The Molecular Mechanism of GhbHLH121 in Response to Iron Deficiency in Cotton Seedlings
Abstract
1. Introduction
2. Results
2.1. Iron Deficiency Induces Differential Expression of Genes Related to Iron Deficiency with Consequent Low Photosynthetic Efficiency in Cotton
2.2. GhbHLH121 Encodes a Transcription Factor Involved in Responding to Iron Deficiency in Root and Shoot Tissues
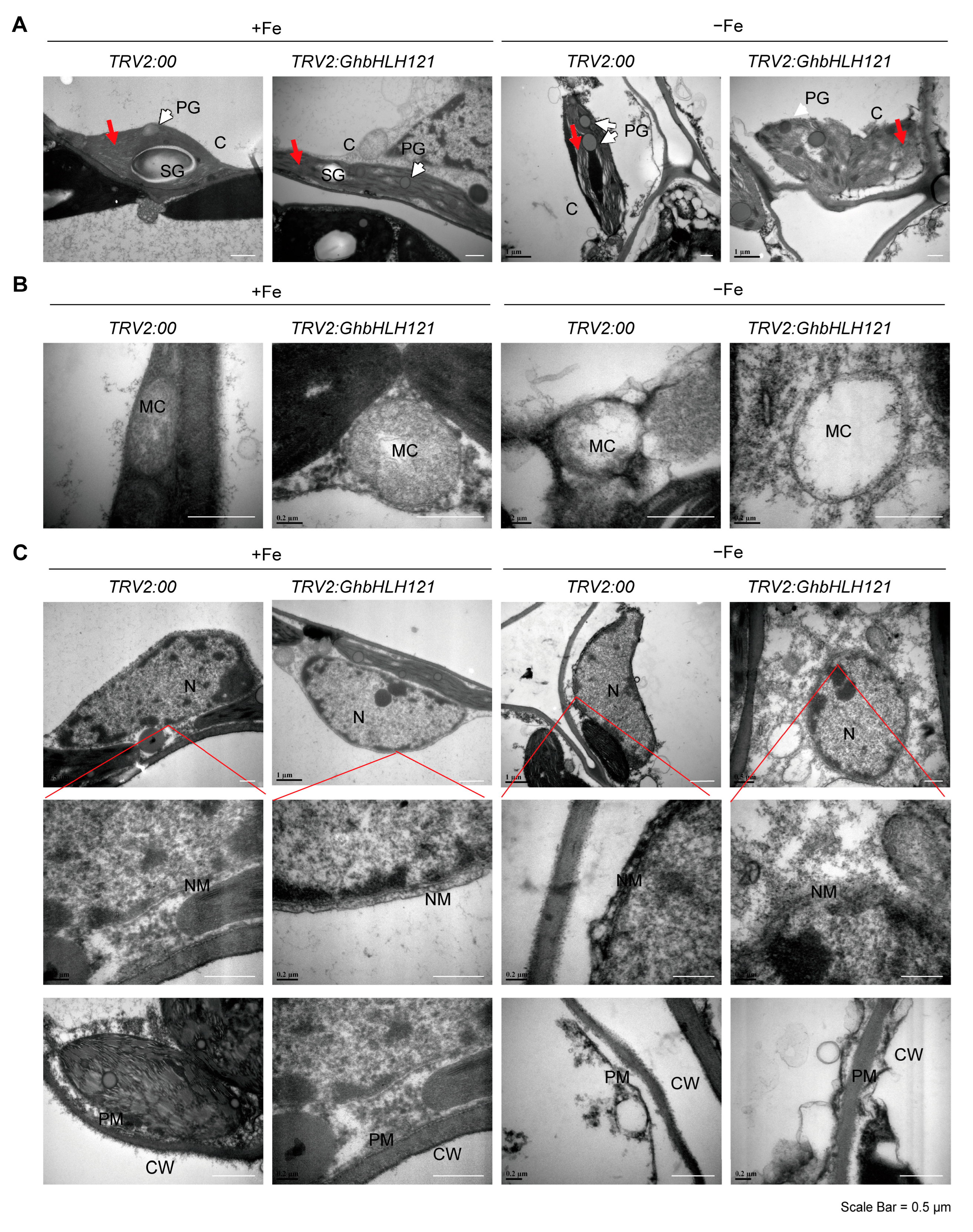
2.3. Suppression of GhbHLH121 in Cotton Seedlings Reduced Tolerance to Iron Deficiency
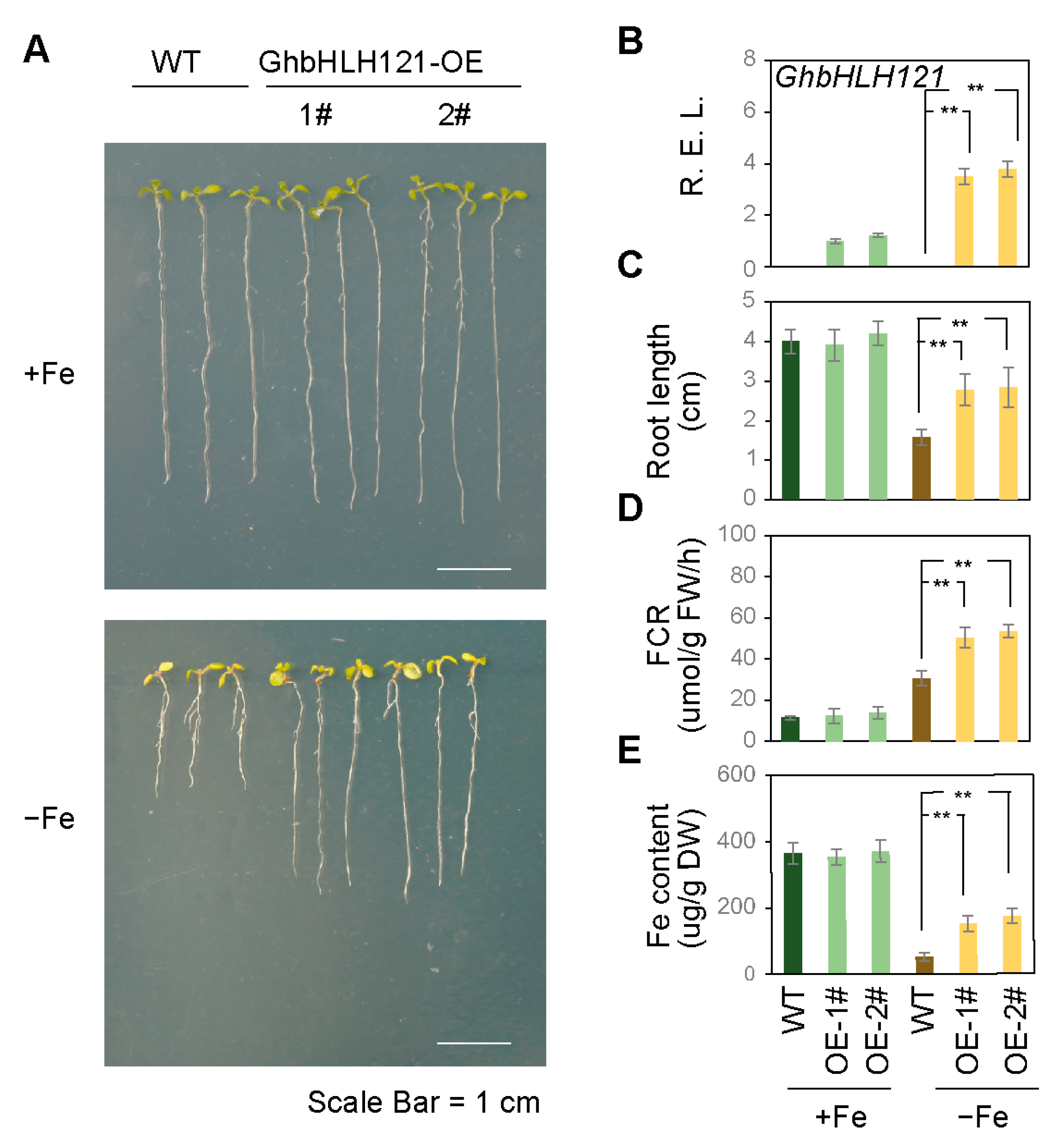
2.4. Ectopic Expression of GhbHLH121 in Arabidopsis Confers Enhanced Tolerance to Iron Deficiency
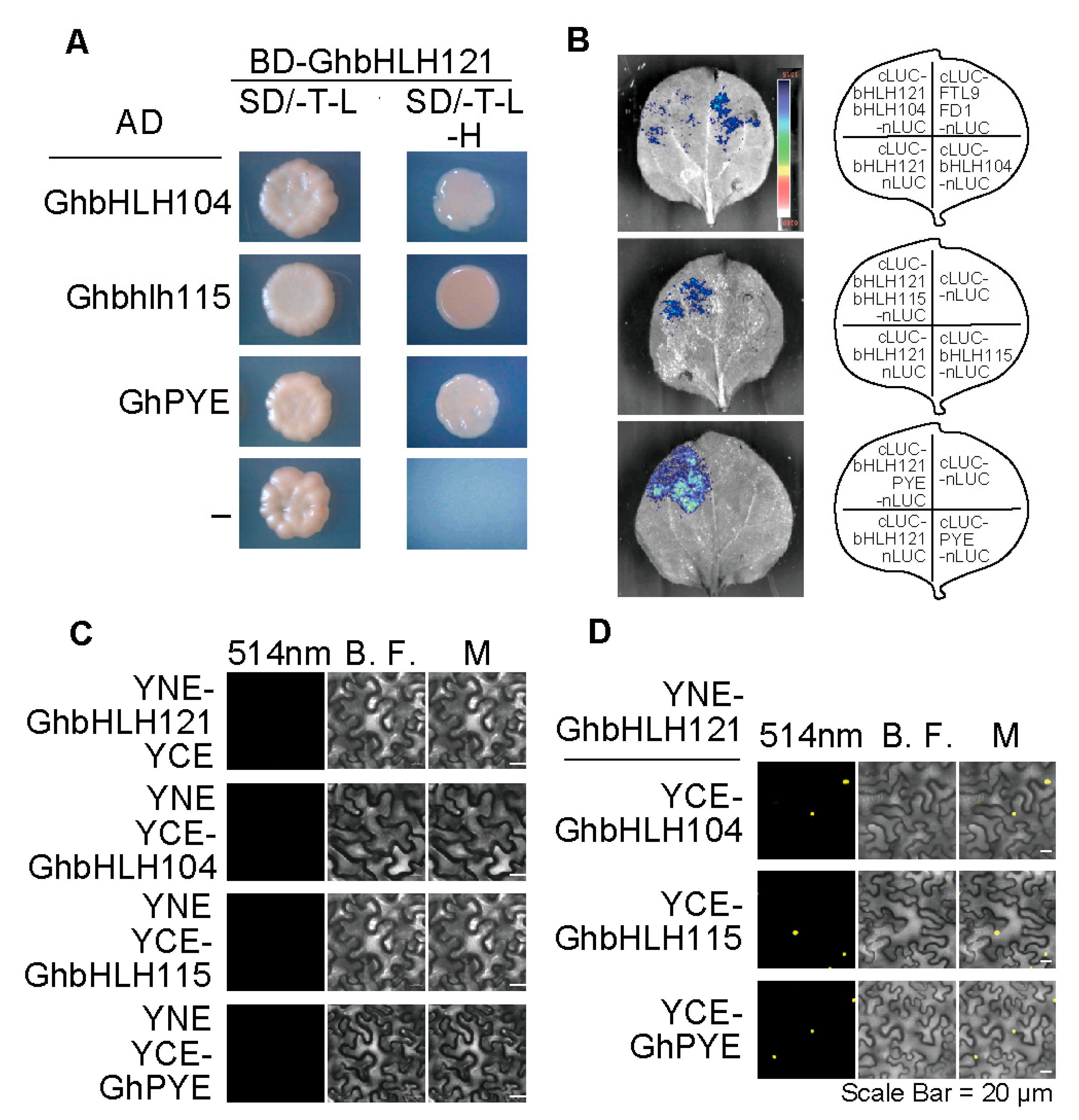
2.5. GhbHLH121 Interacts with GhbHLH IVc Genes and GhPYE
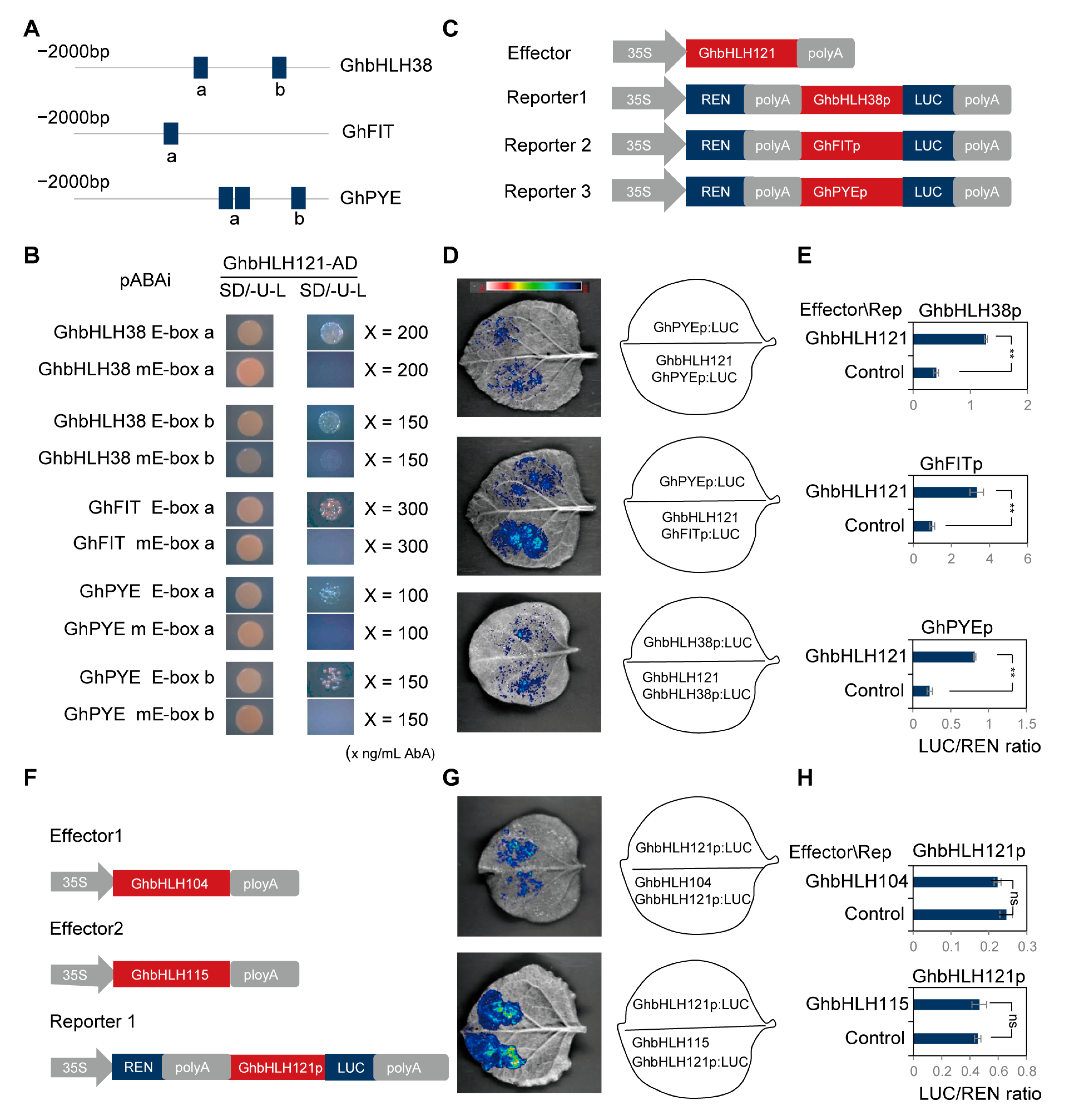
2.6. GhbHLH121 Directly Activates Transcription of GhbHLH38, GhFIT, and GhPYE Independent of GhbHLH104 and GhbHLH115
3. Discussion
3.1. Effects of Iron Deficiency Stress on Cotton Seedling Development
3.2. GhbHLH121 Is a Positive Regulator for the Response to Iron Deficiency in Cotton
3.3. The Transcriptional Regulatory Network of Cotton GhbHLH121
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Construction of Materials for Virus-Induced Gene Silencing (VIGS-GhbHLH121) in TM-1
4.3. Generation of Transgenic Arabidopsis
4.4. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
4.5. Phylogenetic Analysis
4.6. Measurement of Iron Deficiency-Associated Physiological Parameters
4.6.1. Photosynthetic Measurements
4.6.2. Fe Concentration Measurement
4.6.3. Ferric Chelate Reductase Assays
4.6.4. Chlorophyll Measurement
4.6.5. NBT Staining and DAB Staining
4.6.6. Measurements of SOD, POD, and CAT Activity
4.7. Microscopic Observation
4.7.1. Subcellular Localization
