One-Week Scutellar Somatic Embryogenesis in the Monocot Brachypodium distachyon
Abstract
:1. Introduction
2. Results and Discussion
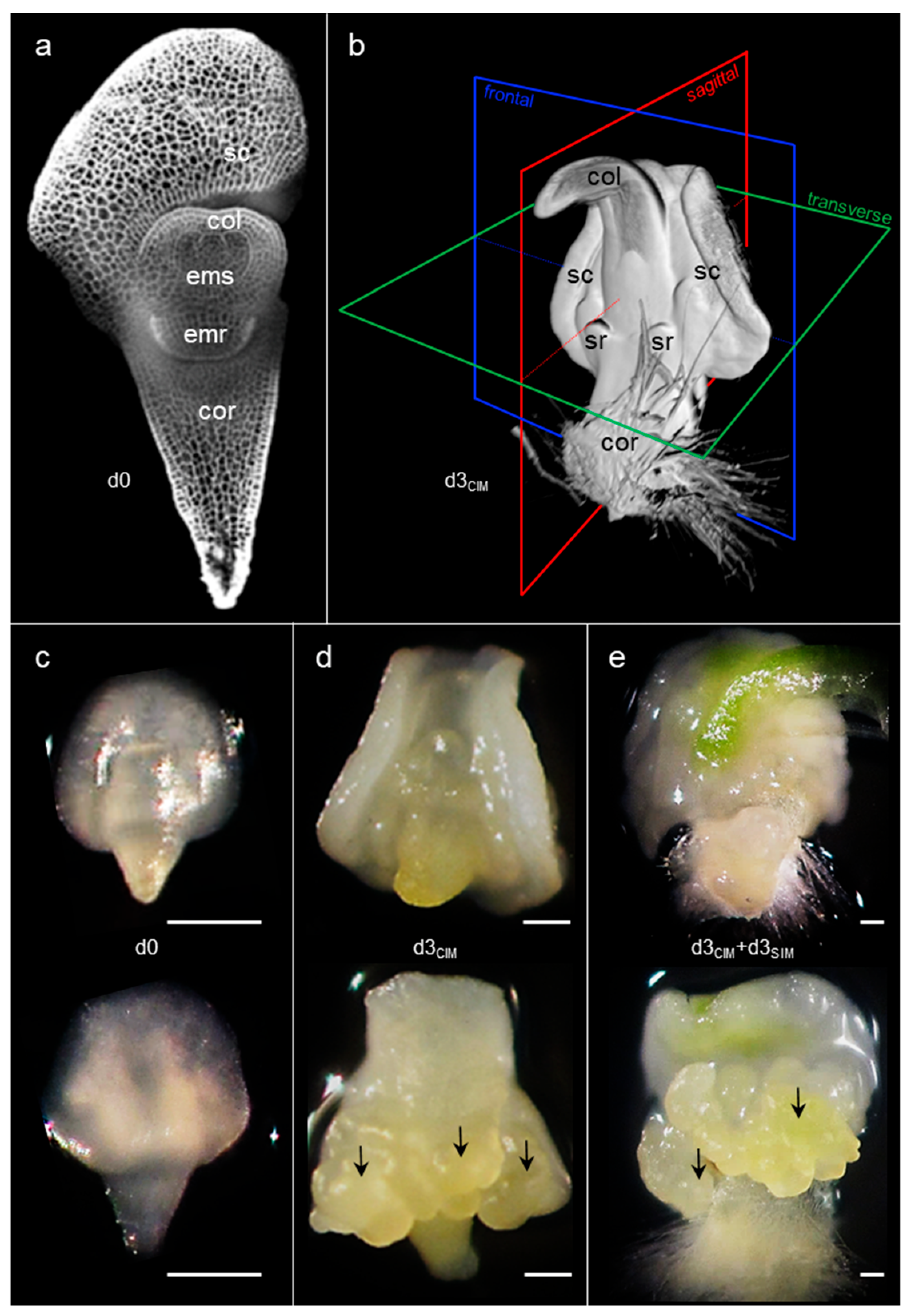
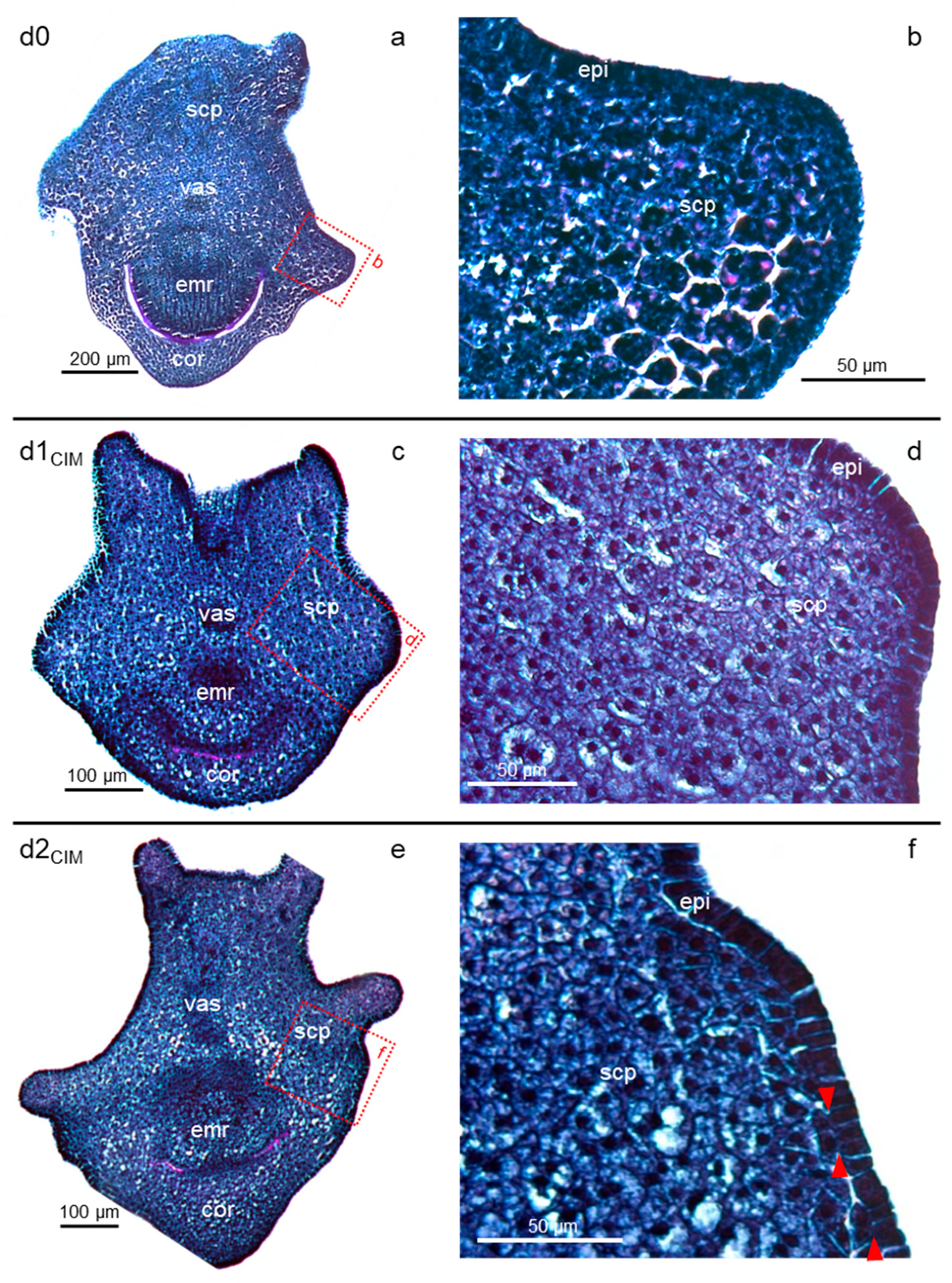
2.1. Cell Proliferation Induced by Exogenous Auxin Initiates Early in Restricted Peripheral Regions of the Scutellum
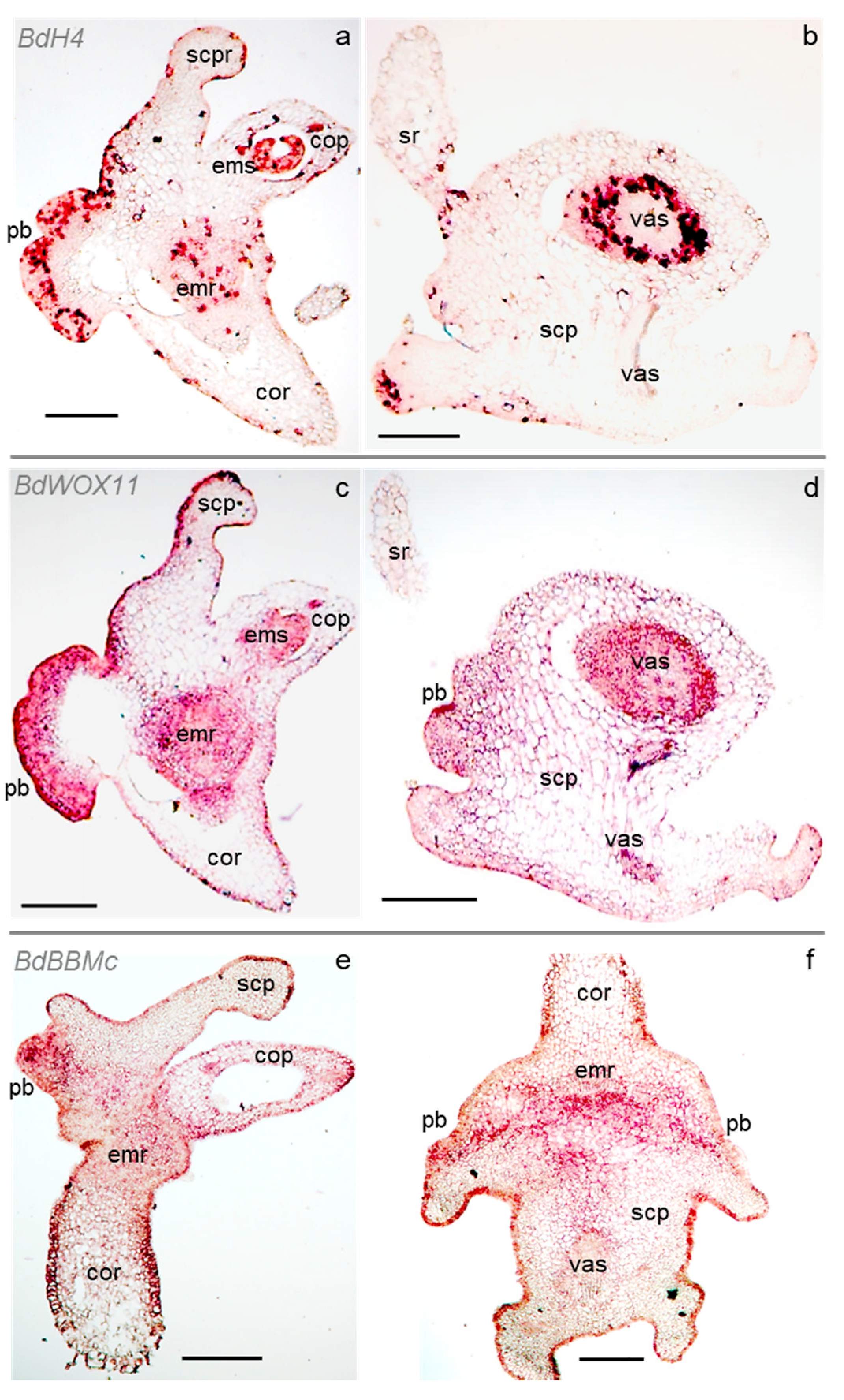
2.2. Cell Division and Developmental Genes Mark Scutellar Embryogenic Tissues
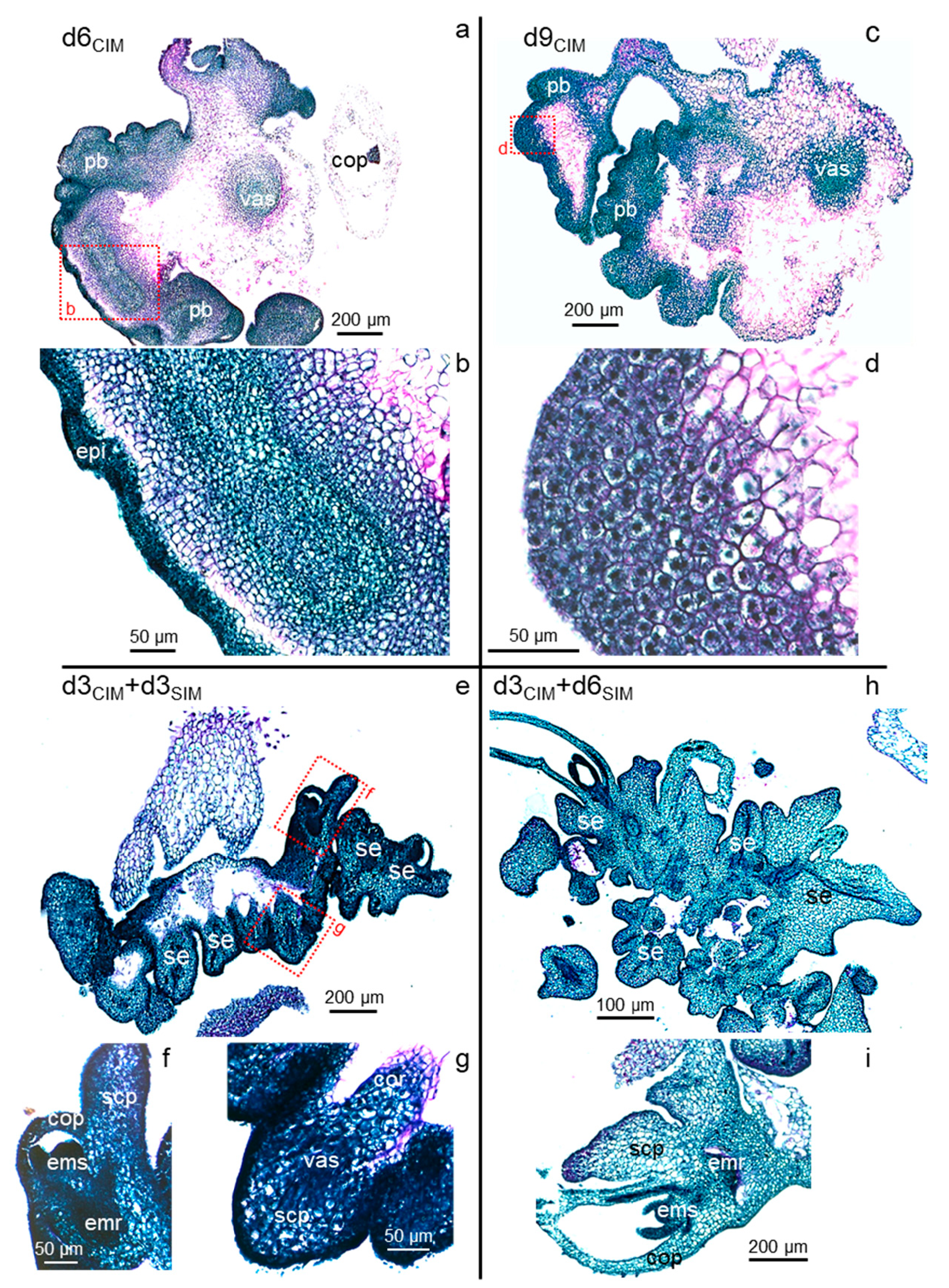
2.3. Prolonged Auxin Exposure Only Promotes the Further Proliferation of Established Scutellar Cell Populations
2.4. Somatic Embryos Can Be Fully Formed within Three Days of Cytokinin Induction and Evolve Rapidly Thereafter
2.5. Brachypodium Zygotic and Somatic Embryogenic Paths Are Very Similar
2.6. Taking Advantage of the Precise Timing of Pluripotency Acquisition and Morphogenesis Induction Following the Application of Exogenous Phytohormones
2.7. Prospective Applications of Express Somatic Embryogenesis
3. Materials and Methods
3.1. Plant Materials
3.2. Histological Analysis
3.3. RNA in Situ Hybridization
4. Supplementary Methods
4.1. Propidium Iodide Staining and Imaging of Bd Immature Embryos
4.2. Characterization of in Situ Hybridization Probes
4.3. RT-PCR Detection of WOX Gene Expression
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Litz, R.E.; Gray, D.J. Somatic embryogenesis for agricultural improvement. World J. Microbiol. Biotechnol. 1995, 4, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Stasolla, C.; Yeung, E.C. Recent advances in conifer somatic embryogenesis: Improving somatic embryo quality. Plant Cell Tissue Organ Cult. 2003, 74, 15–35. [Google Scholar] [CrossRef]
