Centaurea Subsect. Phalolepis (Compositae, Cardueae): A Case Study of Mountain-Driven Allopatric Speciation in the Mediterranean Peninsulas
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Material
2.2. Genetic Analyses
2.2.1. Microsatellite Loci Data
2.2.2. Genetic Structure and Allopatric Speciation Parameters
2.3. Testing the MGH Model
3. Results
3.1. Genetic Structure and Allopatric Speciation Parameters
3.2. Testing the Mountain–Geobiodiversity Hypothesis (MGH) Model
4. Discussion
4.1. Genetic Parameters and Speciation
4.2. Mountains and Speciation: Is the MGH Model Applicable to Our Case Study?
4.3. Climate Niche Breadth (CNB): A New Index for Representing Climate Niche Breadth at the Global Scale
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Barthlott, W.; Mutke, J.; Rafiqpoor, D.; Kier, G.; Kreft, H. Global centers of vascular plant diversity. Nov. Acta Leopoldina 2005, 92, 61–83. [Google Scholar]
- Rahbek, C.; Borregaard, M.K.; Colwell, R.K.; Dalsgaard, B.; Holt, B.G.; Morueta-Holme, N.; Nogués-Bravo, D.; Whittaker, R.J.; Fjeldså, J. Humboldt’s enigma: What causes global patterns of mountain biodiversity? Science 2019, 365, 1108–1113. [Google Scholar] [CrossRef]
- Mosbrugger, V.; Favre, A.; Muellner-Riehl, A.N.; Päckert, M.; Mulch, A. Cenozoic evolution of geo-biodiversity in the Tibeto-Himalayan region. In Mountains, Climate, and Biodiversity; Hoorn, C., Perrigio, A., Antonelli, A., Eds.; Wiley-Blackwell: Chichester, UK, 2018; pp. 429–448. [Google Scholar]
- Muellner-Riehl, A.N.; Schnitzler, J.; Kissling, W.D.; Mosbrugger, V.; Rijsdijk, K.F.; Seijmonsbergen, A.C.; Versteegh, H.; Favre, A. Origins of global mountain plant biodiversity: Testing the ‘mountain-geobiodiversity hypothesis’. J. Biogeogr. 2019, 46, 2826–2838. [Google Scholar] [CrossRef] [Green Version]
- Médail, F.; Quézel, P. Hot-spots analysis for conservation of plant biodiversity in the Mediterranean Basin. Ann. Missouri Bot. Gard. 1997, 84, 112–127. [Google Scholar] [CrossRef]
- Thompson, J.D. Plant Evolution in the Mediterranean; Oxford University Press: Oxford, UK, 2005. [Google Scholar]
- Médail, F.; Diadema, K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J. Biogeog. 2009, 36, 1333–1345. [Google Scholar] [CrossRef]
- Casazza, G.; Barberis, G.; Minuto, L. Ecological characteristics and rarity of endemic plants of the Italian Maritime Alps. Biol. Conserv. 2005, 123, 361–371. [Google Scholar] [CrossRef]
- Kougioumoutzis, K.; Kokkoris, I.P.; Panitsa, M.; Kallimanis, A.; Strid, A.; Dimopoulos, P. Plant endemism centres and biodiversity hotspots in Greece. Biology 2021, 10, 72. [Google Scholar] [CrossRef]
- López-Vinyallonga, S.; López-Pujol, J.; Constantinidis, T.; Susanna, A.; Garcia-Jacas, N. Mountains and refuges: Genetic structure and evolutionary history in closely related, endemic Centaurea in continental Greece. Mol. Phylogenet. Evol. 2015, 92, 243–254. [Google Scholar] [CrossRef]
- López-Pujol, J.; López-Vinyallonga, S.; Susanna, A.; Ertuğrul, K.; Uysal, T.; Tugay, O.; Guetat, A.; Garcia-Jacas, N. Speciation and genetic diversity in Centaurea subsect. Phalolepis in Anatolia. Sci. Rep. 2016, 6, 37818. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Jacas, N.; López-Pujol, J.; López-Vinyallonga, S.; Janaćković, P.; Susanna, A. Centaurea subsect. Phalolepis in Southern Italy: Ongoing speciation or species overestimation? Genetic evidence based on SSRs analyses. Syst. Biodivers. 2019, 17, 93–109. [Google Scholar] [CrossRef]