4.7.2. Transmission Electron Microscopy
4.8. Validation of Protein–Protein Interactions
4.8.1. Yeast Two-Hybrid Screening and Assays
4.8.2. Bimolecular Fluorescence Complementation
4.8.3. Luciferase Complementation Imaging Assay
4.9. Validation of DNA–Protein Interactions
4.9.1. Yeast One-Hybrid Assay
4.9.2. Luciferase Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Barak, P. Iron Nutrition of Plants in Calcareous Soils. Adv. Agron. 1982, 35, 217–240. [Google Scholar]
- Vose, P.B. Iron nutrition in plants: A world overview. J. Plant Nutr. 2008, 5, 233–249. [Google Scholar] [CrossRef]
- Murata, Y.; Itoh, Y.; Iwashita, T.; Namba, K. Transgenic petunia with the iron(III)-phytosiderophore transporter gene acquires tolerance to iron deficiency in alkaline environments. PLoS ONE 2015, 10, e0120227. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Zhang, F. Soil and crop management strategies to prevent iron deficiency in crops. Plant Soil 2010, 339, 83–95. [Google Scholar] [CrossRef]
- Ye, X.; Wang, H.; Cao, X.; Jin, X.; Cui, F.; Bu, Y.; Liu, H.; Wu, W.; Takano, T.; Liu, S. Transcriptome profiling of Puccinellia tenuiflora during seed germination under a long-term saline-alkali stress. BMC Genom. 2019, 20, 589. [Google Scholar] [CrossRef] [PubMed]
- Cesare, F.; Pietrini, F.; Zacchini, M.; Mugnozza, G.S.; Macagnano, A. Catechol-Loading Nanofibrous Membranes for Eco-Friendly Iron Nutrition of Plants. Nanomaterials 2019, 9, 1315–1337. [Google Scholar] [CrossRef]
- Crichton, R.R.; Ward, R.J. Iron species in iron homeostasis and toxicity. Analyst 1995, 120, 693–702. [Google Scholar] [CrossRef]
- Connorton, J.M.; Balk, J.; Rodriguez-Celma, J. Iron homeostasis in plants—A brief overview. Metallomics 2017, 9, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.J. Iron homeostasis and iron acquisition in plants: Maintenance, functions and consequences. Ann. Bot. 2010, 105, 799–800. [Google Scholar] [CrossRef] [PubMed]
