Phytoecdysteroids and Anabolic Effect of Atriplex dimorphostegia: UPLC-PDA-MS/MS Profiling, In Silico and In Vivo Models
Abstract
1. Introduction
2. Results
2.1. Extraction and Fractionation of A. dimorphostegia
2.2. Identification of 20-HE Isolated from EFAD
2.3. Characterization of Total Extract of A. dimorphostegia (TEAD) by UPLC-PDA-ESI-MS/MS
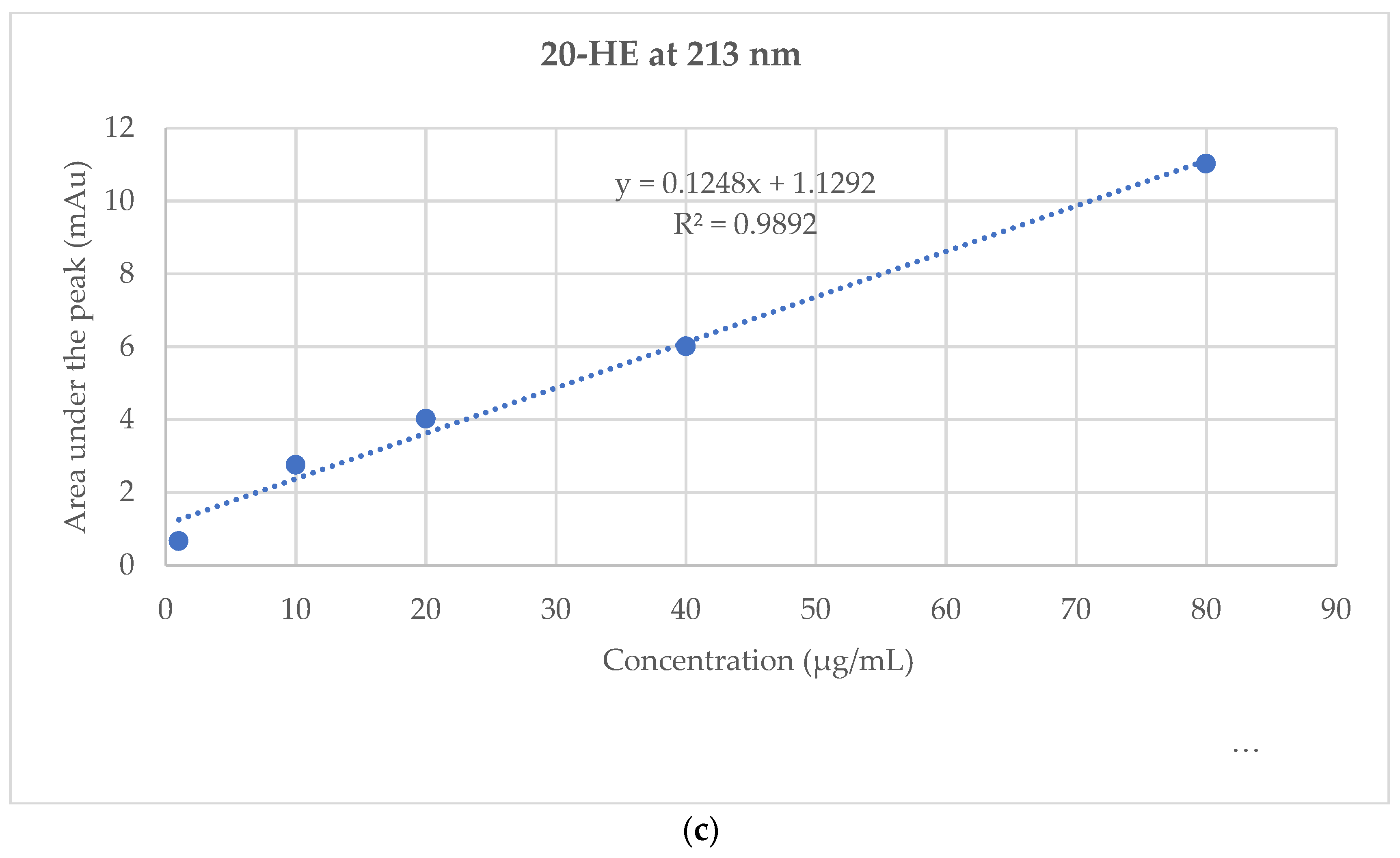
2.4. Standardization of TEAD Using UPLC Analysis
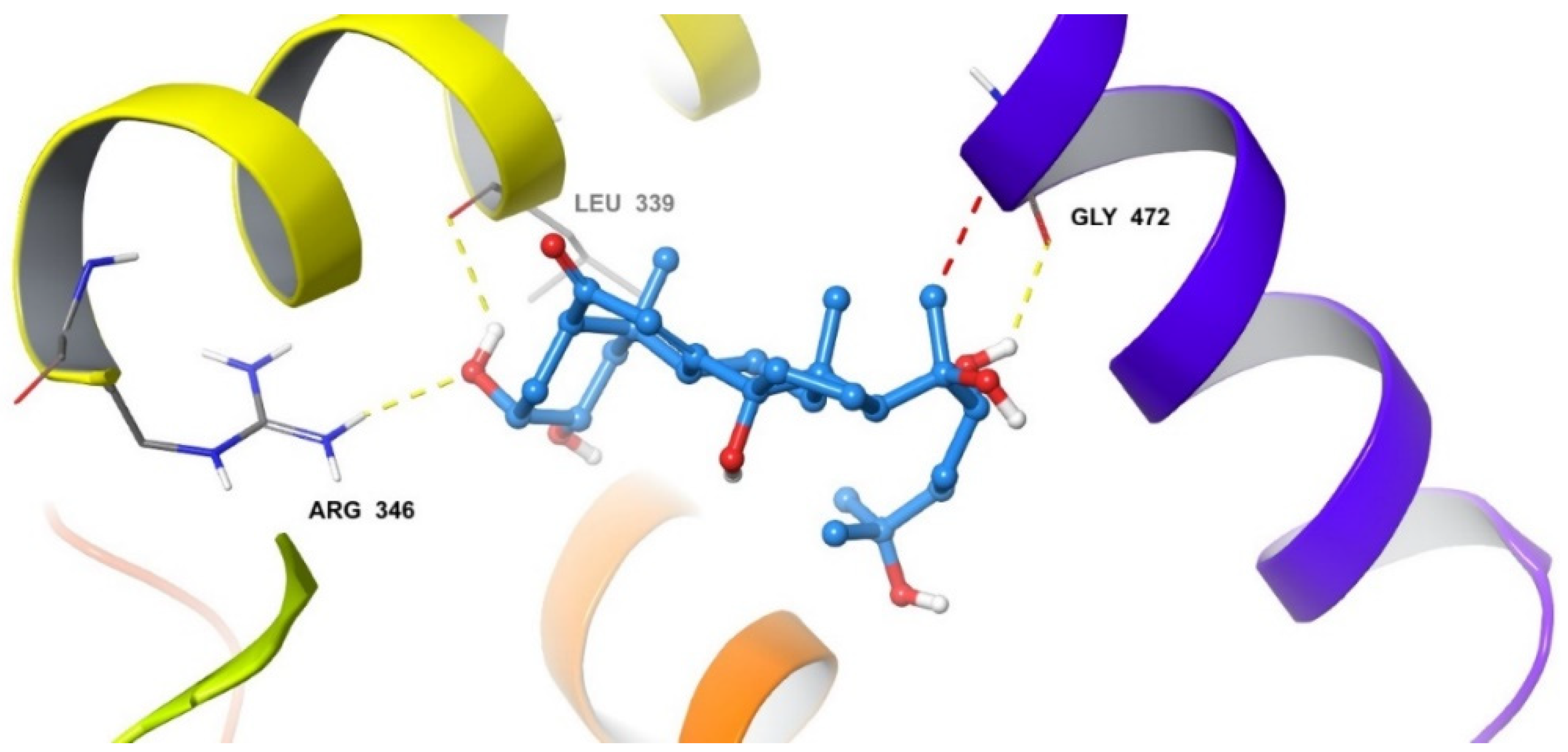
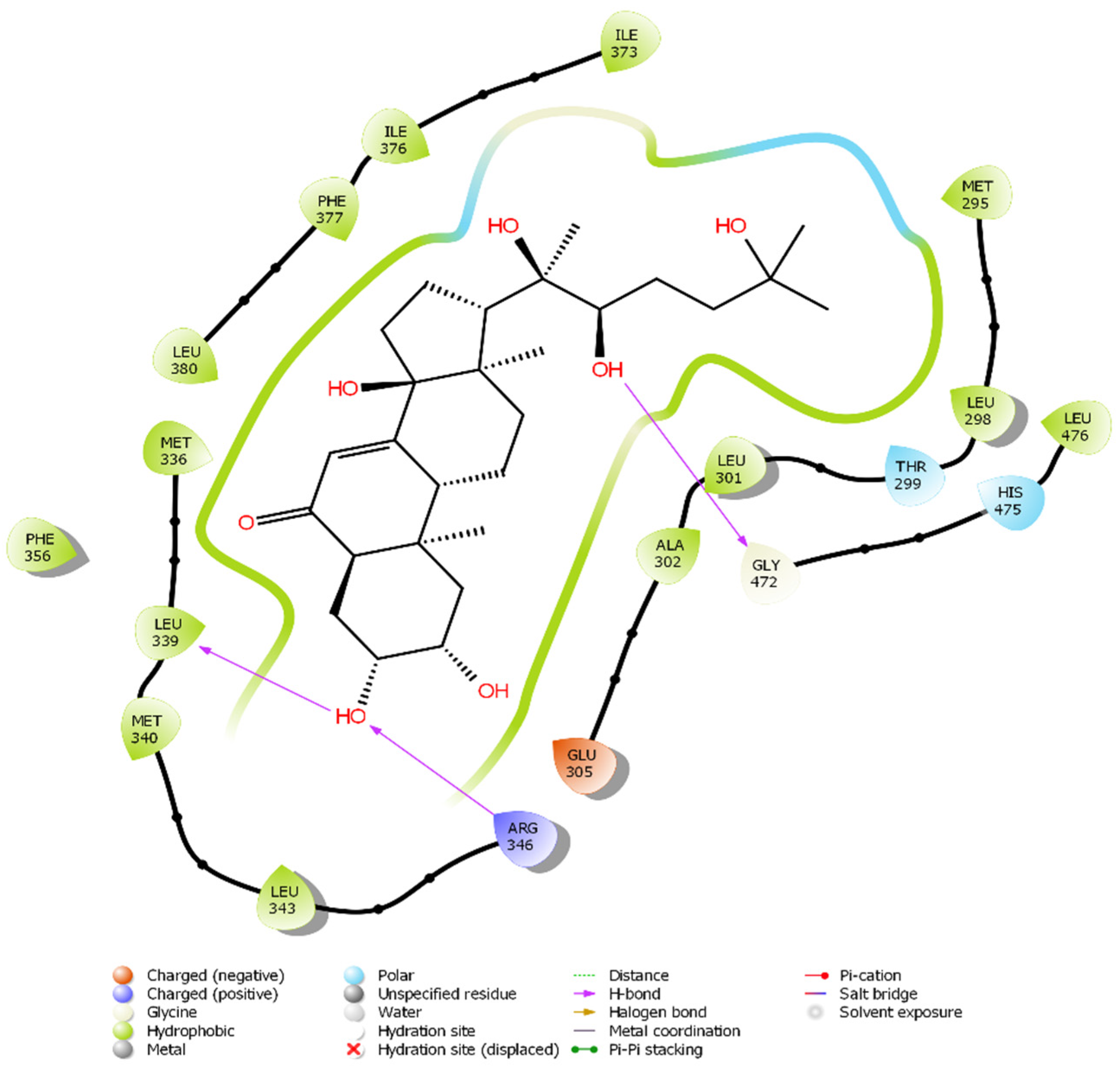
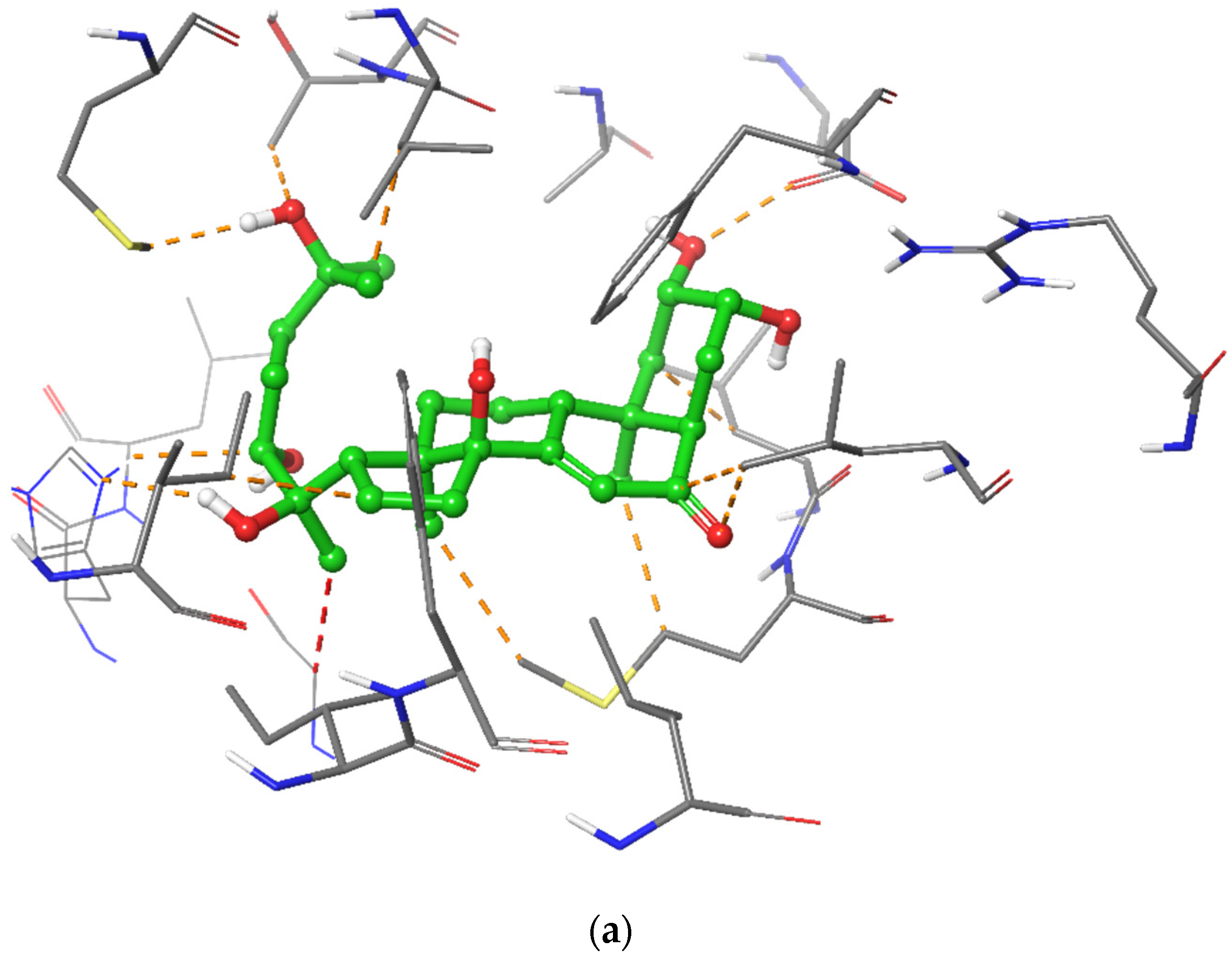
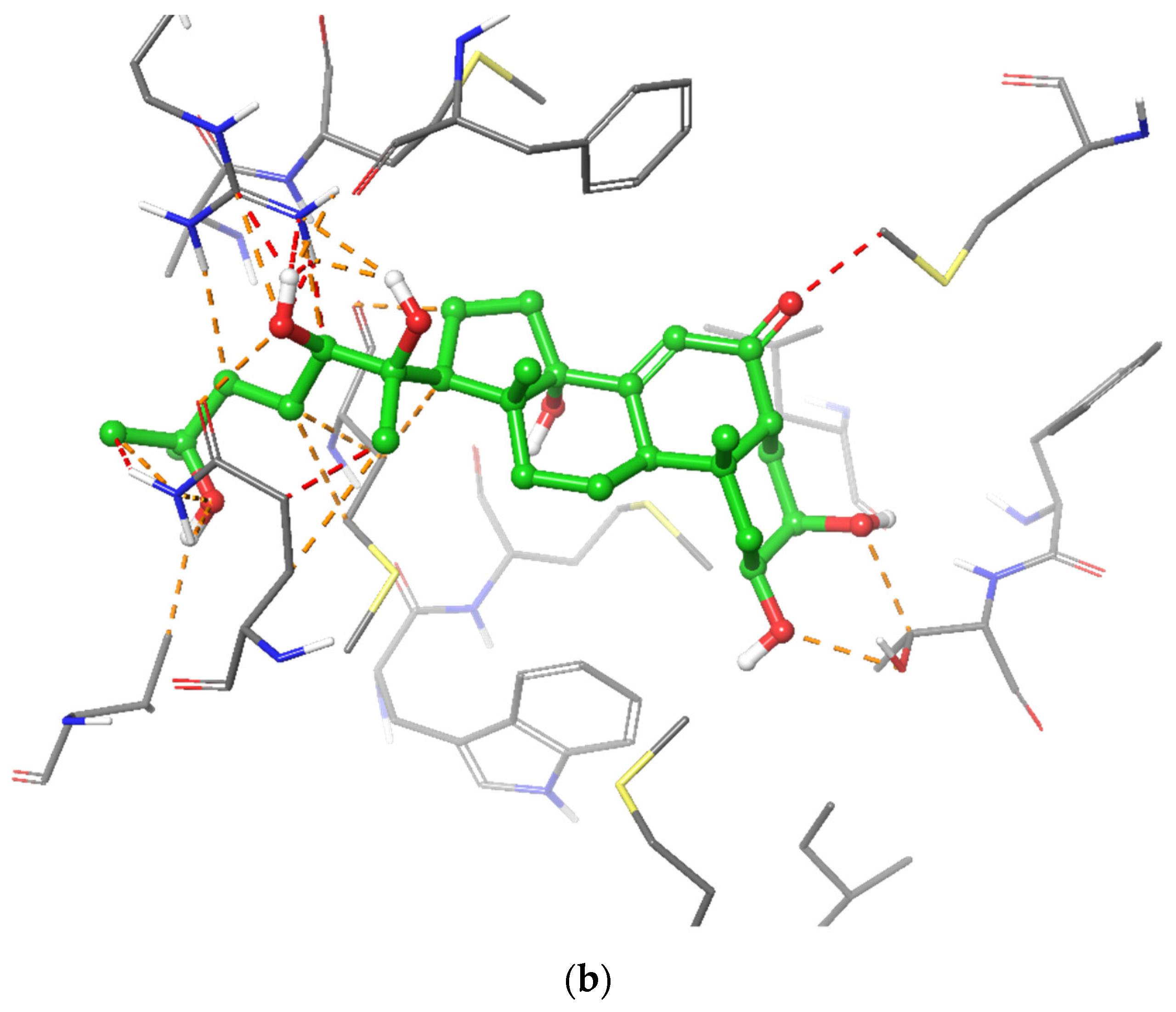
2.5. In Silico Modelling of Steroid Receptor Binding
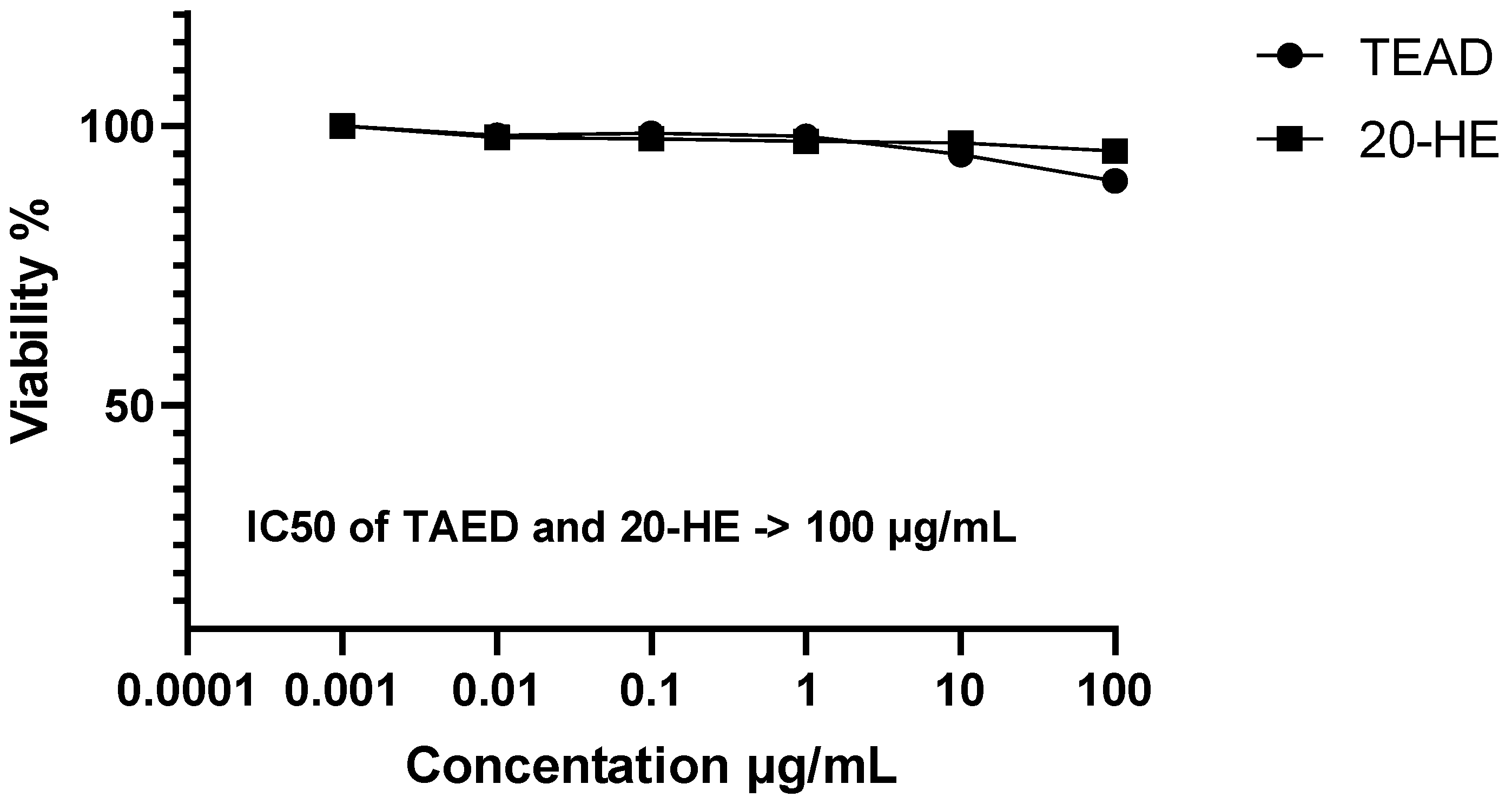
2.6. Evaluation of the Cytotoxic Activity of 20-HE and TEAD (SRB-U Assay)
2.7. In Vivo Anabolic Study
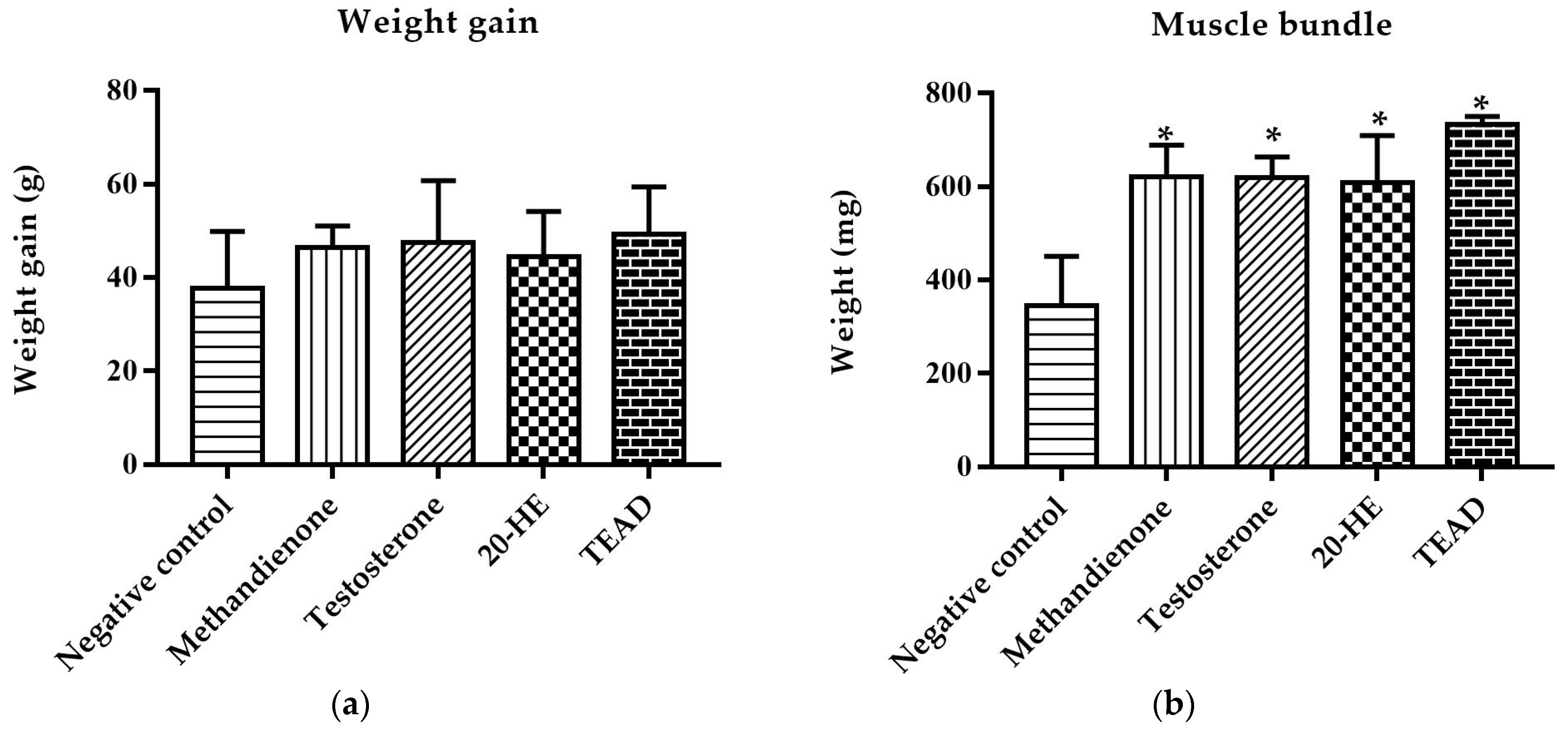
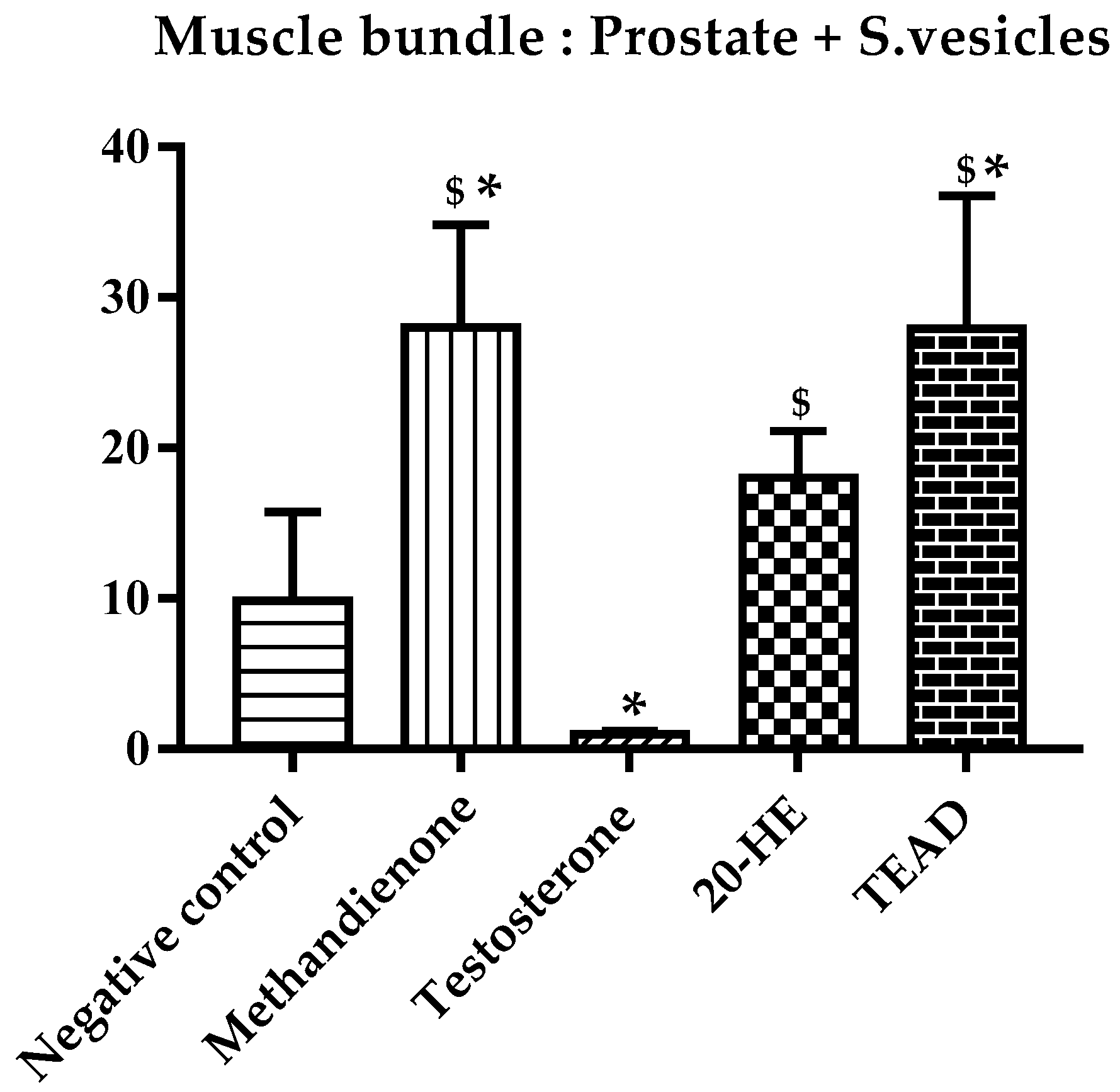
2.7.1. Weight Gain and Mass of Muscle Bundle
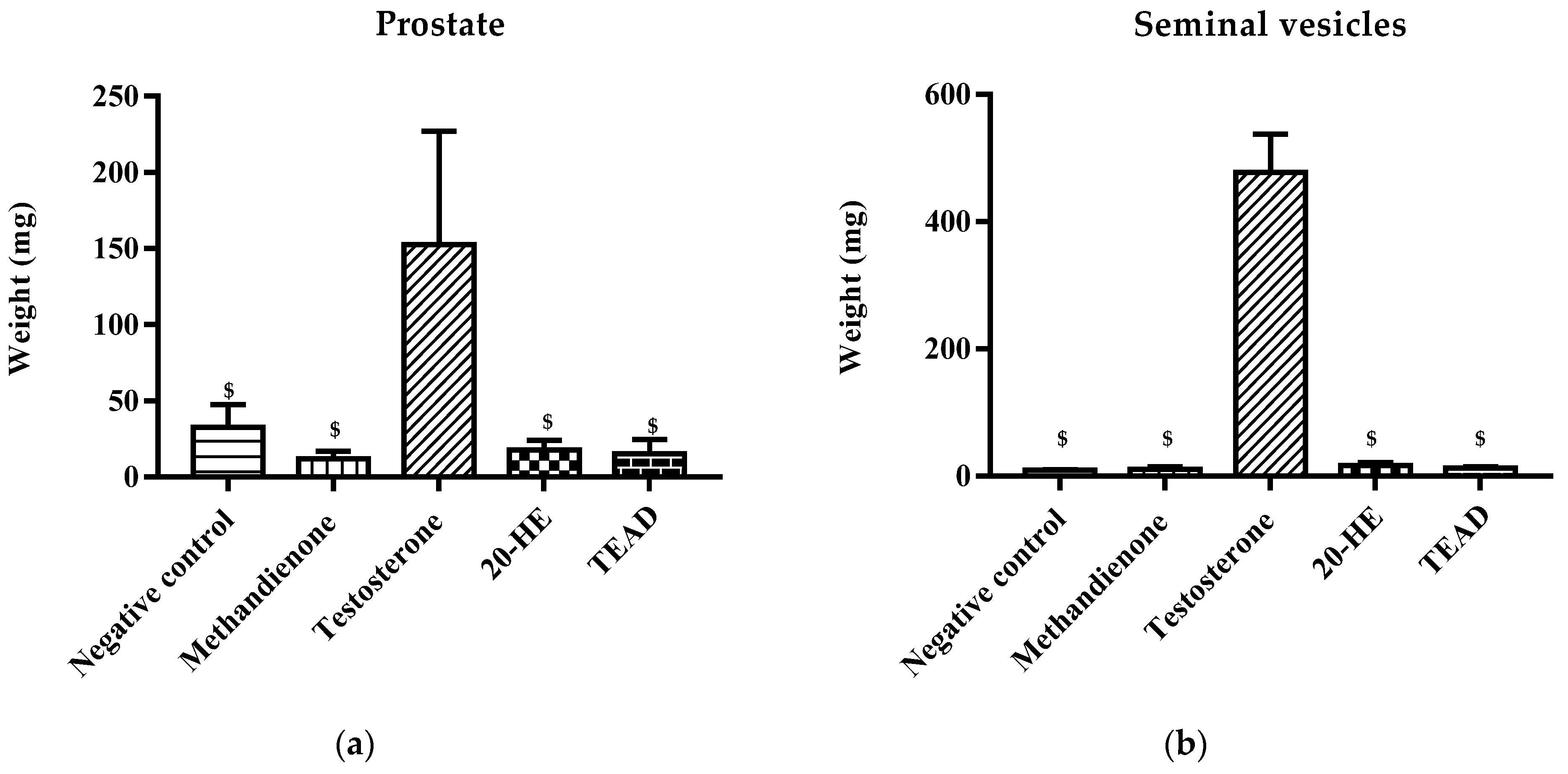
2.7.2. Weight of Secondary Sex Organs
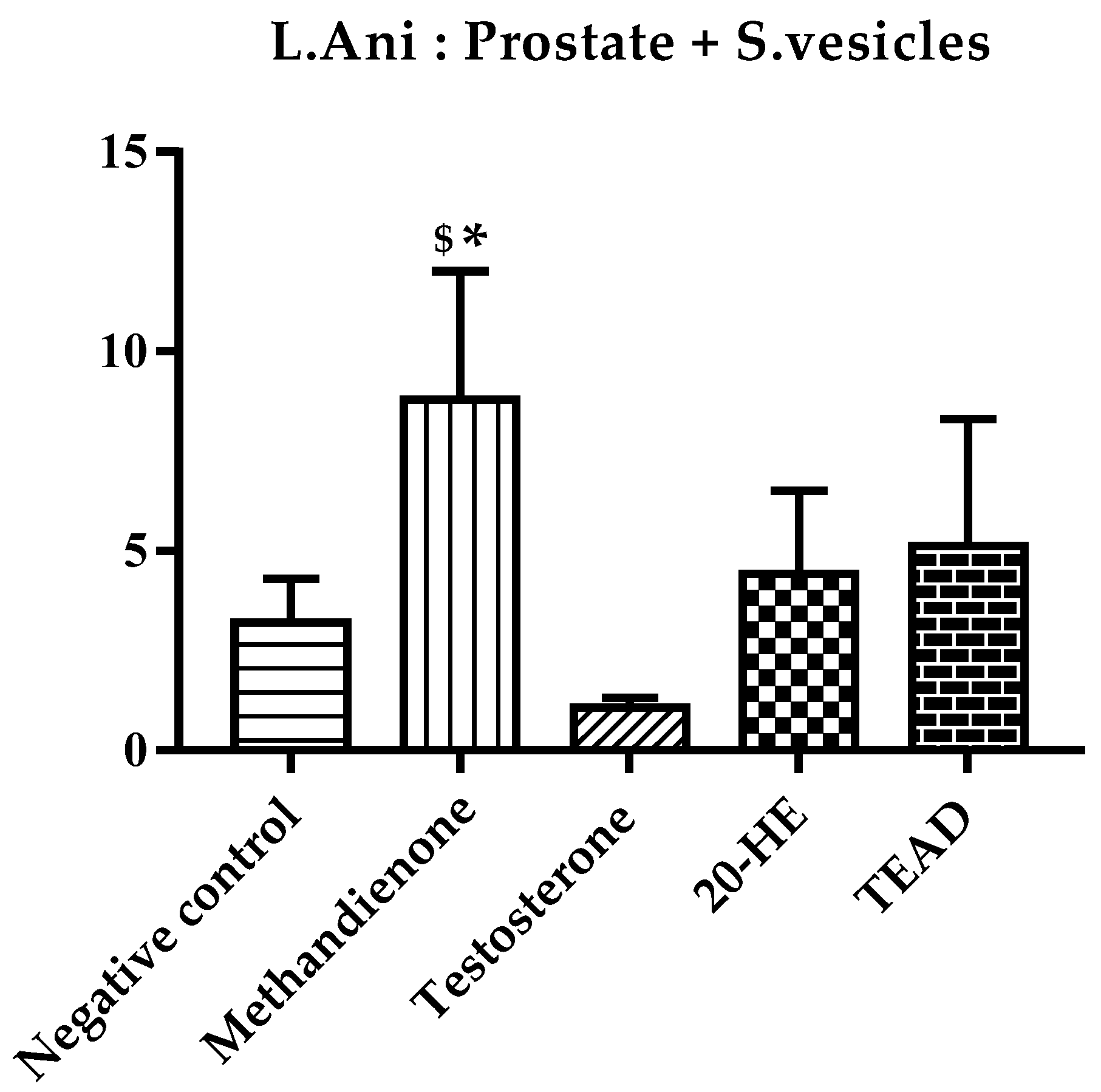
2.7.3. Anabolic to Androgenic Ratios
2.7.4. Muscle Mass Percentage
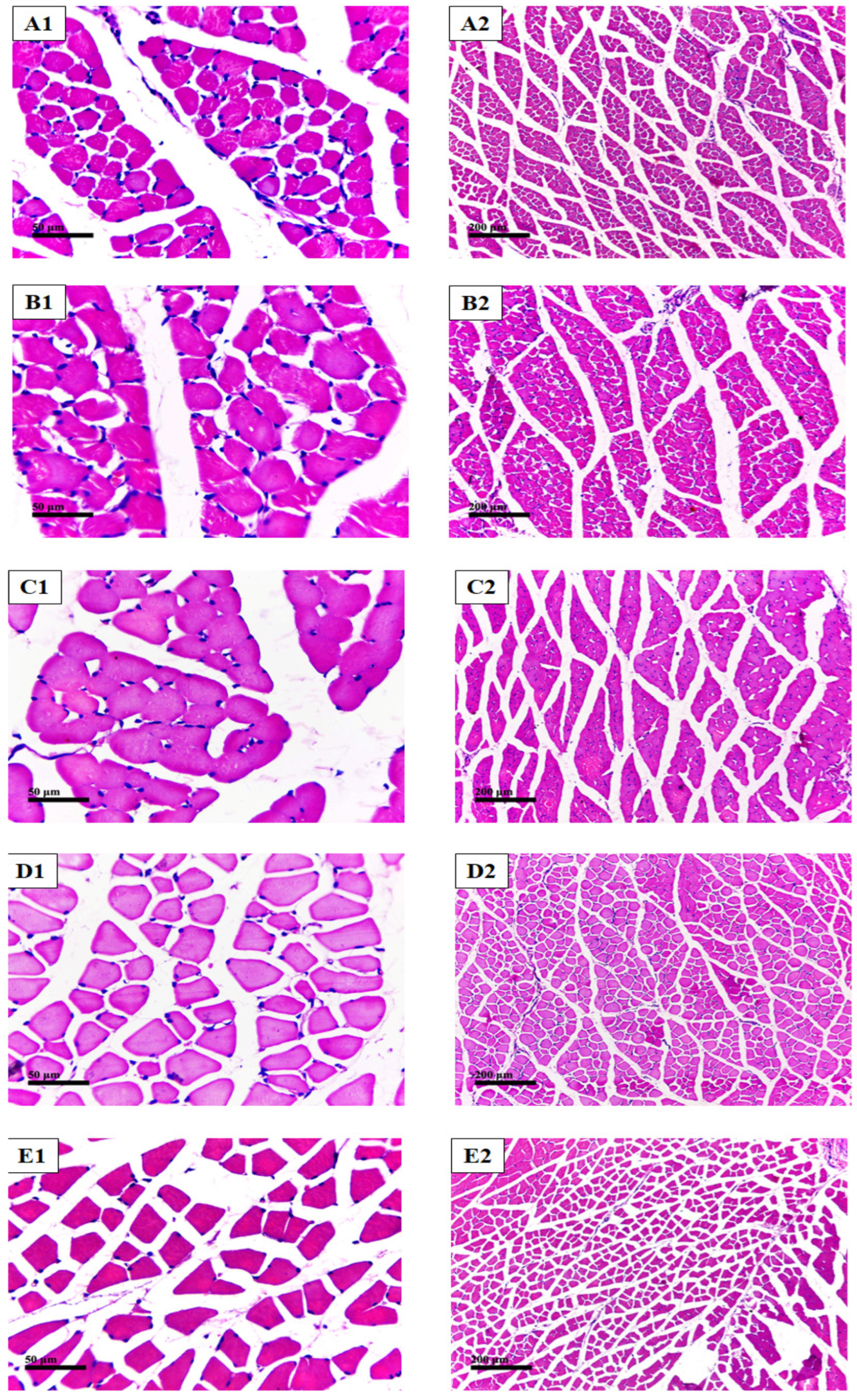
2.7.5. Histopathological Examination with Hematoxylin and Eosin (H&E) Stain
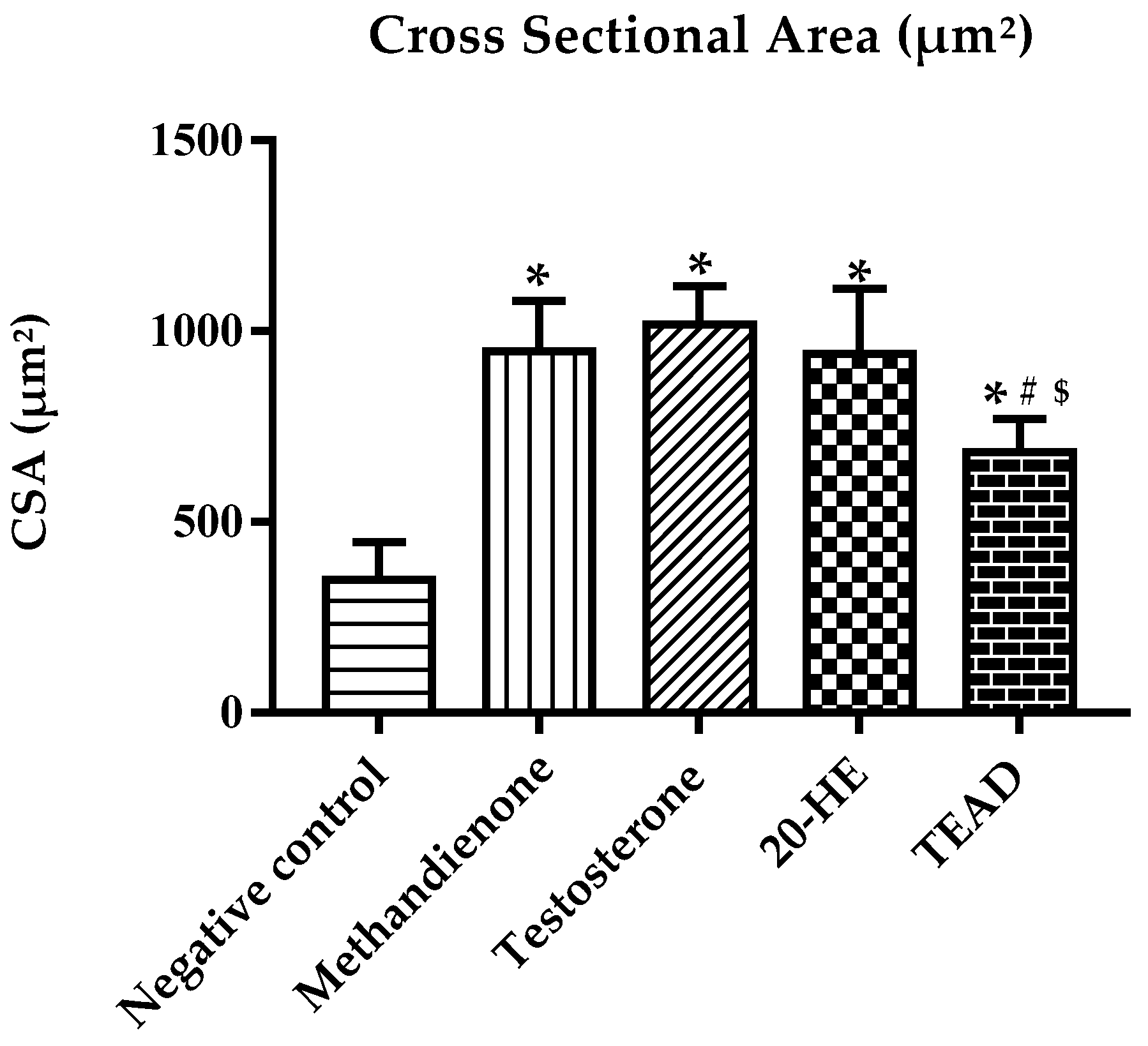
2.7.6. Cross Sectional Area of Muscles
2.7.7. Number of Muscle Fibers
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Extraction and Fractionation of A. dimorphostegia
4.3. Isolation of 20-HE from EFAD
4.4. NMR Spectrometrical Analysis of 20-HE
4.5. LC/ESI/MS Spectrometrical Analysis of 20-HE
4.6. UPLC-PDA-ESI-MS/MS Analysis of TEAD
4.7. Standardisation of TEAD Using UPLC Analysis
4.8. In Silico Modelling of Steroid Receptor Binding
4.9. Evaluation of the Cytotoxic Activity of 20-HE and TEAD (SRB-U Assay)
4.10. In Vivo Anabolic Study
4.10.1. Animals
4.10.2. Orchidectomy
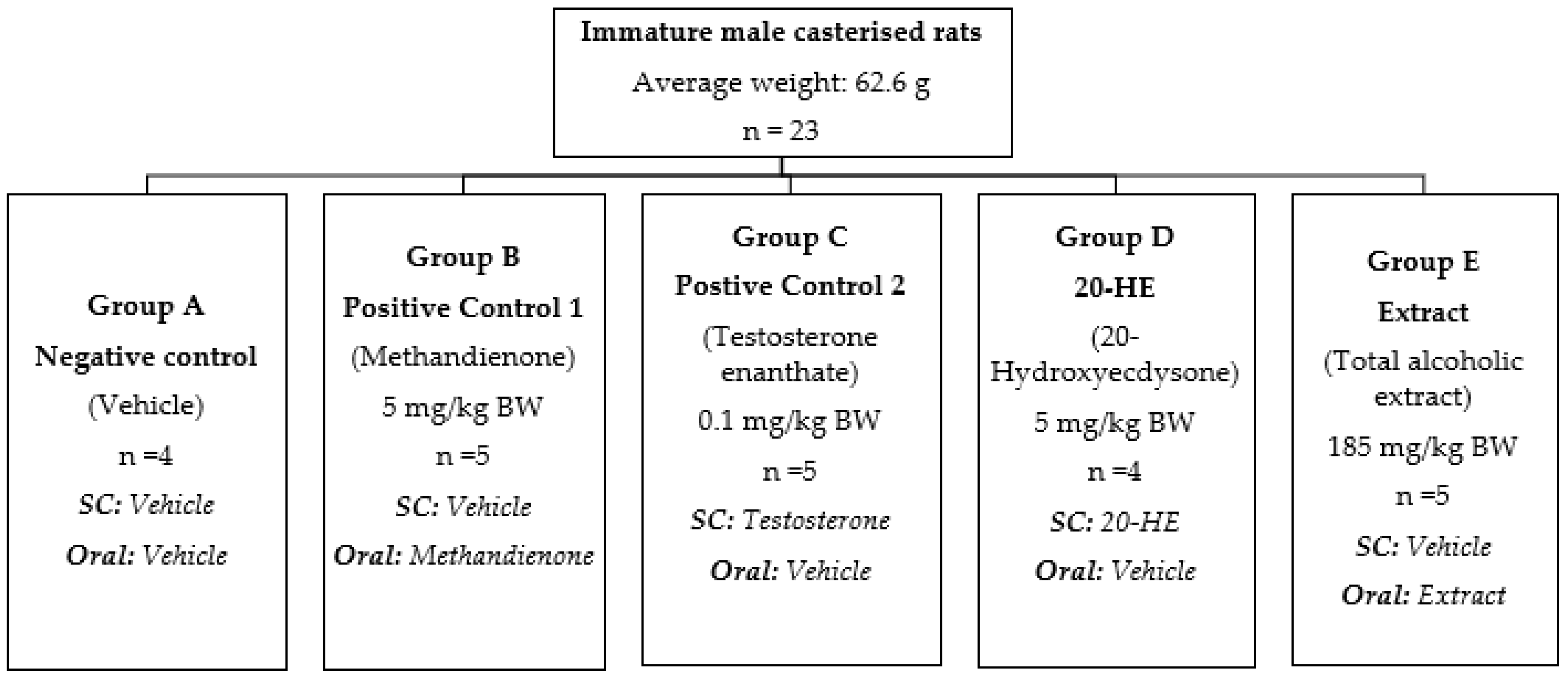
4.10.3. Experimental Design
4.10.4. Sample Collection
4.10.5. Histopathological Examination and Determination of Muscle Fiber Cross-Sectional Area
4.10.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cristina, R.T.; Hanganu, F.; Dumitrescu, E.; Muselin, F.; Butnariu, M.; Constantin, A.; Chiurciu, V. The Impact of Exogenic Testosterone and Nortestosterone-Decanoate Toxicological Evaluation Using a Rat Model. PLoS ONE 2014, 9, e109219. [Google Scholar] [CrossRef] [PubMed]
- Basaria, S.; Wahlstrom, J.T.; Dobs, A.S. Anabolic-Androgenic Steroid Therapy in the Treatment of Chronic Diseases. J. Clin. Endocrinol. Metab. 2001, 86, 5108–5117. [Google Scholar] [CrossRef] [PubMed]
- el Osta, R.; Almont, T.; Diligent, C.; Hubert, N.; Eschwège, P.; Hubert, J. Anabolic Steroids Abuse and Male Infertility. Basic Clin. Androl. 2016, 26, 2. [Google Scholar] [CrossRef] [PubMed]