- Vicient, C.M.; Martínez, F.X. The potential uses of somatic embryogenesis in agroforestry are not limited to synthetic seed technology. Rev. Bras. Fisiol. Veg. 1998, 10, 1–12. [Google Scholar]
- Betekhtin, A.; Rojek, M.; Milewska-Hendel, A.; Gawecki, R.; Karcz, J.; Kurczynska, E.; Hasterok, R. Spatial distribution of selected chemical cell wall components in the embryogenic callus of Brachypodium distachyon. PLoS ONE 2016, 11, e0167426. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, E.J.; Koehler, A.D.; Rocha, D.I.; Vieira, L.M.; Pinheiro, M.V.M.; de Matos, E.M.; da Cruz, A.C.F.; da Silva, T.C.R.; Tanaka, F.A.O.; Nogueira, F.T.S.; et al. Morpho-histological, histochemical, and molecular evidences related to cellular reprogramming during somatic embryogenesis of the model grass Brachypodium distachyon. Protoplasma 2017, 5, 2017–2034. [Google Scholar] [CrossRef] [PubMed]
- Verdeil, J.-L.; Hocher, V.; Huet, C.; Grosdemange, F.; Escoute, J.; Ferrière, N.; Nicole, M. Ultrastructural changes in coconut calli associated with the acquisition of embryogenic competence. Ann. Bot. 2001, 88, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Jones, T.J.; Rost, T.L. The developmental anatomy and ultrastructural of somatic embryos from rice (Oryza sativa L.) scutellum ephitelial cells. Bot. Gaz. 1989, 150, 41–49. [Google Scholar] [CrossRef]
- Gliwicka, M.; Nowak, K.; Balazadeh, S.; Mueller-Roeber, B.; Gaj, M.D. Extensive modulation of the transcription factor transcriptome during somatic embryogenesis in Arabidopsis thaliana. PLoS ONE 2013, 8, e69261. [Google Scholar] [CrossRef] [Green Version]