- Nishio, J.N.; Terry, N. Iron nutrition-mediated chloroplast development. Plant Physiol. 1983, 71, 688–691. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Briat, J.F.; Dubos, C.; Gaymard, F. Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 2015, 20, 33–40. [Google Scholar] [CrossRef]
- Power, F.B.; Chesnut, V.K. Alkaline Reaction of the Cotton Plant. Science 1924, 60, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Saghir, A.; Khan, N.-u.-I.; Iqbal, M.Z.; Hussain, A.; Hassan, M. Salt Tolerance of Cotton (Gossypium hirsutum L.). Asian J. Plant Sci. 2002, 1, 715–719. [Google Scholar]
- Sharif, I.; Aleem, S.; Farooq, J.; Rizwan, M.; Younas, A.; Sarwar, G.; Chohan, S.M. Salinity stress in cotton: Effects, mechanism of tolerance and its management strategies. Physiol. Mol. Biol. Plants 2019, 25, 807–820. [Google Scholar] [CrossRef]
- Vrettako, H.; Kallinis, T.L. Iron Deficiency in Cotton in Relation to Growth and Nutrient Balance. Soil Sci. Soc. Am. J. 1968, 32, 253–258. [Google Scholar]
- Brown, J.C.; Foy, C.D.; Bennett, J.H.; Christiansen, M.N. Light-Sources Differentially Affected Ferric Iron Reduction and Growth of Cotton. Plant Physiol. 1979, 63, 692–695. [Google Scholar] [CrossRef]
- Guerinot, M.L.; Yi, Y. Iron: Nutritious, Noxious, and Not Readily Available. Plant Physiol. 1994, 104, 815–820. [Google Scholar] [CrossRef]
- Nam, H.-I.; Shahzad, Z.; Dorone, Y.; Clowez, S.; Zhao, K.; Bouain, N.; Lay-Pruitt, K.S.; Cho, H.; Rhee, S.Y.; Rouached, H. Interdependent iron and phosphorus availability controls photosynthesis through retrograde signaling. Nat. Commun. 2021, 12, 7211. [Google Scholar] [CrossRef]