- Christou, M.A.; Christou, P.A.; Markozannes, G.; Tsatsoulis, A.; Mastorakos, G.; Tigas, S. Effects of Anabolic Androgenic Steroids on the Reproductive System of Athletes and Recreational Users: A Systematic Review and Meta-Analysis. Sport. Med. 2017, 47, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Hirunsai, M.; Yimlamai, T.; Suksamrarn, A. Effect of 20-Hydroxyecdysone on Proteolytic Regulation in Skeletal Muscle Atrophy. In Vivo 2016, 30, 869–877. [Google Scholar] [CrossRef]
- Parr, M.K.; Botrè, F.; Naß, A.; Hengevoss, J.; Diel, P.; Wolber, G. Ecdysteroids: A Novel Class of Anabolic Agents? Biol. Sport 2015, 32, 169–173. [Google Scholar] [CrossRef]
- Isenmann, E.; Ambrosio, G.; Joseph, J.F.; Mazzarino, M.; de la Torre, X.; Zimmer, P.; Kazlauskas, R.; Goebel, C.; Botrè, F.; Diel, P.; et al. Ecdysteroids as Non-Conventional Anabolic Agent: Performance Enhancement by Ecdysterone Supplementation in Humans. Arch. Toxicol. 2019, 93, 1807–1816. [Google Scholar] [CrossRef]
- Bathori, M.; Toth, N.; Hunyadi, A.; Marki, A.; Zador, E. Phytoecdysteroids and Anabolic-Androgenic Steroids - Structure and Effects on Humans. Curr. Med. Chem. 2008, 15, 75–91. [Google Scholar] [CrossRef]
- Katzenellenbogen, B.S.; Choi, I.; Delage-Mourroux, R.; Ediger, T.R.; Martini, P.G.V.; Montano, M.; Sun, J.; Weis, K.; Katzenellenbogen, J.A. Molecular Mechanisms of Estrogen Action: Selective Ligands and Receptor Pharmacology. J. Steroid. Biochem. Mol. Biol. 2000, 74, 279–285. [Google Scholar] [CrossRef]
- Wiik, A.; Ekman, M.; Johansson, O.; Jansson, E.; Esbjörnsson, M. Expression of Both Oestrogen Receptor Alpha and Beta in Human Skeletal Muscle Tissue. Histochem. Cell Biol. 2009, 131, 181–189. [Google Scholar] [CrossRef]
- Matthews, J.; Gustafsson, J.-Å. Estrogen Receptor and Aryl Hydrocarbon Receptor Signaling Pathways. Nucl. Recept Signal. 2006, 4, nrs.04016. [Google Scholar] [CrossRef] [PubMed]
- Milanesi, L.; Vasconsuelo, A.; de Boland, A.R.; Boland, R. Expression and Subcellular Distribution of Native Estrogen Receptor β in Murine C2C12 Cells and Skeletal Muscle Tissue. Steroids 2009, 74, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Wiik, A.; Glenmark, B.; Ekman, M.; Esbjörnsson-Liljedahl, M.; Johansson, O.; Bodin, K.; Enmark, E.; Jansson, E. Oestrogen Receptor β Is Expressed in Adult Human Skeletal Muscle Both at the MRNA and Protein Level. Acta Physiol. Scand. 2003, 179, 381–387. [Google Scholar] [CrossRef]
- Milanesi, L.; de Boland, A.R.; Boland, R. Expression and Localization of Estrogen Receptor α in the C2C12 Murine Skeletal Muscle Cell Line. J. Cell Biochem. 2008, 104, 1254–1273. [Google Scholar] [CrossRef]
- Velders, M.; Schleipen, B.; Fritzemeier, K.H.; Zierau, O.; Diel, P. Selective Estrogen Receptor-β Activation Stimulates Skeletal Muscle Growth and Regeneration. FASEB J. 2012, 26, 1909–1920. [Google Scholar] [CrossRef] [PubMed]
- Zwetsloot, K.A.; Committee Alan Utter, T.C.; Member, M.; Committee Kelly Cole, T.J.; Poole, M.C.; Williams, C.D. Effects of Phytoecdysteroids on Skeletal Muscle Function and Protein Synthesis after Eccentric Damage. Ph.D. Thesis, Appalachian State University, Boone, NC, USA, 2017. [Google Scholar]