- Requena, J.; López-Pujol, J.; Carnicero, P.; Susanna, A.; Garcia-Jacas, N. The Centaurea alba complex in the Iberian Peninsula: Gene flow, introgression, and blurred genetic boundaries. Pl. Syst. Evol. 2020, 306, 43. [Google Scholar] [CrossRef]
- Hewitt, G.M. Mediterranean Peninsulas: The evolution of hotspots. In Biodiversity Hotspots: Distribution and Protection of Conservation Priority Areas; Zachos, F.E., Habel, J.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 123–147. [Google Scholar]
- Davis, P.H. Distribution patterns in Anatolia with particular references to endemism. In Plant Life of South-West Asia; Davis, P.H., Harper, P.C., Hedge, I.C., Eds.; Botanical Society of Edinburgh: Edinburgh, UK, 1971; pp. 15–27. [Google Scholar]
- Strid, A. The mountain flora of Greece with special reference to the Anatolian element. Proc. Roy. Soc. Edinb. 1986, 89, 59–68. [Google Scholar] [CrossRef]
- Tzedakis, P.C.; Hooghiemstra, H.; Pälike, H. The last 1.35 million years at Tenaghi Philippon: Revised chronostratigraphy and long-term vegetation trends. Quat. Sci. Rev. 2006, 25, 3416–3430. [Google Scholar] [CrossRef]
- Georghiou, K.; Delipetrou, P. Patterns and traits of the endemic plants of Greece. Bot. J. Linn. Soc. 2010, 162, 130–153. [Google Scholar] [CrossRef] [Green Version]
- Tzedakis, P.C.; Lawson, I.T.; Frogley, M.R.; Hewitt, G.M.; Preece, R.C. Buffered tree population changes in a Quaternary refugium: Evolutionary implications. Science 2002, 297, 2044–2047. [Google Scholar] [CrossRef] [Green Version]
- Brullo, S.; Scelsi, F.; Spampinato, G. La Vegetazione del Aspromonte-Studio Fitosociologico; Laruffa Editore: Reggio Calabria, Italy, 2001. [Google Scholar]
- Garcia-Jacas, N.; Soltis, P.S.; Font, M.; Soltis, D.E.; Vilatersana, R.; Susanna, A. The polyploid series of Centaurea toletana: Glacial migrations and introgression revealed by nrDNA and cpDNA sequence analyzes. Mol. Phylogenet. Evol. 2009, 52, 377–394. [Google Scholar] [CrossRef]
- Herrando-Moraira, S.; Nualart, N.; Galbany-Casals, M.; Garcia-Jacas, N.; Ohashi, H.; Matsui, T.; Susanna, A.; Tang, C.Q.; López-Pujol, J. Climate Stability Index maps, a global high resolution cartography of climate stability from Pliocene to 2100. Sci. Data 2022, 9, 48. [Google Scholar] [CrossRef]
- López-Alvarado, J.; Mameli, G.; Farris, E.; Susanna, A.; Filigheddu, R.; Garcia-Jacas, N. Islands as a crossroad of evolutionary lineages: A case study of Centaurea sect Centaurea (Compositae) from Sardinia (Mediterranean Basin). PLoS ONE 2020, 15, e0228776. [Google Scholar] [CrossRef] [Green Version]
- Fréville, H.; Imbert, E.; Justy, F.; Vitalis, R.; Olivieri, I. Isolation and characterization of microsatellites in the endemic species Centaurea corymbosa Pourret (Asteraceae) and other related species. Mol. Ecol. 2000, 9, 1671–1672. [Google Scholar] [CrossRef]
- Marrs, R.A.; Hufbauer, R.A.; Bogdanowicz, S.M.; Sforza, R. Nine polymorphic microsatellite markers in Centaurea stoebe L. subspecies C. s. stoebe and C. s. micranthos (S. G. Gmelin ex Gugler) Hayek. and C. diffusa Lam. (Asteraceae). Mol. Ecol. Notes 2006, 6, 897–899. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation for STRUCTURE Software, Version 2.3; Department of Human Genetics, University of Chicago: Chicago, IL, USA, 2010.