- Lucena, J.J.; Hernandez-Apaolaza, L. Iron nutrition in plants: An overview. Plant Soil. 2017, 418, 1–4. [Google Scholar] [CrossRef]
- Chen, J.; Wu, F.H.; Shang, Y.T.; Wang, W.H.; Hu, W.J.; Simon, M.; Liu, X.; Shangguan, Z.-P.; Zheng, H.-L. Hydrogen sulphide improves adaptation of Zea mays seedlings to iron deficiency. J. Exp. Bot. 2015, 66, 6605–6622. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, G.; Chen, P.; Zhu, H.; Wang, S.; Ding, Y. Shoot-Root Communication Plays a Key Role in Physiological Alterations of Rice (Oryza sativa) Under Iron Deficiency. Front. Plant Sci. 2018, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Liang, G. Iron uptake, signaling, and sensing in plants. Plant Commun. 2022, 3, 100349. [Google Scholar] [CrossRef] [PubMed]
- Mori, S. Iron acquisition by plants. Curr. Opin. Plant Biol. 1999, 2, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Robe, K.; Gaymard, F.; Izquierdo, E.; Dubos, C. The Transcriptional Control of Iron Homeostasis in Plants: A Tale of bHLH Transcription Factors? Front. Plant Sci. 2019, 10, 6. [Google Scholar] [CrossRef]
- Santi, S.; Schmidt, W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009, 183, 1072–1084. [Google Scholar] [CrossRef]
- Connolly, E.L.; Campbell, N.H.; Grotz, N.; Prichard, C.L.; Guerinot, M.L. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 2003, 133, 1102–1110. [Google Scholar] [CrossRef]
- Varotto, C.; Maiwald, D.; Pesaresi, P.; Jahns, P.; Salamini, F.; Leister, D. The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J. 2002, 31, 589–599. [Google Scholar] [CrossRef]
- Shin, L.J.; Lo, J.C.; Chen, G.H.; Callis, J.; Fu, H.Y.; Yeh, K.C. IRT1 DEGRADATION FACTOR1, a RING E3 Ubiquitin Ligase, Regulates the Degradation of IRON-REGULATED TRANSPORTER1 in Arabidopsis. Plant Cell 2013, 25, 3039–3051. [Google Scholar] [CrossRef]