- Bajguz, A.; Bąkała, I.; Talarek, M. Ecdysteroids in Plants and Their Pharmacological Effects in Vertebrates and Humans. In Studies in Natural Products Chemistry; Elsevier B.V.: Amsterdam, The Netherlands, 2015; Volume 45, pp. 121–145. [Google Scholar]
- Adler, J.H.; Grebenok, R.J. Occurrence, Biosynthesis, and Putative Role of Ecdysteroids in Plants. Crit. Rev. Biochem. Mol. Biol. 1999, 34, 253–264. [Google Scholar] [CrossRef]
- Tóth, N.; Szabó, A.; Kacsala, P.; Héger, J.; Zádor, E. 20-Hydroxyecdysone Increases Fiber Size in a Muscle-Specific Fashion in Rat. Phytomedicine 2008, 15, 691–698. [Google Scholar] [CrossRef] [PubMed]
- El-Aasr, M.; Kabbash, A.; Abo El-Seoud, K.A.; Al-Madboly, L.A.; Ikeda, T. Antimicrobial and Immunomodulatory Activities of Flavonol Glycosides Isolated from Atriplex Halimus L. Herb. J. Pharm. Sci. Res. 2016, 8, 1159–1168. [Google Scholar]
- Kumarihamy, M.; Cutler, S.; Tekwani, B.; Chaurasya, N.; Muhammad, I. Chemical and Biological Investigation of Atriplex canescens. Planta Med. 2013, 79, PN35. [Google Scholar] [CrossRef]
- Keckeis, K.; Sarker, S.D.; Dinan, L.N. Phytoecdysteroids from Atriplex nummularia. Fitoterapia 2000, 71, 456–458. [Google Scholar] [CrossRef]
- Walker, D.J.; Lutts, S.; Sánchez-García, M.; Correal, E. Atriplex halimus L.: Its Biology and Uses. J. Arid Environ. 2014; 100, 111–121. [Google Scholar]
- Chikhi, I.; Allali, H.; el Amine Dib, M.; Medjdoub, H.; Tabti, B. Antidiabetic Activity of Aqueous Leaf Extract of Atriplex halimus L. (Chenopodiaceae) in Streptozotocin-Induced Diabetic Rats. Asian Pac. J. Trop. Dis. 2014, 4, 181–184. [Google Scholar] [CrossRef]
- Benhammou, N.; Bekkara, F.A.; Kadifkova Panovska, T. Antioxidant Activity of Methanolic Extracts and Some Bioactive Compounds of Atriplex halimus. Comptes Rendus Chim. 2009, 12, 1259–1266. [Google Scholar] [CrossRef]
- el Souda, S.S.E.D.; Matloub, A.A.; Nepveu, F.; Valentin, A.; Roques, C. Phenolic Composition and Prospective Anti-Infectious Properties of Atriplex lindleyi. Asian Pac. J. Trop. Dis. 2015, 5, 786–791. [Google Scholar] [CrossRef]
- Kamal, Z.; Ullah, F.; Ayaz, M.; Sadiq, A.; Ahmad, S.; Zeb, A.; Hussain, A.; Imran, M. Anticholinesterse and Antioxidant Investigations of Crude Extracts, Subsequent Fractions, Saponins and Flavonoids of Atriplex laciniata L.: Potential Effectiveness in Alzheimer’s and Other Neurological Disorders. Biol. Res. 2015, 48, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Boughalleb, N.; Trabelsi, L.; Harzallah-Skhiri, F. Antifungal Activity from Polar and Non-Polar Extracts of Some Chenopodiaceae Wild Species Growing in Tunisia. Nat. Prod. Res. 2009, 23, 988–997. [Google Scholar] [CrossRef]
