Genetic and Pomological Determination of the Trueness-to-Type of Sweet Cherry Cultivars in the German National Fruit Genebank
Abstract
1. Introduction
2. Results
2.1. Characterization of Trueness-to-Type by Pomological and Molecular Characterization
2.2. Discrimination Power of the SSRs and Genetic Diversity
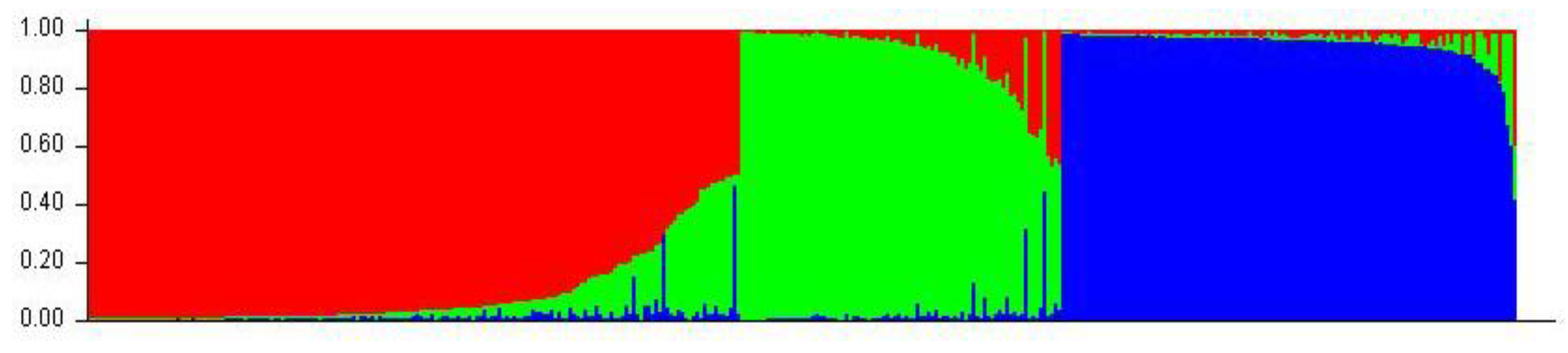
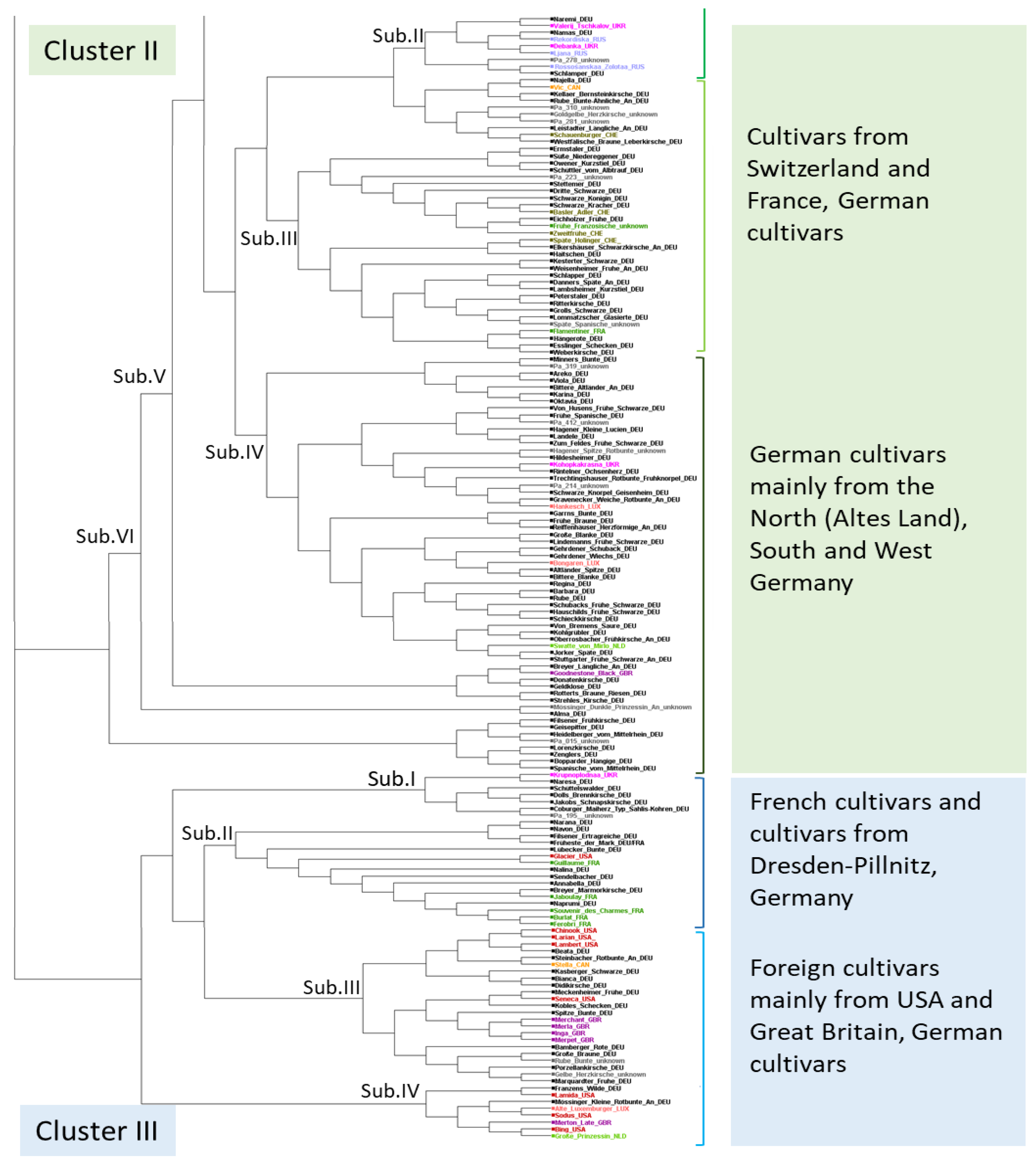
2.3. Genetic Structure within the Unique Cherry Cultivars
2.4. Parentage Analysis
3. Discussion
3.1. Pomological Differentiation of Cultivars
3.2. Choice of Markers
3.3. Genetic Differentiation of Cultivars in the German Cherry Collection
3.4. Diversity Parameters, Genetic Structure, Geographic Origin
3.5. Parentage Analysis
4. Materials and Methods
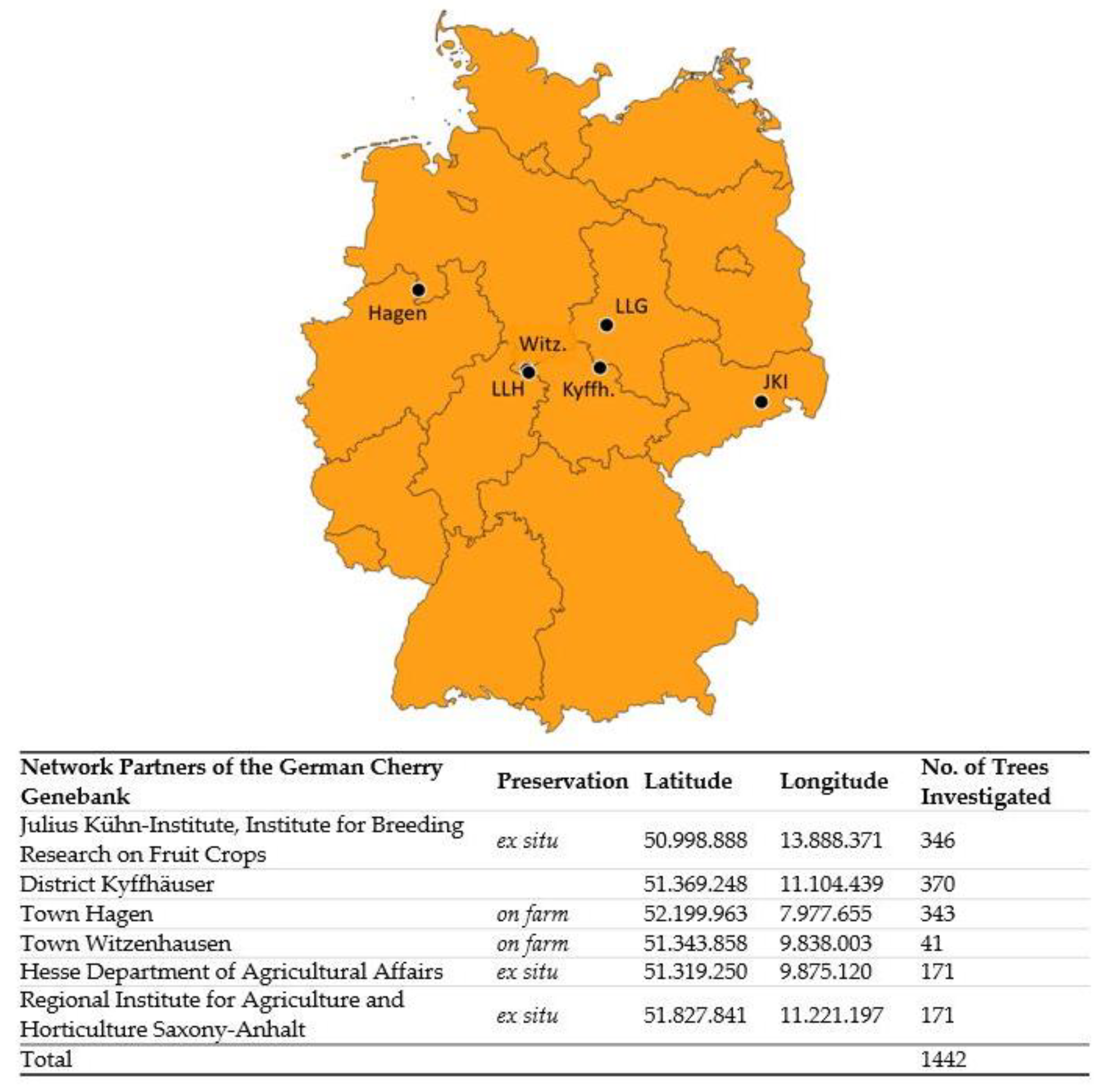
4.1. Plant Material
4.2. Pomological Characterization
4.3. DNA Extraction and Genetic Analysis
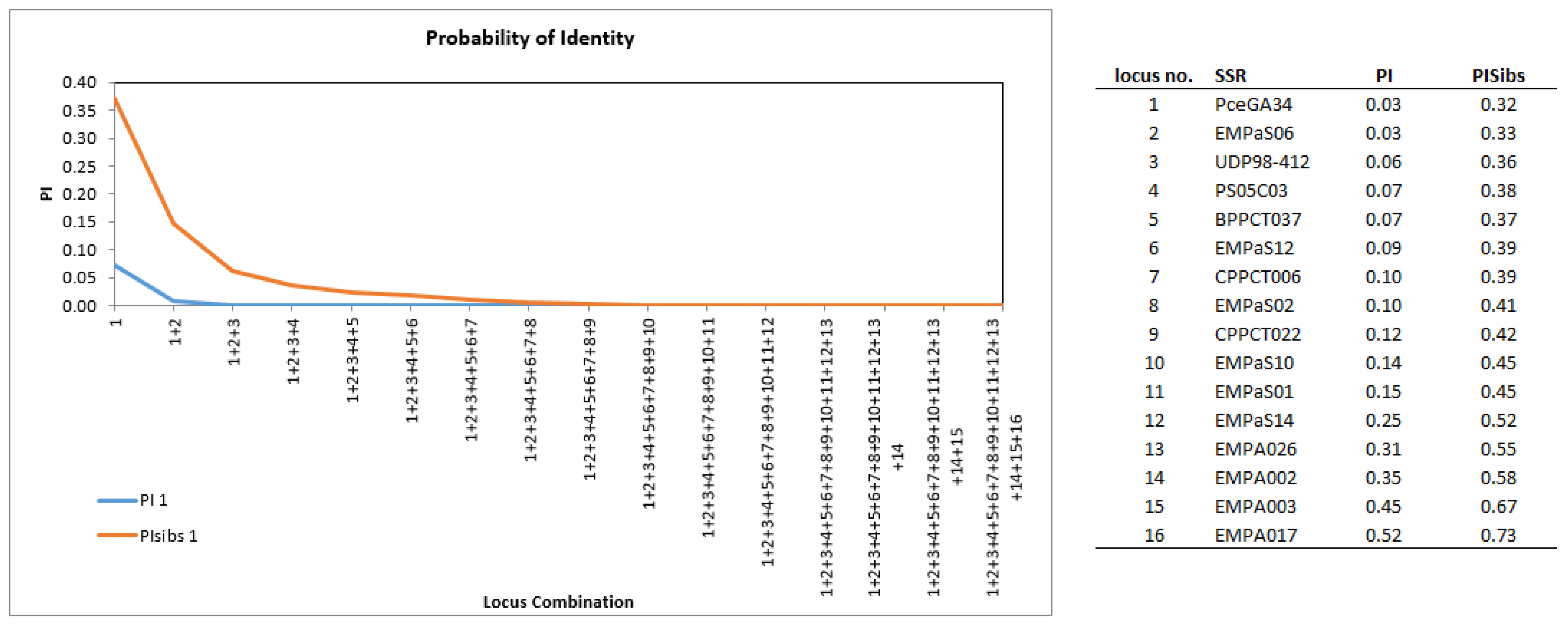
4.4. Probability of Identity (PI) Calculation
4.5. Assessment on Cultivar ‘Trueness-to-Type’
4.6. Diversity Parameter, STRUCTURE Analysis and Phylogenetic Tree Construction
4.7. Parentage Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Serres-Giardi, L.; Dufour, J.; Russell, K.; Buret, C.; Laurens, F.; Santi, F. Natural triploids of wild cherry. Can. J. For. Res. 2010, 40, 1951–1961. [Google Scholar] [CrossRef]
- Dirlewanger, E.; Claverie, J.; Iezzoni, A.; Wünsch, A. Sweet and Sour Cherries: Linkage Maps, QTL Detection and Marker Assisted Selection. In Genetics and Genomics of Rosaceae; Plant Genetics and Genomics: Crops and Models; Folta, K.M., Gardiner, S.E., Eds.; Springer: New York, NY, USA, 2009; Volume 6, pp. 31–291. [Google Scholar] [CrossRef]
- Fernandez i Marti, A.; Athanson, B.; Koepke, T.; Font i Forcada, C.; Dhingra, A.; Oraguzie, N. Genetic Diversity and Relatedness of Sweet Cherry (Prunus Avium L.) Cultivars Based on Single Nucleotide Polymorphic Markers. Front. Plant Sci. 2012, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Truchseß von Wetzhausen, C. Systematische Classification und Beschreibung der Kirschensorten; Friedrich Timotheus Heim: Stuttgard, Germany, 1819; p. 692. [Google Scholar]
- Quero-García, J.; Schuster, M.; López-Ortega, G.; Charlot, G. Sweet cherry varieties and improvement. In Cherries: Botany, Production and Uses; CAB International: Boston, MA, USA, 2017; pp. 60–94. [Google Scholar]
- Braun-Lüllemann, A.; Bannier, H.-J. Alte Süßkirschensorten; Bundesminsterium für Ernährung, Landwirtschaft und Verbraucherschutz: Bonn, Germany, 2010; ISBN 978-3-00-030878-9. [Google Scholar]