- Barberon, M.; Dubeaux, G.; Kolb, C.; Isono, E.; Zelazny, E.; Vert, G. Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 8293–8298. [Google Scholar] [CrossRef]
- Zelazny, E.; Vert, G. Regulation of Iron Uptake by IRT1: Endocytosis Pulls the Trigger. Mol. Plant 2015, 8, 977–979. [Google Scholar] [CrossRef]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2021, 33, 439–440. [Google Scholar] [CrossRef] [PubMed]
- Senoura, T.; Sakashita, E.; Kobayashi, T.; Takahashi, M.; Aung, M.S.; Masuda, H.; Nakanishi, H.; Nishizawa, N.K. The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains. Plant Mol. Biol. 2017, 95, 375–387. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Masuda, H.; Bashir, K.; Inoue, H.; Tsukamoto, T.; Takahashi, M.; Nakanishi, H.; Aoki, N.; Hirose, T.; Ohsugi, R.; et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010, 62, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, Y.; Liang, G. FIT and bHLH Ib transcription factors modulate iron and copper crosstalk in Arabidopsis. Plant Cell Environ. 2021, 44, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Bauer, P.; Ling, H.Q.; Guerinot, M.L. FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol. Biochem. 2007, 45, 260–261. [Google Scholar] [CrossRef]
- Long, T.A.; Tsukagoshi, H.; Busch, W.; Lahner, B.; Salt, D.E.; Benfey, P.N. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 2010, 22, 2219–2236. [Google Scholar] [CrossRef]
- Akmakjian, G.Z.; Riaz, N.; Guerinot, M.L. Photoprotection during iron deficiency is mediated by the bHLH transcription factors PYE and ILR3. Proc. Natl. Acad. Sci. USA 2021, 118, e2024918118. [Google Scholar] [CrossRef]