- Capua, C.J.; Hopson, N.P.; Stewart, C.M.M.; Johnston, G.R.; O’Neill, K.L.; Schaalje, G.B.; Lee, C.M.; Booth, G.M. Cytotoxicity of Atriplex confertifolia. J. Toxicol. 2010, 2010. [Google Scholar] [CrossRef]
- Karim, A.; Fatima, I.; Hussain, S.; Malik, A. Dimorphamides A-C, New Polyphenolic Amides from Atriplex dimorphostagia. Helv. Chim. Acta 2011, 94, 528–533. [Google Scholar] [CrossRef]
- Asilbekova, D.T.; Tursunkhodzhaeva, F.M.; Gazizov, F.Y. Lipids from Atriplex dimorphostegia Leaves. Chem. Nat. Compd. 2008, 44, 764–766. [Google Scholar] [CrossRef]
- Gelin, Z.; Mosyakin, S.L.; Clemants, S.E. CHENOPODIACEAE. Li Ke. Flora China 2003, 5, 351–414. [Google Scholar]
- El-sakhawy, F.S.; Abou-hussein, D.R.; El-kersh, D.M.; Sleem, A.A. Anabolic and Androgenic Effects of Certain Atriplex Species Grown in Egypt. Egypt J. Biomed. Sci. 2012, 4, 97. [Google Scholar]
- Fábián, L.; Argay, G.; Kálmán, A.; Báthori, M. Crystal Structures of Ecdysteroids: The Role of Solvent Molecules in Hydrogen Bonding and Isostructurality. Acta Cryst. B 2002, 58, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Raghava Rao, K.V.; Mani, P.; Satyanarayana, B.; Raghava Rao, T. Purification and Structural Elucidation of Three Bioactive Compounds Isolated from Streptomyces coelicoflavus BC 01 and Their Biological Activity. 3 Biotech. 2017, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- ben Nejma, A.; Nguir, A.; ben Jannet, H.; Hamza, M.A.; Daïch, A.; Othman, M.; Lawson, A.M. New Septanoside and 20-Hydroxyecdysone Septanoside Derivative from Atriplex portulacoides Roots with Preliminary Biological Activities. Bioorg. Med. Chem. Lett. 2015, 25, 1665–1670. [Google Scholar] [CrossRef] [PubMed]
- Olennikov, D.N. Makisterone C-20,22-Acetonide from Rhaponticum uniflorum. Chem. Nat. Compd. 2018, 54, 930–933. [Google Scholar] [CrossRef]
- Yusupova, U.; Usmanov, D.; Azamatov, A.; Ramazonov, N.; Rejepov, J. Phytochemical Constituents and Biological Activities of Dianthus helenae Vved., Growing in Uzbekistan. Nat. Prod. Res. 2022, 36, 3480–3484. [Google Scholar] [CrossRef]
- Girault, J.P.; Lafont, R. The Complete 1H-NMR Assignment of Ecdysone and 20-Hydroxyecdysone. J. Insect Physiol. 1988, 34, 701–706. [Google Scholar] [CrossRef]
- Báthori, M.; Girault, J.P.; Kalasz, H.; Mathé, I.; Dinan, L.N.; Lafont, R. Complex Phytoecdysteroid Cocktail of Silene otites (Caryophyllaceae). Arch. Insect Biochem. Physiol. 1999, 41, 1–8. [Google Scholar] [CrossRef]
- Ayoub, N.; Badr, N.; Al-Ghamdi, S.S.; Alsanosi, S.; Alzahrani, A.R.; Abdel-Naim, A.B.; Nematallah, K.A.; Swilam, N. Hplc/Msn Profiling and Healing Activity of a Muco-Adhesive Formula of Salvadora persica against Acetic Acid-Induced Oral Ulcer in Rats. Nutrients 2022, 14, 28. [Google Scholar] [CrossRef]
- Chen, H.J.; Inbaraj, B.S.; Chen, B.H. Determination of Phenolic Acids and Flavonoids in Taraxacum formosanum Kitam by Liquid Chromatography-Tandem Mass Spectrometry Coupled with a Post-Column Derivatization Technique. Int. J. Mol. Sci. 2012, 13, 260–285. [Google Scholar] [CrossRef]