- Flachowsky, H.; Höfer, M. The German Fruit Genebank, a decentral network for sustainable preservation of fruit genetic resources. J. Für Kult. 2010, 62, 9–16. [Google Scholar] [CrossRef]
- Höfer, M.; Flachowsky, H.; Hanke, M.V. German Fruit Genebank–looking back 10 years after launching a national network for sustainable preservation of fruit genetic resources. J. Für Kult. 2019, 71, 41–51. [Google Scholar] [CrossRef]
- Flachowsky, H.; Hanke, M.-V. The ‘German National Fruit Genebank’, a first review five years after launching. In Proceedings of the Ist International Symposium on Fruit Culture and Its Traditional Knowledge along Silk Road Countries: Tbilisi, Georgia, Yerevan, Armenia, 4–8 November 2013; 2014. Volume 1032, pp. 227–230. [Google Scholar] [CrossRef]
- Clarke, J.B.; Tobutt, K.R. A standard set of accessions, microsatellites and genotypes for harmonising the fingerprinting of cherry collections for the ECPGR. Acta Hortic. 2009, 814, 615–618. [Google Scholar] [CrossRef]
- Höfer, M.; Braun-Lüllemann, A.; Schiffler, J.; Schuster, M.; Flachowsky, H. Pomological and Molecular Characterization of Sweet Cherry Cultivars (Prunus avium L.) of the German Fruit Genebank; OpenAgrar Repository: 2021. Available online: https://www.openagrar.de/: (accessed on 14 November 2022). [CrossRef]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- De Rogatis, A.; Ferrazzini, D.; Ducci, F.; Guerri, S.; Carnevale, S.; Belletti, P. Genetic variation in Italian wild cherry (Prunus avium L.) as characterized by nSSR markers. Forestry 2013, 86, 391–400. [Google Scholar] [CrossRef][Green Version]
- Lassois, L.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; hibrand-saint Oyant, L.; Poncet, C.; Lasserre-Zuber, P.; Feugey, L.; Durel, C.-E. Genetic Diversity, Population Structure, Parentage Analysis, and Construction of Core Collections in the French Apple Germplasm Based on SSR Markers. Plant Mol. Biol. Report. 2016, 34, 827–844. [Google Scholar] [CrossRef]
- Patzak, J.; Henychova, A.; Paprstein, F.; Sedlak, J. Evaluation of genetic variability within sweet cherry (Prunus avium L.) genetic resources by molecular SSR markers. Acta Sci. Pol.-Hortoru 2019, 18, 157–165. [Google Scholar] [CrossRef]
- Liu, C.L.; Qi, X.L.; Song, L.L.; Li, Y.H.; Li, M. Species identification, genetic diversity and population structure of sweet cherry commercial cultivars assessed by SSRs and the gametophytic selfincompatibility locus. Sci. Hortic. 2018, 237, 28–35. [Google Scholar] [CrossRef]