- Li, Y.; Lu, C.K.; Li, C.Y.; Lei, R.H.; Pu, M.N.; Zhao, J.H.; Peng, F.; Ping, H.Q.; Wang, D.; Liang, G. IRON MAN interacts with BRUTUS to maintain iron homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2109063118. [Google Scholar] [CrossRef]
- Hindt, M.N.; Akmakjian, G.Z.; Pivarski, K.L.; Punshon, T.; Baxter, I.; Salt, D.E.; Guerinot, M.L. BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 2017, 9, 876–890. [Google Scholar] [CrossRef]
- Rodriguez-Celma, J.; Chou, H.; Kobayashi, T.; Long, T.A.; Balk, J. Hemerythrin E3 Ubiquitin Ligases as Negative Regulators of Iron Homeostasis in Plants. Front. Plant Sci. 2019, 10, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Wu, H.; Wang, N.; Li, J.; Zhao, W.; Du, J.; Wang, D.; Ling, H.-Q. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008, 18, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, N.; Noshi, M.; Mori, D.; Nozawa, K.; Tamoi, M.; Shigeoka, S. The basic helix-loop-helix transcription factor, bHLH11 functions in the iron-uptake system in Arabidopsis thaliana. J. Plant Res. 2019, 132, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lei, R.; Pu, M.; Cai, Y.; Lu, C.; Li, Z.; Liang, G. bHLH11 inhibits bHLH IVc proteins by recruiting the TOPLESS/TOPLESS-RELATED corepressors. Plant Physiol. 2021, 188, 1335–1349. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; LaCroix, I.S.; Gerber, S.A.; Guerinot, M.L. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc. Natl. Acad. Sci. USA 2019, 116, 24933–24942. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.; Li, Y.; Cai, Y.; Li, C.; Pu, M.; Lu, C.; Yang, Y.; Liang, G. bHLH121 Functions as a Direct Link that Facilitates the Activation of FIT by bHLH IVc Transcription Factors for Maintaining Fe Homeostasis in Arabidopsis. Mol. Plant 2020, 13, 634–649. