to Reveal the Landscape of Crossovers in an F1 Hybrid Population of Populus deltoides and Populus simonii. Plants, 11(8), 1046. https://doi.org/10.3390/plants11081046