- Farsad, A.; Esna-Ashari, M. Genetic diversity of some Iranian sweet cherry (Prunus avium) cultivars using microsatellite markers and morphological traits. Cytol. Genet. 2016, 50, 8–19. [Google Scholar] [CrossRef]
- Ivanovych, Y.; Volkov, R. Genetic relatedness of sweet cherry (Prunus avium L.) cultivars from Ukraine determined by microsatellite markers. J. Hortic. Sci. Biotechnol. 2018, 93, 64–72. [Google Scholar] [CrossRef]
- Krmpot, T.; Rados, L.; Vokurka, A. Genetic characterisation of autochthonous sweet cherry genotypes (Prunus avium L.) using ssr markers. Genet.-Belgrade 2020, 52, 43–53. [Google Scholar] [CrossRef]
- Ordidge, M.; Litthauer, S.; Venison, E.; Blouin-Delmas, M.; Fernandez-Fernandez, F.; Höfer, M.; Kaegi, C.; Kellerhals, M.; Marchese, A.; Mariette, S.; et al. Towards a Joint International Database: Alignment of SSR Marker Data for European Collections of Cherry Germplasm. Plants 2021, 10, 1243. [Google Scholar] [CrossRef] [PubMed]
- Lacis, G.; Rashal, I.; Ruisa, S.; Trajkovski, V.; Iezzoni, A.F. Assessment of genetic diversity of Latvian and Swedish sweet cherry (Prunus avium L.) genetic resources collections by using SSR (microsatellite) markers. Sci. Hortic. 2009, 121, 451–457. [Google Scholar] [CrossRef]
- Vaughan, S.P.; Russell, K. Characterization of novel microsatellites and development of multiplex PCR for large-scale population studies in wild cherry, Prunus avium. Mol. Ecol. Notes 2004, 4, 429–431. [Google Scholar] [CrossRef]
- Waits, L.P.; Luikart, G.; Taberlet, P. Estimating the probability of identity among genotypes in natural populations: Cautions and guidelines. Mol. Ecol. 2001, 10, 249–256. [Google Scholar] [CrossRef]
- Liang, W.; Dondini, L.; De Franceschi, P.; Paris, R.; Sansavini, S.; Tartarini, S. Genetic Diversity, Population Structure and Construction of a Core Collection of Apple Cultivars from Italian Germplasm. Plant Mol. Biol. Rep. 2015, 33, 458–473. [Google Scholar] [CrossRef]
- Gross, B.L.; Henk, A.D.; Richards, C.M.; Fazio, G.; Volk, G.M. Genetic diversity in Malus ×domestica (Rosaceae) through time in response to domestication. Am. J. Bot. 2014, 101, 1770–1779. [Google Scholar] [CrossRef]
- Zarouri, B.; Vargas, A.M.; Gaforio, L.; Aller, M.; de Andrés, M.T.; Cabezas, J.A. Whole-genome genotyping of grape using a panel of microsatellite multiplex PCRs. Tree Genet. Genomes 2015, 11, 17. [Google Scholar] [CrossRef]