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 4, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Molec. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Berg, E.E.; Hamrick, J.L. Quantification of genetic diversity at allozyme loci. Can. J. For. Res. 1997, 27, 415–424. [Google Scholar] [CrossRef]
- Chapuis, M.-P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinf. Online 2005, 1, 47–50. [Google Scholar] [CrossRef] [Green Version]
- Wilson, G.A.; Rannala, B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 2003, 163, 1177–1191. [Google Scholar] [CrossRef]
- Beerli, P. Comparison of Bayesian and maximum likelihood inference of population genetic parameters. Bioinformatics 2006, 22, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Beerli, P.; Felsenstein, J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl. Acad. Sci. USA 2001, 98, 4563–4568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.L.; Hill, D.J.; Dolan, A.M.; Carnaval, A.C.; Haywood, A.M. PaleoClim, high spatial resolution paleoclimate surfaces for global land areas. Sci. Data 2018, 5, 180254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barres, L.; Sanmartín, I.; Anderson, C.L.; Susanna, A.; Buerki, S.; Galbany-Casals, M.; Vilatersana, R. Reconstructing the evolution and biogeographic history of tribe Cardueae (Compositae). Am. J. Bot. 2013, 100, 867–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilpold, A.; Vilatersana, R.; Susanna, A.; Meseguer, A.S.; Boršić, I.; Constantinidis, T.; Filigheddu, R.; Romaschenko, K.; Suárez-Santiago, V.N.; Tugay, O.; et al. Phylogeny of the Centaurea group (Centaurea, Compositae)—Geography is a better predictor than morphology. Mol. Phylogenet. Evol. 2014, 77, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling. R package version 3.5–15. 2022. Available online: https://CRAN.R-project.org/package=raster (accessed on 8 August 2022).