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Robe, K.; Bettembourg, M.; Navarro, N.; Rofidal, V.; Santoni, V.; Gaymard, F.; Vignols, F.; Roschzttardtz, H.; Izquierdo, E.; et al. The Transcription Factor bHLH121 Interacts with bHLH105 (ILR3) and Its Closest Homologs to Regulate Iron Homeostasis in Arabidopsis. Plant Cell 2020, 32, 508–524. [Google Scholar] [CrossRef]
- Sivitz, A.B.; Hermand, V.; Curie, C.; Vert, G. Arabidopsis bHLH100 and bHLH101 Control Iron Homeostasis via a FIT-Independent Pathway. PLoS ONE 2012, 7, e44843. [Google Scholar] [CrossRef]
- Gao, F.; Robe, K.; Dubos, C. Further insights into the role of bHLH121 in the regulation of iron homeostasis in Arabidopsis thaliana. Plant Signal. Behav. 2020, 15, 1795582. [Google Scholar] [CrossRef]
- Gao, F.; Dubos, C. Transcriptional integration of plant responses to iron availability. J. Exp. Bot. 2021, 72, 2056–2070. [Google Scholar] [CrossRef]
- Riaz, N.; Guerinot, M.L. All together now: Regulation of the iron deficiency response. J. Exp. Bot. 2021, 72, 2045–2055. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Tu, L.; Wang, P.; Du, X.; Ye, S.; Luo, M.; Zhang, X. Ascorbate Alleviates Fe Deficiency-Induced Stress in Cotton (Gossypium hirsutum) by Modulating ABA Levels. Front. Plant Sci. 2016, 7, 1997. [Google Scholar] [CrossRef] [PubMed]
- Hamdani, S.; Wang, H.; Zheng, G.; Perveen, S.; Qu, M.; Khan, N.; Khan, N.; Khan, W.; Jiang, J.; Li, M.; et al. Genome-wide association study identifies variation of glucosidase being linked to natural variation of the maximal quantum yield of photosystem II. Plant Physiol. 2019, 166, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Essemine, J.; Hamdani, S.; Qu, M.N.; Lyu, M.J.A.; Perveen, S.; Stirbet, A.; Govindjee, G.; Zhu, X.G. Natural variation in the fast phase of chlorophyll a fluorescence induction curve (OJIP) in a global rice minicore panel. Photosynth Res. 2020, 150, 137–158. [Google Scholar] [CrossRef]