- Boutaoui, N.; Zaiter, L.; Benayache, F.; Benayache, S.; Cacciagrano, F.; Cesa, S.; Secci, D.; Carradori, S.; Giusti, A.M.; Campestre, C.; et al. Atriplex Mollis Desf. Aerial Parts: Extraction Procedures, Secondary Metabolites and Color Analysis. Molecules 2018, 23, 1962. [Google Scholar] [CrossRef]
- Simirgiotis, M.J.; Benites, J.; Areche, C.; Sepu, B. Antioxidant Capacities and Analysis of Phenolic Compounds in Three Endemic Nolana Species by HPLC-PDA-ESI-MS. Molecules 2015, 20, 11490–11507. [Google Scholar] [CrossRef] [PubMed]
- Emam, S.S. Bioactive Constituents of Atriplex halimus Plant Shalabia. J. Nat. Prod. 2011, 4, 25–41. [Google Scholar]
- Ibrahima, R.M.; El-Halawany, A.M.; Saleh, D.O.; el Naggar, E.M.B.; EL-Shabrawy, A.E.R.O.; El-Hawary, S.S. HPLC-DAD-MS/MS Profiling of Phenolics from Securigera securidaca Flowers and Its Anti-Hyperglycemic and Anti-Hyperlipidemic Activities. Rev. Bras. Farmacogn. 2015, 25, 134–141. [Google Scholar] [CrossRef]
- Patel, N.; Sharath Kumar, L.; Gajera, J.; Jadhav, A.; Muguli, G.; Babu, U. Isolation and Characterization of Flavonoid C-Glycosides from Prosopis glandulosa Torr. Leaves. Pharm. Mag. 2018, 14, 451–454. [Google Scholar] [CrossRef]
- ben Said, R.; Hamed, A.I.; Mahalel, U.A.; Al-Ayed, A.S.; Kowalczyk, M.; Moldoch, J.; Oleszek, W.; Stochmal, A. Tentative Characterization of Polyphenolic Compounds in the Male Flowers of Phoenix dactylifera by Liquid Chromatography Coupled with Mass Spectrometry and DFT. Int. J. Mol. Sci. 2017, 18, 512. [Google Scholar] [CrossRef]
- Awaad, A.S.; Maitland, D.J.; Donia, A.E.R.M.; Alqasoumi, S.I.; Soliman, G.A. Novel Flavonoids with Antioxidant Activity from a Chenopodiaceous Plant. Pharm. Biol. 2012, 50, 99–104. [Google Scholar] [CrossRef]
- Beelders, T.; de Beer, D.; Stander, M.A.; Joubert, E. Comprehensive Phenolic Profiling of Cyclopia genistoides (L.) Vent. by LC-DAD-MS and -MS/MS Reveals Novel Xanthone and Benzophenone Constituents. Molecules 2014, 19, 11760–11790. [Google Scholar] [CrossRef]
- Ağalar, H.G.; Çiftçi, G.A.; Göger, F.; Kırımer, N. Activity Guided Fractionation of Arum italicum Miller Tubers and the LC/MS-MS Profiles. Rec. Nat. Prod. 2018, 12, 64–75. [Google Scholar] [CrossRef]
- Barros, L.; Dueñas, M.; Dias, M.I.; Sousa, M.J.; Santos-Buelga, C.; Ferreira, I.C.F.R. Phenolic Profiles of Cultivated, in Vitro Cultured and Commercial Samples of Melissa officinalis L. Infusions. Food Chem. 2013, 136, 1–8. [Google Scholar] [CrossRef]
- Ye, M.; Yang, W.Z.; di Liu, K.; Qiao, X.; Li, B.J.; Cheng, J.; Feng, J.; Guo, D.A.; Zhao, Y.Y. Characterization of Flavonoids in Millettia nitida Var. Hirsutissima by HPLC/DAD/ESI-MSn. J. Pharm. Anal. 2012, 2, 35–42. [Google Scholar] [CrossRef]