- Cheng, W.H.; Zhu, H.G.; Tian, W.G.; Zhu, S.H.; Xiong, X.P.; Sun, Y.Q.; Zhu, Q.H.; Sun, J. De novo transcriptome analysis reveals insights into dynamic homeostasis regulation of somatic embryogenesis in upland cotton (G. hirsutum L.). Plant Mol. Biol. 2016, 92, 279–292. [Google Scholar] [CrossRef] [Green Version]
- Indoliya, Y.; Tiwari, P.; Chauhan, A.S.; Goel, R.; Shri, M.; Bag, S.K.; Chakrabarty, D. Decoding regulatory landscape of somatic embryogenesis reveals differential regulatory networks between japonica and indica rice subspecies. Sci. Rep. 2016, 6, 23050. [Google Scholar] [CrossRef] [Green Version]
- Gaj, M.D. Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul. 2004, 43, 27–47. [Google Scholar] [CrossRef]
- Jiménez, V.M.; Thomas, C. Participation of plant hormones in determination and progression of somatic embryogenesis. Plant Cell Monogr. 2005, 2, 103–118. [Google Scholar] [CrossRef]
- Francis, D.; Sorrell, D.A. The interface between the cell cycle and plant growth regulators: A mini review. Plant Growth Regul. 2001, 33, 1–12. [Google Scholar] [CrossRef]
- Moubayidin, L.; Di Mambro, R.; Sabatini, S. Cytokinin–auxin crosstalk. Trends Plant Sci. 2009, 14, 557–562. [Google Scholar] [CrossRef]
- Su, Y.H.; Liu, Y.B.; Zhang, X.S. Auxin–cytokinin interaction regulates meristem development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef]
- Kumar, S.; Ruggles, A.; Logan, S.; Mazarakis, A.; Tyson, T.; Bates, M.; Grosse, C.; Reed, D.; Li, Z.; Grimwood, J.; et al. Comparative transcriptomics of non-embryogenic and embryogenic callus in semi-recalcitrant and non-recalcitrant upland cotton lines. Plants 2021, 10, 1775. [Google Scholar] [CrossRef]