- Blais, J.; Lavoie, S.B.; Giroux, S.; Bussières, J.; Lindsay, C.; Dionne, J.; Laroche, M.; Giguère, Y.; Rousseau, F. Risk of Misdiagnosis Due to Allele Dropout and False-Positive PCR Artifacts in Molecular Diagnostics: Analysis of 30,769 Genotypes. J. Mol. Diagn. 2015, 17, 505–514. [Google Scholar] [CrossRef]
- Cummings, S.M.; McMullan, M.; Joyce, D.A.; van Oosterhout, C. Solutions for PCR, cloning and sequencing errors in population genetic analysis. Conserv. Genet. 2010, 11, 1095–1097. [Google Scholar] [CrossRef]
- van de Wouw, M.; van Treuren, R.; van Hintum, T. Authenticity of Old Cultivars in Genebank Collections: A Case Study on Lettuce. Crop Sci. 2011, 51, 736–746. [Google Scholar] [CrossRef]
- Cipriani, G.; Spadotto, A.; Jurman, I.; Di Gaspero, G.; Crespan, M.; Meneghetti, S.; Frare, E.; Vignani, R.; Cresti, M.; Morgante, M.; et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 2010, 121, 1569–1585. [Google Scholar] [CrossRef]
- Reim, S.; Lochschmidt, F.; Proft, A.; Hofer, M. Genetic integrity is still maintained in natural populations of the indigenous wild apple speciesMalus sylvestris(Mill.) in Saxony as demonstrated with nuclear SSR and chloroplast DNA markers. Ecol. Evol. 2020, 10, 11798–11809. [Google Scholar] [CrossRef] [PubMed]
- Reim, S.; Lochschmidt, F.; Proft, A.; Wolf, H.; Wolf, H. Species delimitation, genetic diversity and structure of the European indigenous wild pear (Pyrus pyraster) in Saxony, Germany. Genet. Resour. Crop Evol. 2017, 64, 1075–1085. [Google Scholar] [CrossRef]
- Wolko, Ł.; Antkowiak, W.; Lenartowicz, E.; Bocianowski, J. Genetic diversity of European pear cultivars (Pyrus communis L.) and wild pear (Pyrus pyraster (L.) Burgsd.) inferred from microsatellite markers analysis. Genet. Resour. Crop Evol. 2010, 57, 801–806. [Google Scholar] [CrossRef]
- Marchese, A.; Giovannini, D.; Leone, A.; Mafrica, R.; Palasciano, M.; Cantini, C.; Di Vaio, C.; De Salvador, F.R.; Giacalone, G.; Caruso, T.; et al. S-genotype identification, genetic diversity and structure analysis of Italian sweet cherry germplasm. Tree Genet. Genomes 2017, 13, 93. [Google Scholar] [CrossRef]
- Barreneche, T.; de la Concepcion, M.C.; Blouin-Delmas, M.; Ordidge, M.; Nybom, H.; Lacis, G.; Feldmane, D.; Sedlak, J.; Meland, M.; Kaldmae, H.; et al. SSR-Based Analysis of Genetic Diversity and Structure of Sweet Cherry (Prunus avium L.) from 19 Countries in Europe. Plants 2021, 10, 1983. [Google Scholar] [CrossRef]