- Amatulli, G.; Domisch, S.; Tuanmu, M.-N.; Parmentier, B.; Ranipeta, A.; Malczyk, J.; Jetz, W. A suite of global, cross-scale topographic variables for environmental and biodiversity modeling. Sci. Data 2018, 5, 180040. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 15 February 2022).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Software 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A.; Mundt, F. factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.7. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 8 August 2022).
- Jolliffe, I.T. Discarding variables in a principal component analysis. I: Artificial data. J. R. Stat. Soc. 1972, 21, 160–173. [Google Scholar] [CrossRef]
- López-Pujol, J.; Garcia-Jacas, N.; Susanna, A.; Vilatersana, R. Should we conserve pure species or hybrid species? Delimiting hybridization and introgression in the Iberian endemic Centaurea podospermifolia. Biol. Conserv. 2012, 152, 271–279. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Wendel, J.F. Introgression and its consequences in plants. In Hybrid Zones and the Evolutionary Process; Harrison, R.G., Ed.; Oxford University Press: New York, NY, USA, 1993; pp. 70–109. [Google Scholar]
- Ellstrand, N.C. Gene flow by pollen: Implications for plant conservation genetics. Oikos 1992, 63, 77–86. [Google Scholar] [CrossRef]
- Stap, L.B.; de Boer, B.; Ziegler, M.; Bintanja, R.; Lourens, L.J.; van de Wal, R.S.W. CO2 over the past 5 million years: Continuous simulation and new δ11B-based proxy data. Earth Planet. Sci. Lett. 2016, 439, 1–10. [Google Scholar] [CrossRef]
- Willeit, M.; Ganopolski, A.; Calov, R.; Brovkin, V. Mid-Pleistocene transition in glacial cycles explained by declining CO2 and regolith removal. Sci. Adv. 2019, 5, eaav7337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rull, V. Microrefugia. J. Biogeogr. 2009, 36, 481–484. [Google Scholar] [CrossRef]