- Zhai, N.; Xu, L. Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nat. Plants 2021, 7, 1453–1460. [Google Scholar] [CrossRef]
- Chugh, A.; Khurana, P. Gene expression during somatic embryogenesis-recent advances. Curr. Sci. 2002, 86, 715–730. [Google Scholar]
- Karami, O.; Aghavaisi, B.; Pour, A.M. Molecular aspects of somatic-to-embryogenic transition in plants. J. Chem. Biol. 2009, 2, 177–190. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Zhang, X. Regulation of somatic embryogenesis in higher plants. Crit. Rev. Plant Sci. 2010, 29, 36–57. [Google Scholar] [CrossRef]
- Smertenko, A.; Bozhkov, P.V. Somatic embryogenesis: Life and death processes during apical and basal patterning. J. Exp. Bot. 2014, 65, 1343–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delporte, F.; Muhovski, Y.; Pretova, A.; Watillon, B. Analysis of expression profiles of selected genes associated with the regenerative property and the receptivity to gene transfer during somatic embryogenesis in Triticum aestivum L. Mol. Biol. Rep. 2013, 40, 5883–5906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horstman, A.; Bemer, M.; Boutilier, K. A transcriptional view on somatic embryogenesis. Regeneration 2017, 4, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Betekhtin, A.; Rojek, M.; Nowak, K.; Pinski, A.; Milewska-Hendel, A.; Kurczynska, E.; Doonan, J.H.; Hasterok, R. Cell wall epitopes and endoploidy as reporters of embryogenic potential in Brachypodium distachyon callus culture. Int. J. Mol. Sci. 2018, 19, 3811. [Google Scholar] [CrossRef] [Green Version]