- Francescato, L.N.; Debenedetti, S.L.; Schwanz, T.G.; Bassani, V.L.; Henriques, A.T. Identification of Phenolic Compounds in Equisetum giganteum by LC-ESI-MS/MS and a New Approach to Total Flavonoid Quantification. Talanta 2013, 105, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Felipe, D.F.; Brambilla, L.Z.S.; Porto, C.; Pilau, E.J.; Cortez, D.A.G. Phytochemical Analysis of Pfaffia glomerata Inflorescences by LC-ESI-MS/MS. Molecules 2014, 19, 15720–15734. [Google Scholar] [CrossRef] [PubMed]
- Chernonosov, A.A.; Karpova, E.A.; Lyakh, E.M. Identification of Phenolic Compounds in Myricaria bracteata Leaves by High-Performance Liquid Chromatography with a Diode Array Detector and Liquid Chromatography with Tandem Mass Spectrometry. Rev. Bras. Farmacogn. 2017, 27, 576–579. [Google Scholar] [CrossRef]
- Clauser, M.; Dall’Acqua, S.; Loi, M.C.; Innocenti, G. Phytochemical Investigation on Atriplex halimus L. from Sardinia. Nat. Prod. Res. 2013, 27, 1940–1944. [Google Scholar] [CrossRef] [PubMed]
- Salih, E.Y.A.; Fyhrquist, P.; Abdalla, A.M.A.; Abdelgadir, A.Y.; Kanninen, M.; Sipi, M.; Luukkanen, O.; Fahmi, M.K.M.; Elamin, M.H.; Ali, H.A. LC-MS/MS Tandem Mass Spectrometry for Analysis of Phenolic Compounds and Pentacyclic Triterpenes in Antifungal Extracts of Terminalia brownii (Fresen). Antibiotics 2017, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Bystrom, L.M.; Lewis, B.A.; Brown, D.L.; Rodriguez, E.; Obendorf, R.L. Characterisation of Phenolics by LC-UV/Vis, LC-MS/MS and Sugars by GC in Melicoccus bijugatus Jacq. “Montgomery” Fruits. Food Chem. 2008, 111, 1017–1024. [Google Scholar] [CrossRef]
- Brito, A.; Ramirez, J.E.; Areche, C.; Sepúlveda, B.; Simirgiotis, M.J. HPLC-UV-MS Profiles of Phenolic Compounds and Antioxidant Activity of Fruits from Three Citrus Species Consumed in Northern Chile. Molecules 2014, 19, 17400–17421. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.M. Studies on the Chemical Constituents Hi Seeds of Atriplex centralasiatica. Zhongguo Zhongyao Zazhi 2005, 30, 679–681. [Google Scholar]
- El-Askary, H.; Handoussa, H.; Badria, F.; El-Khatib, A.H.; Alsayari, A.; Linscheid, M.W.; Abdel Motaal, A. Characterization of Hepatoprotective Metabolites from Artemisia annua and Cleome droserifolia Using HPLC/PDA/ESI/MS–MS. Rev. Bras. Farmacogn. 2019, 29, 213–220. [Google Scholar] [CrossRef]
- Ahmed, R.; Elkhrisy, E.; EL-kashak, W.; el Raey, M.; Nassar, M.; Aboutabl, E.-S. Structural Characterization of Polyphenolics in Livistona chinensis Using HPLC-PDA-MS. J. Adv. Pharm. Res. 2019, 3, 23–29. [Google Scholar] [CrossRef]
- Khalaf, I.; Corciovă, A.N.D.R.E.I.A.; Vlase, L.; Ivănescu, B.; Lazăr, D. LC/MS Analysis of Sterolic Compounds from Glycyrrhiza glabra. Stud. UBB Chem. 2011, 56, 97–102. [Google Scholar]
- Göger, F.; Köse, Y.B.; Göger, G.; Demirci, F. Phytochemical Characterization of Phenolics by LC-MS/MS and Biological Evaluation of Ajuga orientalis from Turkey Fatih. Available online: https://www.banglajol.info/index.php/BJP/article/view/23500/16518 (accessed on 28 June 2022).