- Irl, S.D.; Harter, D.E.; Steinbauer, M.J.; Gallego-Puyol, D.; Fernández-Palacios, J.M.; Jentsch, A.; Beierkuhnlein, C. Climate vs. topography–spatial patterns of plant species diversity and endemism on a high-elevation island. J. Ecol. 2015, 103, 621–1633. [Google Scholar] [CrossRef]
- Zhao, J.-L.; Gugger, P.F.; Xia, Y.-M.; Li, Q.-J. Ecological divergence of two closely related Roscoea species associated with late Quaternary climate change. J. Biogeog. 2016, 43, 1990–2001. [Google Scholar] [CrossRef]
- López-Pujol, J.; Zhang, F.-M.; Sun, H.-Q.; Ying, T.-S.; Ge, S. Mountains of southern China as ‘plant museums’ and ‘plant cradles’: Evolutionary and conservation insights. Mount. Res. Devel. 2011, 31, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Sosa, V.; De-Nova, J.A.; Vásquez-Cruz, M. Evolutionary history of the flora of Mexico: Dry forests cradles and museums of endemism. J. Syst. Evol. 2018, 56, 523–536. [Google Scholar] [CrossRef]
- Dagallier, L.P.M.; Janssens, S.B.; Dauby, G.; Blach-Overgaard, A.; Mackinder, B.A.; Droissart, V.; Svenning, J.C.; Sosef, M.S.M.; Stévart, T.; Harris, D.J.H.; et al. Cradles and museums of generic plant diversity across tropical Africa. New Phytol. 2020, 225, 2196–2213. [Google Scholar] [CrossRef] [Green Version]
- Valencia, B.G.; Matthews-Bird, F.; Urrego, D.H.; Williams, J.J.; Gosling, W.D.; Bush, M. Andean microrefugia: Testing the Holocene to predict the Anthropocene. New Phytol. 2016, 212, 510–522. [Google Scholar] [CrossRef]
- Gubler, M.; Henne, P.D.; Schwörer, C.; Boltshauser-Kaltenrieder, P.; Lotter, A.F.; Brönnimann, S.; Tinner, W. Microclimatic gradients provide evidence for a glacial refugium for temperate trees in a sheltered hilly landscape of Northern Italy. J. Biogeogr. 2018, 45, 2564–2575. [Google Scholar] [CrossRef]