- Zuo, J.; Niu, Q.W.; Frugis, G.; Chua, N.H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002, 30, 349–359. [Google Scholar] [CrossRef]
- Gallois, J.L.; Nora, F.R.; Mizukami, Y.; Sablowski, R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 2004, 18, 375–380. [Google Scholar] [CrossRef] [Green Version]
- Bouchabke-Coussa, O.; Obellianne, M.; Linderme, D.; Montes, E.; Maia-Grondard, A.; Vilaine, F.; Pannetier, C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013, 32, 675–686. [Google Scholar] [CrossRef]
- Arroyo-Herrera, A.; Ku Gonzalez, A.; Canche Moo, R.; Quiroz-Figueroa, F.R.; Loyola-Vargas, V.M.; Rodriguez-Zapata, L.C.; Burgeff D’Hondt, C.; Suárez-Solís, V.M.; Castaño, E. Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases in somatic embryogenesis. Plant Cell Tissue Organ Cult. 2008, 94, 171–180. [Google Scholar] [CrossRef]
- Kadri, A.; Grenier De March, G.; Guerineau, F.; Cosson, V.; Ratet, P. WUSCHEL Overexpression promotes callogenesis and somatic embryogenesis in Medicago truncatula Gaertn. Plants 2021, 10, 715. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Iwase, A.; Ito, T.; Tanaka, H.; Favero, D.S.; Kawamura, A.; Sakamoto, S.; Wakazaki, M.; Tameshige, T.; Fujii, H.; et al. Wound-inducible WUSCHEL-RELATED HOMEOBOX 13 is required for callus growth and organ reconnection. Plant Physiol. 2021, 188, kiab510. [Google Scholar] [CrossRef]
- Palovaara, J.; Hallberg, H.; Stasolla, C.; Hakman, I. Comparative expression pattern analysis of WUSCHEL-related homeobox 2 (WOX2) and WOX8/9 in developing seeds and somatic embryos of the gymnosperm Picea abies. N. Phytol. 2010, 188, 122–135. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [Green Version]