- Stehr, R. Screening of Sweet Cherry Cultivars in Northern Germany; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2005; pp. 65–68. [Google Scholar]
- Lacombe, T.; Boursiquot, J.-M.; Laucou, V.; Di Vecchi-Staraz, M.; Péros, J.-P.; This, P. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor. Appl. Genet. 2013, 126, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.; Boursiquot, J.M.; This, P.; Chu, K.; Johansson, H.; Meredith, C. Historical Genetics: The Parentage of Chardonnay, Gamay, and Other Wine Grapes of Northeastern France. Science 1999, 285, 1562–1565. [Google Scholar] [CrossRef] [PubMed]
- Dirlewanger, E.; Cosson, P.; Tavaud, M.; Aranzana, J.; Poizat, C.; Zanetto, A.; Arús, P.; Laigret, F. Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Appl. Genet. 2002, 105, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Aranzana, M.; Garcia-Mas, J.; Carbó, J.; Arús PAranzana, M.J.; Garcia-Mas, J.; Carbo, J.; Arus, P. Development and variability analysis of microsatellite markers in peach. Plant Breeding, 121: 87-92. Plant Breed. 2002, 121, 87–92. [Google Scholar] [CrossRef]
- Downey, S.L.; Iezzoni, A.F. Polymorphic DNA Markers in Black Cherry (Prunus serotina) Are Identified Using Sequences from Sweet Cherry, Peach, and Sour Cherry. J. Am. Soc. Hortic. Sci. 2000, 125, 76–80. [Google Scholar] [CrossRef]
- Sosinski, B.; Gannavarapu, M.; Hager, L.; Beck, L.; King, G.; Dunsdon, C.; Rajapakse, S.; Baird, W.; Ballard, R.; Abbott, A.G. Characterization of microsatellite markers in peach [Prunus persica (L.) Batsch]. TAG Theor. Appl. Genet. 2000, 101, 421–428. [Google Scholar] [CrossRef]
- Testolin, R.; Marrazzo, T.; Cipriani, G.; Quarta, R.; Verde, I.; Dettori, M.T.; Pancaldi, M.; Sansavini, S. Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome 2000, 43, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Slotta, T.A.B.; Brady, L.; Chao, S. High throughput tissue preparation for large-scale genotyping experiments. Mol. Ecol. Resour. 2008, 8, 83–87. [Google Scholar] [CrossRef]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef]
- Marshall, T.C.; Slate, J.; Kruuk, L.E.; Pemberton, J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Szpiech, Z.A.; Jakobsson, M.; Rosenberg, N.A. ADZE: A rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 2008, 24, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kalinowki, S. hp-rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software; Online Database. 2006. Available online: https://darwin.cirad.fr/ (accessed on 14 November 2022).