- Llorent-Martínez, E.J.; Gouveia, S.; Castilho, P.C. Analysis of Phenolic Compounds in Leaves from Endemic Trees from Madeira Island. A Contribution to the Chemotaxonomy of Laurisilva forest Species. Ind. Crops Prod. 2015, 64, 135–151. [Google Scholar] [CrossRef]
- Siddiqui, B.S.; Ahmed, S.; Ghiasuddin; Khan, M.A.U. Triterpenoids of Atriplex stocksii. Phytochemistry 1994, 37, 1123–1125. [Google Scholar] [CrossRef]
- Guo, H.; Liu, A.H.; Ye, M.; Yang, M.; Guo, D.A. Characterization of Phenolic Compounds in the Fruits of Forsythia suspensa by High-Performance Liquid Chromatography Coupled with Electrospray Ionization Tandem Mass Spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Novotny, L.; Abdel-Hamid, M.E.; Hamza, H.; Masterova, I.; Grancai, D. Development of LC-MS Method for Determination of Ursolic Acid: Application to the Analysis of Ursolic Acid in Staphylea holocarpa Hemsl. J. Pharm. Biomed. Anal. 2003, 31, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Carballo-Arce, A.F.; Singh, R.; Saleem, A.; Rocha, M.; Mullally, M.; Otarola-Rojas, M.; Alvarrez, L.P.; Sanchez-Vindas, P.; Garcia, M.; et al. A Selective Ion HPLC-APCI-MS Method for the Quantification of Pentacyclic Triterpenes in an Anxiolytic Botanical Dietary Supplement for the Animal Health Market. Nat. Prod. Commun. 2019, 14, 11–14. [Google Scholar] [CrossRef]
- Rybin, V.; Boltenkov, E.; Novozhilova, E. Application of High-Performance Liquid Chromatography for Simultaneous Identification of Integristerone A, 20-Hydroxyecdysone, Ecdysone and 2-Deoxy-20-Hydroxyecdysone. Nat. Prod. Commun. 2007, 2, 1101–1104. [Google Scholar] [CrossRef]
- Kosyakov, D.S.; Ul’yanovskii, N.V.; Falev, D.I. Determination of Triterpenoids from Birch Bark by Liquid Chromatography-Tandem Mass Spectrometry. J. Anal. Chem. 2014, 69, 1264–1269. [Google Scholar] [CrossRef]
- Hamed, M.; Article, O.; Matloub, A.A.; Hamed, M.A.; Souda, S.S.M. el Chemo-protective effect on hepato-renal toxcicity and cytotoxic activity of lipoidal matter of atriplex lindleyi moq. Orig. Artic. Artic. Int. J. Pharm. Pharm. Sci. 2014, 6, 187–196. [Google Scholar]
- Rozenberg, R.; Ruibal-Mendieta, N.L.; Petitjean, G.; Cani, P.; Delacroix, D.L.; Delzenne, N.M.; Meurens, M.; Quetin-Leclercq, J.; Habib-Jiwan, J.L. Phytosterol Analysis and Characterization in Spelt (Triticum aestivum Ssp. Spelta L.) and Wheat (T. Aestivum L.) Lipids by LC/APCI-MS. J. Cereal Sci. 2003, 38, 189–197. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, Y.; Zhang, W.; Chen, Z. Identification and Quantification of Oleanolic Acid and Ursolic Acid in Chinese Herbs by Liquid Chromatography-Ion Trap Mass Spectrometry. Biomed. Chromatogr. 2011, 25, 1381–1388. [Google Scholar] [CrossRef] [PubMed]
- Chermnykh, N.S.; Shimanovskiĭ, N.L.; Shutko, G.V.; Syrov, V.N. The Action of Methandrostenolone and Ecdysterone on the Physical Endurance of Animals and on Protein Metabolism in the Skeletal Muscles. Farm. Toksikol. 1988, 51, 57–60. [Google Scholar]
- Wang, D.; Yang, Y.; Zou, X.; Zhang, J.; Zheng, Z.; Wang, Z. Antioxidant Apigenin Relieves Age-Related Muscle Atrophy by Inhibiting Oxidative Stress and Hyperactive Mitophagy and Apoptosis in Skeletal Muscle of Mice. J. Gerontol. -Ser. A Biol. Sci. Med. Sci. 2020, 75, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.J.; Son, H.J.; Choi, Y.M.; Ahn, J.; Jung, C.H.; Ha, T.Y. Apigenin Enhances Skeletal Muscle Hypertrophy and Myoblast Differentiation by Regulating Prmt7. Oncotarget 2017, 8, 78300–78311. [Google Scholar] [CrossRef]
- Kanzaki, N.; Takemoto, D.; Ono, Y.; Izumo, T.; Shibata, H.; Ye, X.; Ingram, D.K.; Ye, J. Quercetin Glycosides Improve Motor Performance and Muscle Weight in Adult Mice. J. Nutr. Food Sci. 2019, 09, 1–7. [Google Scholar] [CrossRef][Green Version]
- ABEER, F. MOSTAFA, M.D., SHEREEN M. SAMIR, M.D. Ferulic Acid Promotes Growth of Both Fast Glycolytic and Slow Oxidative Skeletal Muscles in Corticosteroid-Induced Rat Myopathy. Med. J. Cairo Univ. 2019, 87, 1703–1715. [Google Scholar] [CrossRef]
- Ommati, M.M.; Farshad, O.; Mousavi, K.; Khalili, M.; Jamshidzadeh, A.; Heidari, R. Chlorogenic Acid Supplementation Improves Skeletal Muscle Mitochondrial Function in a Rat Model of Resistance Training. Biologia 2020, 75, 1221–1230. [Google Scholar] [CrossRef]
- Verma, S.K.; Jain, V.; Katewa, S.S. Anabolic effect of Bombax ceiba linn. root in idiopathic involuntary weight loss—A case study. J. Herb. Med. Toxicol. 2011, 5, 1–5. [Google Scholar]
- Kunkel, S.D.; Suneja, M.; Ebert, S.M.; Bongers, K.S.; Fox, D.K.; Malmberg, S.E.; Alipour, F.; Shields, R.K.; Adams, C.M. MRNA Expression Signatures of Human Skeletal Muscle Atrophy Identify a Natural Compound That Increases Muscle Mass. Cell Metab. 2011, 13, 627–638. [Google Scholar] [CrossRef]
- Pereira de Jésus-Tran, K.; Côté, P.-L.; Cantin, L.; Blanchet, J.; Labrie, F.; Breton, R. Comparison of Crystal Structures of Human Androgen Receptor Ligand-Binding Domain Complexed with Various Agonists Reveals Molecular Determinants Responsible for Binding Affinity. Protein Sci. 2006, 15, 987–999. [Google Scholar] [CrossRef]
- Möcklinghoff, S.; Rose, R.; Carraz, M.; Visser, A.; Ottmann, C.; Brunsveld, L. Synthesis and Crystal Structure of a Phosphorylated Estrogen Receptor Ligand Binding Domain. ChemBioChem 2010, 11, 2251–2254. [Google Scholar] [CrossRef] [PubMed]
- Delfosse, V.; Grimaldi, M.; Pons, J.L.; Boulahtouf, A.; le Maire, A.; Cavailles, V.; Labesse, G.; Bourguet, W.; Balaguer, P. Structural and Mechanistic Insights into Bisphenols Action Provide Guidelines for Risk Assessment and Discovery of Bisphenol A Substitutes. Proc. Natl. Acad. Sci. USA 2012, 109, 14930–14935. [Google Scholar] [CrossRef] [PubMed]
- Gorelick-Feldman, J.; MacLean, D.; Ilic, N.; Poulev, A.; Lila, M.A.; Cheng, D.; Raskin, I. Phytoecdysteroids Increase Protein Synthesis in Skeletal Muscle Cells. J. Agric. Food Chem. 2008, 56, 3532–3537. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, A.R.; Haque, M. Preparation of Medicinal Plants: Basic Extraction and Fractionation Procedures for Experimental Purposes. J. Pharm. Bioallied Sci. 2020, 12, 1–10. [Google Scholar] [CrossRef]
- El-Zahar, H.; Menze, E.T.; Handoussa, H.; Osman, A.K.; El-Shazly, M.; Mostafa, N.M.; Swilam, N. UPLC-PDA-MS/MS Profiling and Healing Activity of Polyphenol-Rich Fraction of Alhagi maurorum against Oral Ulcer in Rats. Plants 2022, 11, 455. [Google Scholar] [CrossRef]
- Swilam, N.; Nematallah, K.A. Polyphenols Profile of Pomegranate Leaves and Their Role in Green Synthesis of Silver Nanoparticles. Sci. Rep. 2020, 10, 14851. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; Mcmahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Allam, R.M.; Al-Abd, A.M.; Khedr, A.; Sharaf, O.A.; Nofal, S.M.; Khalifa, A.E.; Mosli, H.A.; Abdel-Naim, A.B. Fingolimod Interrupts the Cross Talk between Estrogen Metabolism and Sphingolipid Metabolism within Prostate Cancer Cells. Toxicol. Lett. 2018, 291, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Mazur, J.E. Review: Theory and Practice of Histological Techniques, by John D. Bancroft and Alan Stevens. Am. Biol. Teach. 1978, 40, 446. [Google Scholar] [CrossRef]
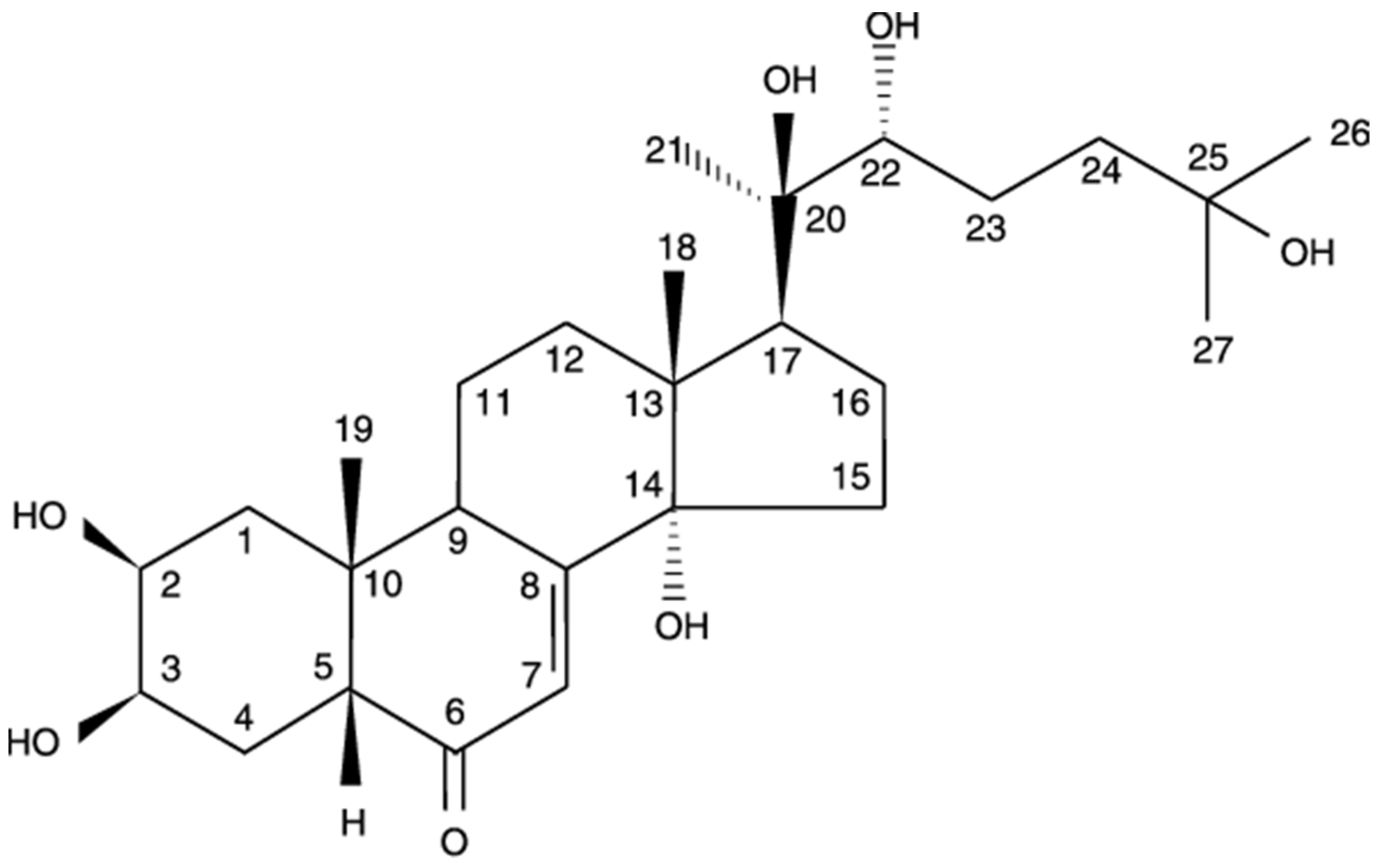



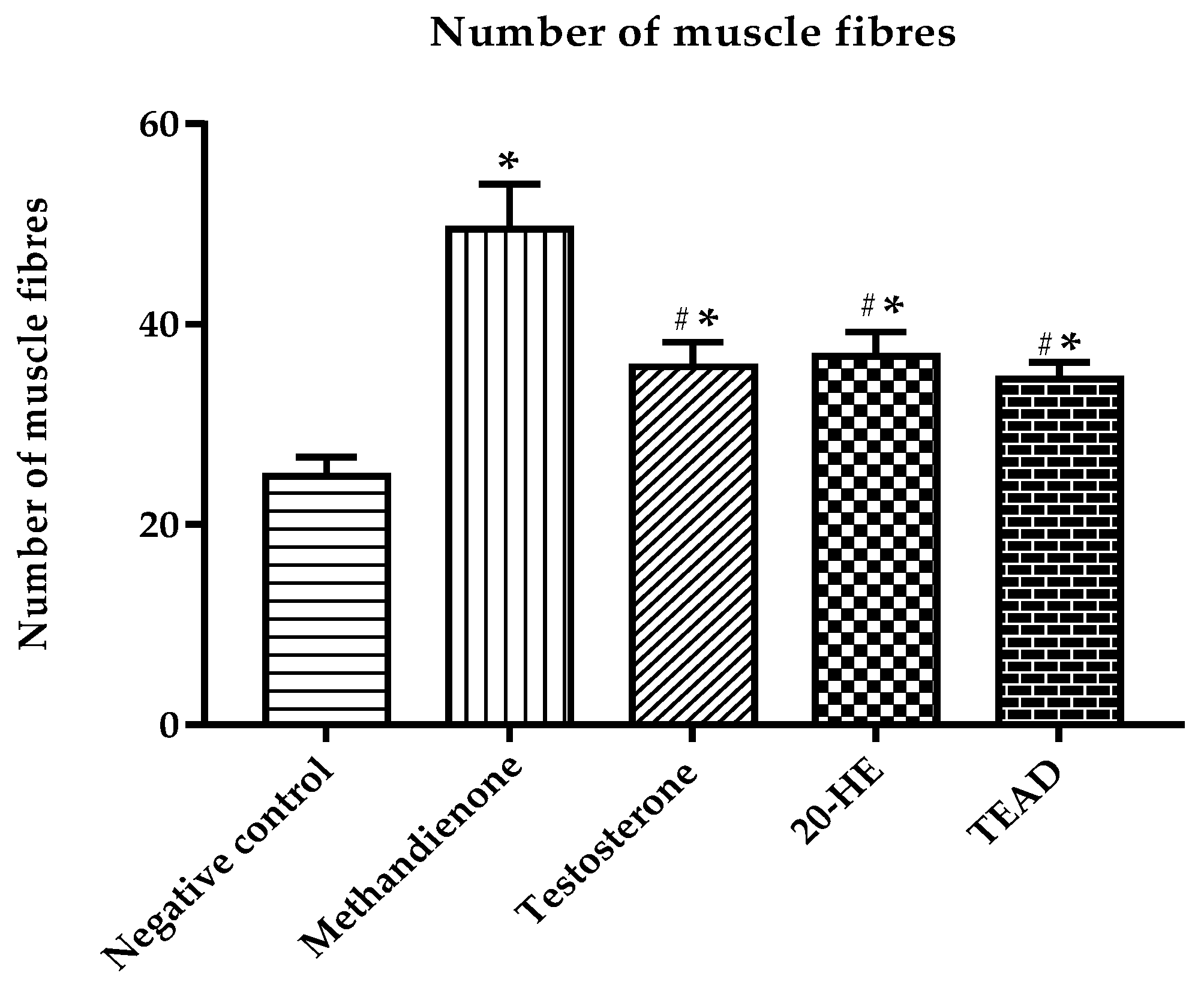
| Position | δ H | δ C |
|---|---|---|
| 1α | 1.97 | 38.89 |
| 1β | 1.48 | |
| 2 | 3.8 | 67.22 |