- Passarinho, P.; Ketelaar, T.; Xing, M.; van Arkel, J.; Maliepaard, C.; Hendriks, M.W.; Joosen, R.; Lammers, M.; Herdies, L.; den Boer, B.; et al. BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol. Biol. 2008, 68, 225–237. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 2016, 9, 1998–2015. [Google Scholar] [CrossRef] [Green Version]
- Jha, P.; Kumar, V. BABY BOOM (BBM): A candidate transcription factor gene in plant biotechnology. Biotechnol. Lett. 2018, 40, 1467–1475. [Google Scholar] [CrossRef]
- Ouakfaoui, S.E.; Schnell, J.; Abdeen, A.; Colville, A.; Labbe, H.; Han, S.; Baum, B.; Laberge, S.; Miki, B. Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol. Biol. 2010, 74, 313–326. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Wrobel-Marek, J.; Heidmann, I.; Horstman, A.; Chen, B.; Reis, R.; Angenent, G.C.; Boutilier, K. Auxin biosynthesis maintains embryo identity and growth during BABY BOOM-induced somatic embryogenesis. Plant Physiol. 2021, 188, kiab558. [Google Scholar] [CrossRef]
- Guillon, F.; Larré, C.; Petipas, F.; Berger, A.; Moussawi, J.; Rogniaux, H.; Santoni, A.; Saulnier, L.; Jamme, F.; Miquel, M.; et al. A comprehensive overview of grain development in Brachypodium distachyon variety Bd21. J. Exp. Bot. 2012, 2, 739–755. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.; Begcy, K.; Dresselhaus, T.; Sun, M. Does early embryogenesis in eudicots and monocots involve the same mechanism and molecular players? Plant Physiol. 2017, 173, 130–142. [Google Scholar] [CrossRef] [Green Version]
- Bablak, P.; Draper, J.; Davey, M.R.; Lynch, P.T. Plant regeneration and micropropagation of Brachypodium distachyon. Plant Cell Tissue Organ Cult. 1995, 42, 97–107. [Google Scholar] [CrossRef]