- Gascuel, O.; Mirkin, B.; McMorris, F.; Roberts, F.; Rzhetsky, A. Concerning the NJ algorithm and its unweighted version, UNJ. In Mathematical Hierarchies and Biology; DIMACS series in discrete mathematics and theoretical computer science; RI American Mathematical Society: Providence, RI, USA, 1997; pp. 149–170. [Google Scholar]
- Huson, D.H.; Richter, D.C.; Rausch, C.; Dezulian, T.; Franz, M.; Rupp, R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinform. 2007, 8, 460. [Google Scholar] [CrossRef]
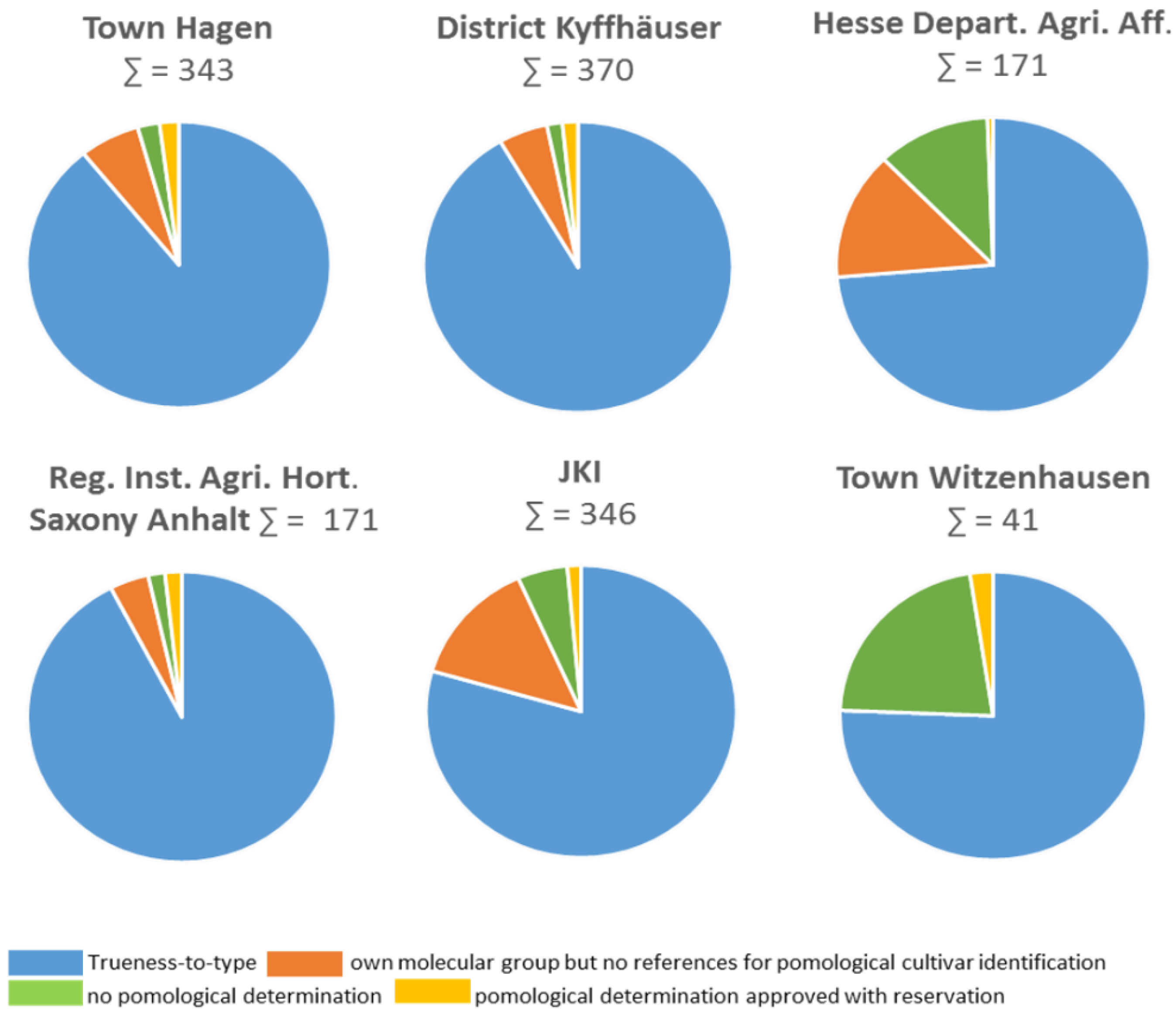

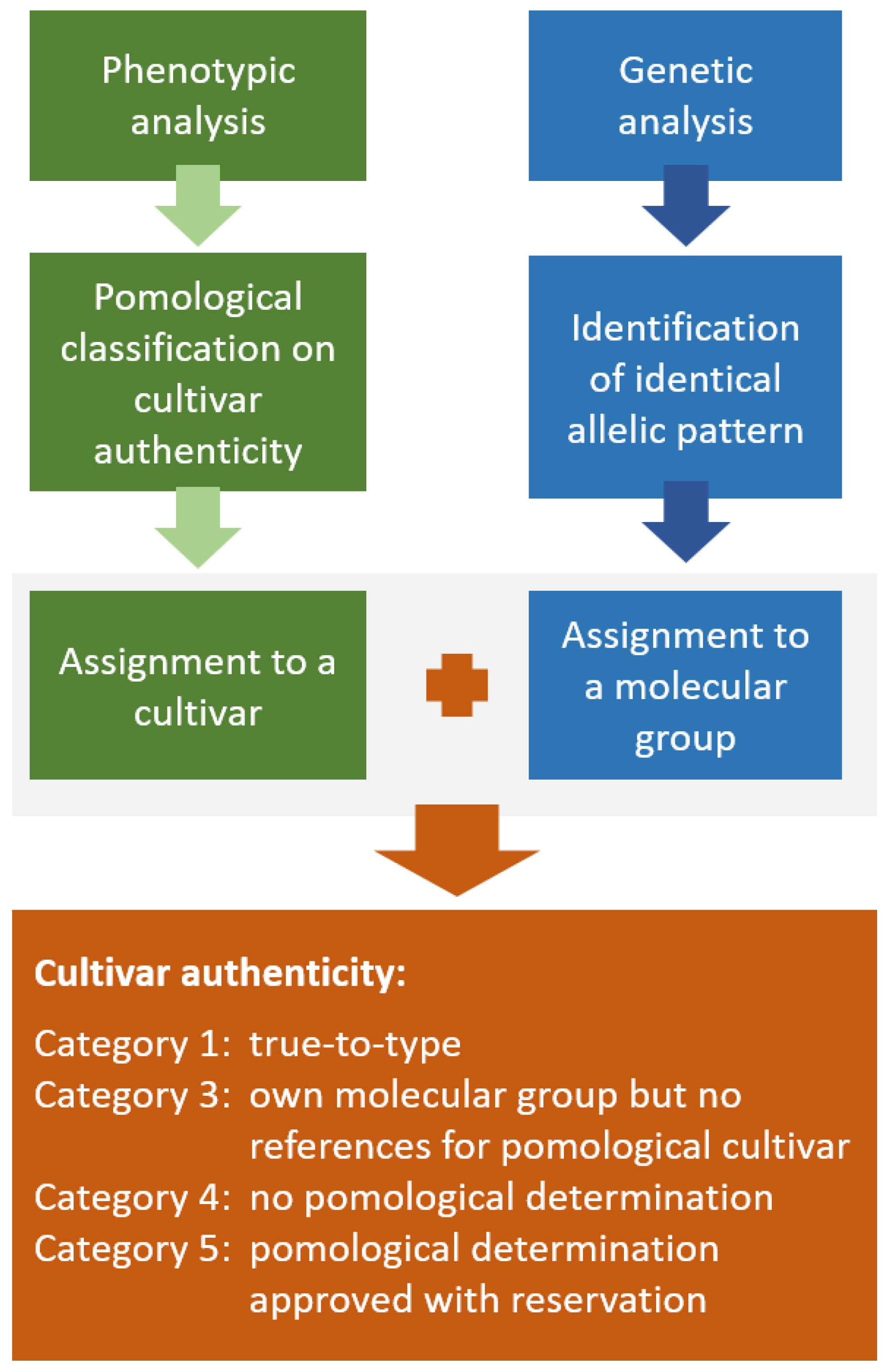
| Result of Assessment | No. Trees | % Trees |
|---|---|---|
| True-to-type | 1235 | 85.64 |
| Own molecular group but no references for pomological cultivar identification | 120 | 8.32 |
| No pomological determination | 64 | 4.4 |
| Pomological determination approved with reservation | 23 | 1.6 |
| Total | 1442 |
| Locus | N | Na | Ne | Ho | He | Ar |
|---|---|---|---|---|---|---|
| BPPCT037 | 383 | 15.00 | 4.87 | 0.83 | 0.79 | 9.51 |
| CPPCT006 | 383 | 15.00 | 4.17 | 0.78 | 0.76 | 9.56 |
| CPPCT022 | 383 | 14.00 | 3.56 | 0.75 | 0.72 | 9.89 |
| EMPA002 | 383 | 11.00 | 2.07 | 0.61 | 0.52 | 9.89 |
| EMPA003 | 383 | 5.00 | 1.62 | 0.40 | 0.38 | 3.11 |
| EMPA017 | 383 | 13.00 | 1.41 | 0.27 | 0.29 | 6.94 |
| EMPA026 | 382 | 14.00 | 2.20 | 0.51 | 0.55 | 8.02 |
| EMPaS01 | 383 | 11.00 | 3.13 | 0.71 | 0.68 | 11.89 |
| EMPaS02 | 383 | 15.00 | 3.71 | 0.75 | 0.73 | 10.55 |
| EMPaS06 | 383 | 23.00 | 7.43 | 0.86 | 0.87 | 15.56 |
| EMPaS10 | 383 | 20.00 | 3.09 | 0.61 | 0.68 | 11.71 |
| EMPaS12 | 383 | 11.00 | 4.27 | 0.77 | 0.77 | 12.89 |
| EMPaS14 | 383 | 9.00 | 2.43 | 0.61 | 0.59 | 10.89 |
| PceGA34 | 256 | 24.00 | 7.47 | 0.88 | 0.87 | 13.89 |
| PS05C03 | 258 | 20.00 | 4.61 | 0.78 | 0.78 | 14.97 |
| UDP98-412 | 383 | 23.00 | 5.08 | 0.62 | 0.80 | 16.27 |
| Mean | 15.19 | 3.82 | 0.67 | 0.67 | 10.97 |
| Cultivar Name | Assumed Mother | Assumed Father | Trio Confidence 1 |