| 3 | 4.11 | 67.02 |
| 4α | 1.77 | 30.21 |
| 4β | 1.61 | |
| 5 | 2.39 | 50.52 |
| 6 | - | 203.8 |
| 7 | 5.82 | 120.85 |
| 8 | - | 166.09 |
| 9 | 3.13 | 38.09 |
| 10 | - | 39.73 |
| 11α | 1.81 | 22.79 |
| 11β | 1.61 | |
| 12α | 2.10 | 30.11 |
| 12β | 1.90 | |
| 13 | - | 49.12 |
| 14 | - | 83.55 |
| 15α | 2.17 | 29.28 |
| 15β | 1.58 | |
| 16α | 1.81 | 23.69 |
| 16β | 1.97 | |
| 17 | 2.51 | 49.12 |
| 18 | 0.89 | 17.54 |
| 19 | 1.05 | 24.20 |
| 20 | - | 76.34 |
| 21 | 1.22 | 21.27 |
| 22 | 3.49 | 76.76 |
| 23α | 1.73 | 24.91 |
| 23β | 1.32 | |
| 24α | 1.83 | 41.65 |
| 24β | 1.46 | |
| 25 | - | 69.49 |
| 26 | 1.22 | 26.44 |
| 27 | 1.25 | 28.76 |
| Peak # | Retention Time (min) | Identified Compound | UV-Vis (λmax) | [M−H]− (m/z) | [M+H]+ (m/z) | Fragment Ions (m/z) | Peak Area (%) | Occurrence | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.205 | Caffeoyl hexose-deoxyhexoside | 232, 318 | 487.01 | 307, 179 | 0.02 | - | [42] | |
| 2 | 0.306 | Chlorogenic acid derivative | 220, 325 | 451.12 | 353, 191 | 0.95 | A. mollis | [43,44] | |
| 3 | 0.37 | Isoorientin | 238, 349 | 448.2 | 357, 327 | 0.89 | A. halimus | [45,46] | |
| 4 | 0.42 | Apigenin dihexoside | 242, 334 | 593.4 | 269 | 1.29 | A. halimus | [45,47] | |
| 5 | 0.56 | Liquiritin | 239, 335 | 417.01 | 255 | 0.78 | - | [44] | |
| 6 | 0.65 | Ferulic acid | 235 | 193.1 | 178, 161, 134 | 0.75 | A. mollis | [43,48] | |
| 7 | 0.78 | Kaempferol deoxyhexoside | 245, 348 | 431.04 | 285 | 0.52 | A. lentiformis | [49] | |
| 8 | 0.795 | Dihydroxy benzoic acid | 247, 314 | 153.0 | 108 | 2.25 | A. hortensis | [50] | |
| 9 | 0.92 | Caffeic acid | 233 | 179.01 | 135, 107 | 0.99 | A. lindelyi | [26,51] | |
| 10 | 0.95 | Caffeoyl hexose-deoxyhexoside isomer | 233, 322 | 487.08 | 308, 179 | 0.36 | - | [42] | |
| 11 | 1.01 | Quercetin pentosyl hexoside | 235, 355 | 595.2 | 463, 301 | 0.99 | A. lindelyi | [26] | |
| 12 | 1.021 | Rosmarinic acid hexoside | 232, 318 | 521.1 | 359, 197 | 0.45 | - | [52] | |
| 13 | 1.138 | Retusin methyl ether | 230, 282 | 297.3 | 282, 239, 211 | 0.69 | - | [53] | |
| 14 | 1.2 | Caffeic acid derivative | 244, 327 | 295 | 135, 179 | 1.06 | - | [54] | |
| 15 | 1.3 | Ferulic acid ester derivative | 241, 327 | 309.53 | 192, 177, 115 | 0.63 | - | [54] | |
| 16 | 1.5 | Tetrahydroxyflavan (Afzelechin) | 242, 323 | 273.1 | 255, 179 | 0.76 | - | [48] | |
| 17 | 2.12 | Kaempferol | 250, 370 | 285.10 | 153 | 0.65 | A. halimus | [49,55] | |
| 18 | 2.2 | Isorhamnetin | 244, 368 | 315.2 | 301,271, 151 | 0.34 | A. lindelyi | [26,56] | |
| 19 | 2.23 | Myricetin | 243, 372 | 317.1 | 179, 151 | 0.42 | A. halimus | [56,57] | |
| 20 | 2.24 | Quercetin | 238, 374 | 301.0 | 179, 151 | 0.09 | A. halimus A. lindelyi | [26,57] | |
| 21 | 2.25 | Dihydroxybenzoyl hexose | nd | 316.1 | 153,108 | 1.08 | A. lindelyi | [26] | |
| 22 | 3.58 | Coumaroyl hydroxy-palmitic acid | 223 | 418.5 | 163, 145, 119, | 2.50 | - | [48] | |
| 23 | 4.23 | Quercetin-galloyl-pentoside | 236, 354 | 585 | 301, 179, 153 | 2.80 | - | [58] | |
| 24 | 4.52 | Coumaroyl-hexose | 228, 315 | 325.65 | 163 | 0.07 | - | [59] | |
| 25 | 4.52 | Kaempferol–dideoxyhexoside (Kaempferitrin) | 246, 354 | 577 | 285 | 1.33 | [49] | ||
| 26 | 4.71 | Dicaffeoyl-spermidine | 232, 309 | 468.2 | 332, 135 | 0.98 | - | [51] | |
| 27 | 4.75 | Syringetin rutinoside | 222, 312 | 579.2 | 345 | 1.02 | A. halimus | [20,52] | |
| 28 | 4.79 | Acetylated kaempferol deoxyhexosyl hexoside | 243, 350 | 635.2 | 299, 284 | 3.59 | - | [52] | |
| 29 | 4.92 | Kaempferol deoxyhexosyl hexoside | 262, 355 | 593.1 | 447, 285 | 0.55 | A. lentiformis | [49] | |
| 30 | 5.3 | Isorhamnetin hexoside | 268, 355 | 477 | 315, 271 | 1.07 | - | [60] | |
| 31 | 5.95 | Quercetin deoxyhexoside | 225, 358 | 447 | 301,179 | 1.20 | A. centralasiatica | [61,62] | |
| 32 | 6.07 | Apigenin-C-hexoside | 269, 334 | 431.01 | 311, 341 | 0.77 | - | [61,63] | |
| 33 | 6.52 | Daucosterol (Sitogluside) | 255, 278 | 577 | 413, 369 | 1.96 | A. centralasiatica | [64] | |
| 34 | 6.52 | Isorhamnetin deoxhexosyl hexoside | 268, 356 | 623.3 | 315, 301, 179, 151 | 2.52 | A. halimus | [20,60] | |
| 35 | 6.78 | Kaempferol glucuronide | 247, 354 | 513.1 | 461, 285, 175,135 | 0.08 | - | [65] | |
| 36 | 6.92 | Sinapic acid hexoside | 228 | 385 | 223 | 1.04 | - | [66] | |
| 37 | 7.3 | Tamerexetin deoxyhexosyl hexoside | 250, 368 | 623.2 | 325, 315 | 1.71 | A. halimus | [20] | |
| 38 | 7.5 | Apigenin-C-hexoside | 270, 336 | 431 | 353, 341, 311, 269 | 0.59 | - | [61,63] | |
| 39 | 7.9 | Caffeoyl-coumaroyl spermidine | 223 | 452.4 | 332, 316, 306, 289, 135 | 2.90 | - | [51] | |
| 40 | 8.75 | β-Sitosterol | 415.5 | 397, 175, 257 | 5.66 | A. stocksii | [64,67] | ||
| 41 | 9.01 | Caffeoyl pinoresinol | 232, 309 | 519.5 | 357, 151 | 0.85 | - | [68] | |
| 42 | 9.23 | Tetrahydroxyflavan (Afzelechin) | 242, 323 | 273.1 | 255, 179 | 0.92 | - | [48] | |
| 43 | 9.85 | Kaempferol | 250, 370 | 285.10 | 153 | 1.60 | A. lentiformis | [49] | |
| 44 | 10.01 | Portulasoid butoxyseptanoside | 265 | /237 | 253 | 1.52 | A. portulacoides | [36] | |
| 45 | 10.2 | Isorhamnetin | 244, 368 | 315.2 | 301, 271, 151 | 1.71 | A. halimus | [56,57] | |
| 46 | 11.05 | Myricetin | 243, 372 | 317.1 | 179, 151 | 1.91 | A. halimus | [56,57] | |
| 47 | 11.51 | Quercetin | 238, 374 | 301.0 | 179, 151 | 2.02 | A. lindelyi | [26] | |
| 48 | 11.59 | Tetrahydroxyflavan (Afzelechin) | 242, 323 | 273.1 | 255, 179 | 0.73 | - | [48] | |
| 49 | 11.84 | Ursolic acid | 226 | 457 | 407, 391 | 1.68 | A. stocksii | [67,69] | |
| 50 | 12.05 | β/α-amyrin | 255, 270 | 427.2 | 218, 203, 189 | 2.08 | A. stocksii | [67,70] | |
| 51 | 12.5 | 20-hydroxyecdysone | 248 | 481 | 445, 427, 409, 391 | 11.91 | A. portulacoides | [36,71] | |
| 52 | 12.8 | β/α-amyrin | 270 | 427.2 | 218, 203, 189 | 1.25 | A. stocksii | [67,70] | |
| 53 | 13.2 | Lupeol | 265 | 427.5 | 409, 397 | 0.52 | A. lindelyi | [72,73] | |
| 54 | 13.55 | Stigmasterol | 285 | 413.2 | 395, 411 | 3.02 | A. lindelyi | [73,74] | |
| 55 | 14.57 | Oleonolic acid | 260 | 457 | 439, 411, 393 | 1.36 | A. stocksii | [67,75] | |
| 56 | 14.89 | Septanoecdysone | 255 | 657.2 | 521, 439 | 0.22 | A. portulacoides | [36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaghloul, E.; Handousa, H.; Singab, A.N.B.; Elmazar, M.M.; Ayoub, I.M.; Swilam, N. Phytoecdysteroids and Anabolic Effect of Atriplex dimorphostegia: UPLC-PDA-MS/MS Profiling, In Silico and In Vivo Models. Plants 2023, 12, 206. https://doi.org/10.3390/plants12010206
Zaghloul E, Handousa H, Singab ANB, Elmazar MM, Ayoub IM, Swilam N. Phytoecdysteroids and Anabolic Effect of Atriplex dimorphostegia: UPLC-PDA-MS/MS Profiling, In Silico and In Vivo Models. Plants. 2023; 12(1):206. https://doi.org/10.3390/plants12010206
Chicago/Turabian StyleZaghloul, Eman, Heba Handousa, Abdel Nasser B. Singab, Mohey M. Elmazar, Iriny M. Ayoub, and Noha Swilam. 2023. "Phytoecdysteroids and Anabolic Effect of Atriplex dimorphostegia: UPLC-PDA-MS/MS Profiling, In Silico and In Vivo Models" Plants 12, no. 1: 206. https://doi.org/10.3390/plants12010206
APA StyleZaghloul, E., Handousa, H., Singab, A. N. B., Elmazar, M. M., Ayoub, I. M., & Swilam, N. (2023). Phytoecdysteroids and Anabolic Effect of Atriplex dimorphostegia: UPLC-PDA-MS/MS Profiling, In Silico and In Vivo Models. Plants, 12(1), 206. https://doi.org/10.3390/plants12010206