- Draper, J.; Mur, L.A.J.; Jenkins, G.; Ghosh-Biswas, G.C.; Bablak, P.; Hasterok, R.; Routledge, A.P.M. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001, 127, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Hill, T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 2008, 27, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Brandstädter, J.; Rossbach, C.; Theres, K. The pattern of histone H4 expression in the tomato shoot apex changes during development. Planta 1994, 92, 69–74. [Google Scholar] [CrossRef]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91–95. [Google Scholar] [CrossRef]
- Hu, W.W.; Gong, H.; Pua, E.C. The pivotal roles of the plant S-adenosylmethionine decarboxylase 5’ untranslated leader sequence in regulation of gene expression at the transcriptional and posttranscriptional levels. Plant Physiol. 2005, 138, 276–286. [Google Scholar] [CrossRef] [Green Version]
- Halperin, H.; Wetherell, D.F. Adventive embryony in tissue cultures of the wild carrot, Daucus carota. Am. J. Bot. 1964, 51, 274–283. [Google Scholar] [CrossRef]
- Hao, Z.; Zhang, Z.; Xiang, D.; Venglat, P.; Chen, J.; Gao, P.; Datla, R.; Weijers, D. Conserved, divergent and heterochronic gene expression during Brachypodium and Arabidopsis embryo development. Plant Reprod. 2021, 34, 207–224. [Google Scholar] [CrossRef]
- Alves, S.C.; Worland, B.; Thole, V.; Snape, J.W.; Bevan, M.W.; Vain, P. A protocol for Agrobacterium-mediated transformation of Brachypodium distachyon community standard line Bd21. Nat. Protoc. 2009, 4, 638–649. [Google Scholar] [CrossRef]