|---|---|---|---|
| Alma | Rube | Allers Späte | + |
| Annabella | Rube | Allers Späte Knorpel | + |
| Areko | Kordia | Regina | + |
| Bianca | Rube | Allers Späte | + |
| Erika | Rube | Stechmanns Bunte | + |
| Ferbolus | Hedelfinger Riesenkirsche | Reverchon | + |
| Ferobri | Burlat | Fercer | + |
| Glacier | Stella | Burlat | + |
| Habunt | Valeska | Sunburst | + |
| Johanna | Schneiders Späte Knorpelkirsche | Rube | + |
| Karina | Schneiders Späte Knorpelkirsche | Rube | + |
| Lapins | Van | Stella | + |
| Merton Bounty | Elton | Schreckenskirsche | * |
| Müncheberger Fruehernte | Flamentiner | Früheste der Mark | + |
| Nafrina | Werdersche Frühe | Büttners Rote Knorpelkirsche | - |
| Namada | Badeborner Schwarze Knorpelkirsche | Rivers Frühe | + |
| New Star | Van | Stella | + |
| Oktavia | Schneiders Späte Knorpelkirsche | Rube | + |
| Rainier | Bing | Van | + |
| Regina | Schneiders Späte Knorpelkirsche | Rube | + |
| Ria | Kordia | Vic | + |
| Somerset | Van | Vic | + |
| Summit | Van | Sam | + |
| Sunburst | Van | Stella | + |
| Sylvia | Van | Sam; Van | + |
| Techlovan | Van | Kordia | * |
| Valeska | Rube | Stechmanns Bunte | + |
| Vanda | Van | Kordia | * |
| Vega | Bing | Victor | * |
| Viola | Schneiders Späte Knorpelkirsche | Rube | + |
| Vista | Hedelfinger Riesenkirsche | Victor | + |
| Cultivar name | Assumed Father # | Assumed Mother | Pair confidence 2 |
| Beata | open pollinated | Lambert | * |
| Chelan | Beaulieu | Stella | * |
| Chinook | Gil Peck | Bing | * |
| Ferprime | open pollinated | Fercer | * |
| Katalin | Podjebrad sarga | Schneiders Späte Knorpelkirsche | - |
| Lambert | Black Heart | Große Prinzessin | * |
| Lamida | open pollinated | Lambert | + |
| Larian | UCD 50 (Bing x Bush Tartarian) | Lambert | * |
| Linda | Germersdorfer | Hedelfinger | * |
| Magda | open pollinated | Basler Adlerkirsche | * |
| Merchant | open pollinated | Merton Glory | + |
| Merla | open pollinated | Merton Late | * |