- Rull, V. Pantepui. In Encyclopedia of Islands; Gillespie, R., Clague, D., Eds.; University of California Press: Berkeley, CA, USA, 2009; pp. 717–720. [Google Scholar]


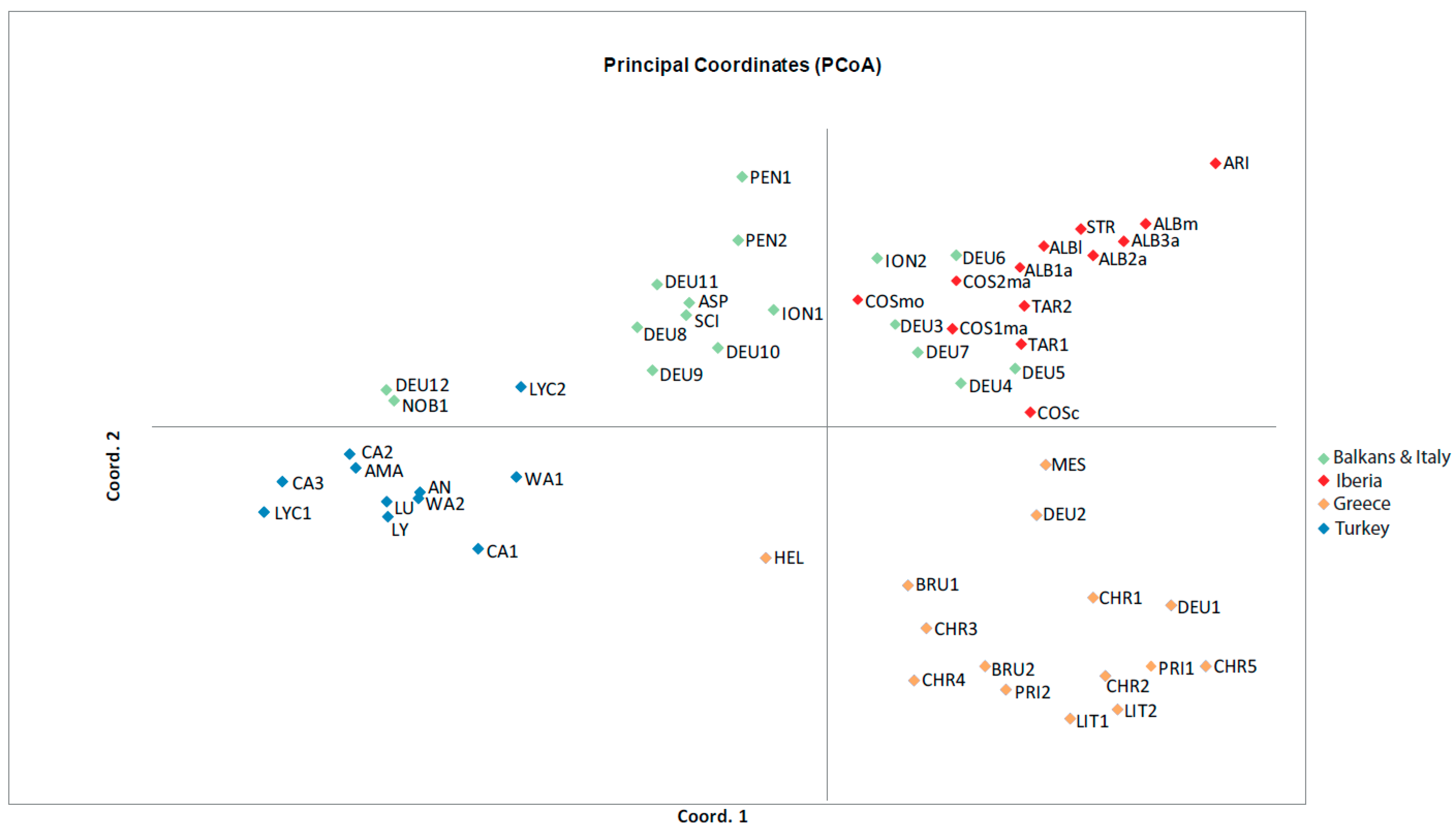

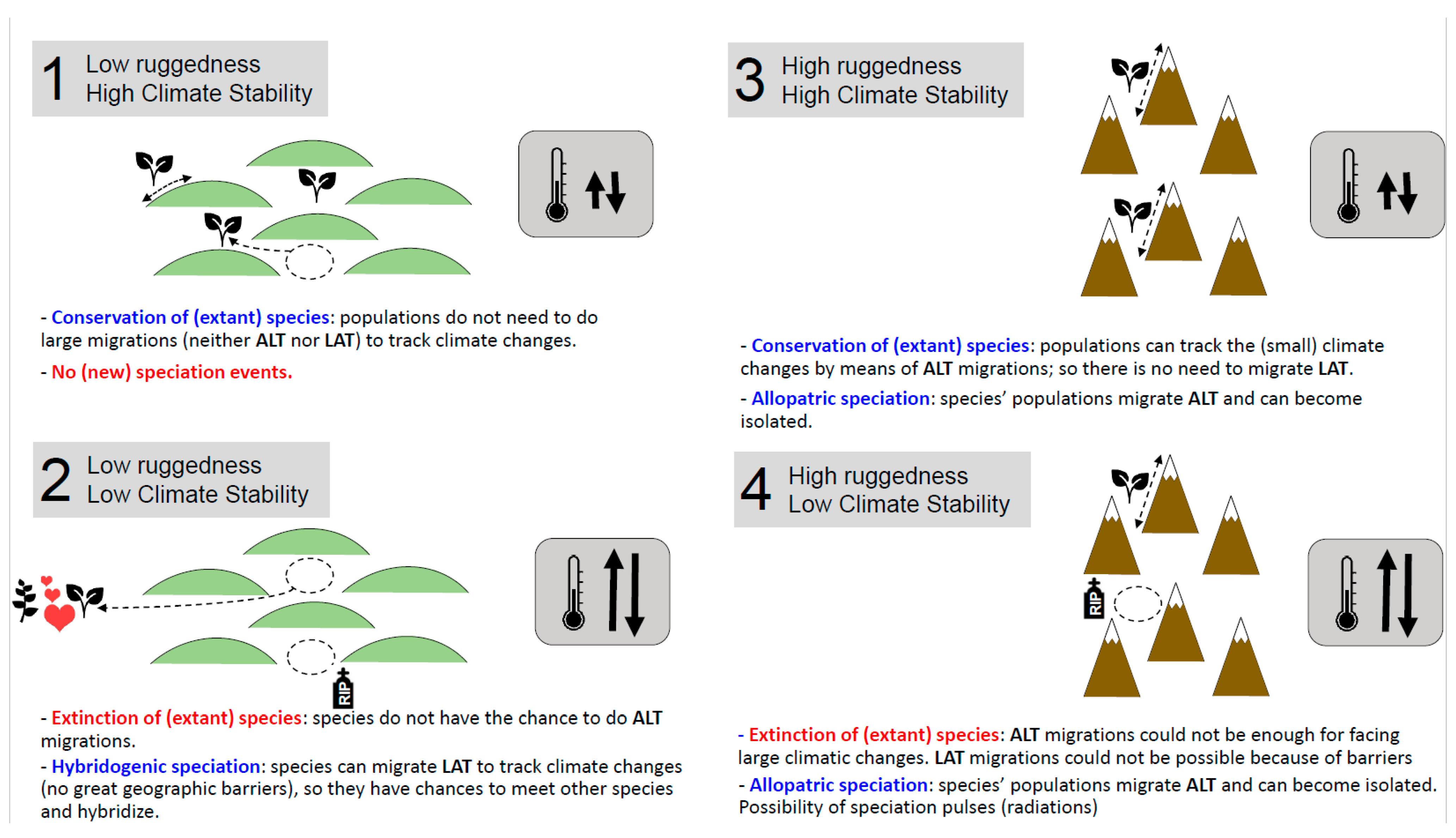

| Regions | Species n° | TRI ** | CSI ** | CNB ** | Genetic Diversity (He) | Genetic Differentiation (FST) | Past Gene Flow (Nm) | Recent Gene Flow (m) |
|---|---|---|---|---|---|---|---|---|
| Turkey | 9(7) * | 38.53/33.71 | 0.128/0.129 | 0.124/0.108 | 0.580 | 0.198 | 0.466 | 0.0044 |
| Greece | 22(7) * | 47.89/38.83 | 0.129/0.130 | 0.136/0.123 | 0.587 | 0.243 | 0.534 | 0.0024 |
| Italian Peninsula/Northern Balkans | 10(6) * | 32.45/28.69 | 0.172/0.171 | 0.125/0.103 | 0.662 | 0.232 | 0.645 | 0.0084 |
| Iberian Peninsula | 2(2) * | 27.86/29.41 | 0.157/0.157 | 0.096/0.099 | 0.645 | 0.176 | 7.475 | 0.0526 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Jacas, N.; López-Pujol, J.; Nualart, N.; Herrando-Moraira, S.; Romaschenko, K.; Ren, M.-X.; Susanna, A. Centaurea Subsect. Phalolepis (Compositae, Cardueae): A Case Study of Mountain-Driven Allopatric Speciation in the Mediterranean Peninsulas. Plants 2023, 12, 11. https://doi.org/10.3390/plants12010011
Garcia-Jacas N, López-Pujol J, Nualart N, Herrando-Moraira S, Romaschenko K, Ren M-X, Susanna A. Centaurea Subsect. Phalolepis (Compositae, Cardueae): A Case Study of Mountain-Driven Allopatric Speciation in the Mediterranean Peninsulas. Plants. 2023; 12(1):11. https://doi.org/10.3390/plants12010011
Chicago/Turabian StyleGarcia-Jacas, Núria, Jordi López-Pujol, Neus Nualart, Sonia Herrando-Moraira, Konstantin Romaschenko, Ming-Xun Ren, and Alfonso Susanna. 2023. "Centaurea Subsect. Phalolepis (Compositae, Cardueae): A Case Study of Mountain-Driven Allopatric Speciation in the Mediterranean Peninsulas" Plants 12, no. 1: 11. https://doi.org/10.3390/plants12010011