- Christiansen, P.; Andersen, C.H.; Didion, T.; Folling, M.; Nielsen, K.K. A rapid and efficient transformation protocol for the grass Brachypodium distachyon. Plant Cell Rep. 2005, 23, 751–758. [Google Scholar] [CrossRef]
- Truernit, E.; Bauby, H.; Dubreucq, B.; Grandjean, O.; Runions, J.; Barthélémy, J.; Palauqui, J.C. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 2008, 20, 1494–1503. [Google Scholar] [CrossRef] [Green Version]
- Fisher, D.B. Protein staining of ribboned epon sections for light microscopy. Histochemie 1968, 16, 92–96. [Google Scholar] [CrossRef]
- Coen, O.; Lu, J.; Xu, W.; De Vos, D.; Péchoux, C.; Domergue, F.; Grain, D.; Lepiniec, L.; Magnani, E. Deposition of a cutin apoplastic barrier separating seed maternal and zygotic tissues. BMC Plant Biol. 2019, 19, 304. [Google Scholar] [CrossRef] [Green Version]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wehbi, H.; Soulhat, C.; Morin, H.; Bendahmane, A.; Hilson, P.; Bouchabké-Coussa, O. One-Week Scutellar Somatic Embryogenesis in the Monocot Brachypodium distachyon. Plants 2022, 11, 1068. https://doi.org/10.3390/plants11081068
Wehbi H, Soulhat C, Morin H, Bendahmane A, Hilson P, Bouchabké-Coussa O. One-Week Scutellar Somatic Embryogenesis in the Monocot Brachypodium distachyon. Plants. 2022; 11(8):1068. https://doi.org/10.3390/plants11081068
Chicago/Turabian StyleWehbi, Houssein, Camille Soulhat, Halima Morin, Abdelhafid Bendahmane, Pierre Hilson, and Oumaya Bouchabké-Coussa. 2022. "One-Week Scutellar Somatic Embryogenesis in the Monocot Brachypodium distachyon" Plants 11, no. 8: 1068. https://doi.org/10.3390/plants11081068