| Mermat | open pollinated | Merton Glory | * |
| Merpet | open pollinated | Merton Glory | * |
| Nalitta | open pollinated | Querfurter Königskirsche | * |
| Sodus | Giant | Große Prinzessin | * |
| Stella | JI 2420 (Emperor Francis x Napoleon X-rayed pollen) | Lambert | * |
| Vic | Schmidt | Bing | + |
| Cultivar name | Assumed Mother # | Assumed Father | Pair confidence 3 |
| Bing | Black Republican | Große Prinzessin | * |
| Fernier | Tardif de Vignola | Rainier | * |
| Merton Favourite | Knight’s Early Black | Schreckenskirsche | * |
| Merton Glory | Ursula Rivers | Noble | * |
| Merton Late | Hildesheim (Belle Agathe) | Große Prinzessin | * |
| Sandra Rose | 2C-61-18 (Star x Van) | Sunburst | * |
| Swing | Nabigos | Stella | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reim, S.; Schiffler, J.; Braun-Lüllemann, A.; Schuster, M.; Flachowsky, H.; Höfer, M. Genetic and Pomological Determination of the Trueness-to-Type of Sweet Cherry Cultivars in the German National Fruit Genebank. Plants 2023, 12, 205. https://doi.org/10.3390/plants12010205
Reim S, Schiffler J, Braun-Lüllemann A, Schuster M, Flachowsky H, Höfer M. Genetic and Pomological Determination of the Trueness-to-Type of Sweet Cherry Cultivars in the German National Fruit Genebank. Plants. 2023; 12(1):205. https://doi.org/10.3390/plants12010205
Chicago/Turabian StyleReim, Stefanie, Juliane Schiffler, Annette Braun-Lüllemann, Mirko Schuster, Henryk Flachowsky, and Monika Höfer. 2023. "Genetic and Pomological Determination of the Trueness-to-Type of Sweet Cherry Cultivars in the German National Fruit Genebank" Plants 12, no. 1: 205. https://doi.org/10.3390/plants12010205
APA StyleReim, S., Schiffler, J., Braun-Lüllemann, A., Schuster, M., Flachowsky, H., & Höfer, M. (2023). Genetic and Pomological Determination of the Trueness-to-Type of Sweet Cherry Cultivars in the German National Fruit Genebank. Plants, 12(1), 205. https://doi.org/10.3390/plants12010205