- Rodríguez-Celma, J.; Connorton, J.M.; Kruse, I.; Green, R.T.; Franceschetti, M.; Chen, Y.-T.; Cui, Y.; Ling, H.-Q.; Yeh, K.-C.; Balk, J. Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. Proc. Natl. Acad. Sci. USA 2019, 116, 17584–17591. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Irrcher, I.; Ljubicic, V.; Kirwan, A.F.; Hood, D.A. AMP-activated protein kinase-regulated activation of the PGC-1alpha promoter in skeletal muscle cells. PLoS ONE 2008, 3, e3614. [Google Scholar] [CrossRef]
- Nan, L.; Guo, Q.; Cao, S. Archaeal community diversity in different types of saline-alkali soil in arid regions of Northwest China. J. Biosci. Bioeng. 2020, 130, 382–389. [Google Scholar] [CrossRef]
- Ben-Shem, A.; Frolow, F.; Nelson, N. Crystal structure of plant photosystem I. Nature 2003, 426, 630–635. [Google Scholar] [CrossRef]
- Umena, Y.; Kawakami, K.; Shen, J.R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Terry, N. Limiting Factors in Photosynthesis: IV. Iron Stress-Mediated Changes in Light-Harvesting and Electron Transport Capacity and its Effects on Photosynthesis in Vivo. Plant Physiol. 1983, 71, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.X.; Prentice, I.C.; Medlyn, B.E. Bridging Drought Experiment and Modeling: Representing the Differential Sensitivities of Leaf Gas Exchange to Drought. Front. Plant Sci. 2018, 9, 1965. [Google Scholar] [CrossRef] [PubMed]
- Bjorn, M.; Ruiz-Torres, N.A. Effects of Water-Deficit Stresson Photosynthesis, Its Components and Component Limitations, and on Water Use Efficiency in Wheat (Triticum aestivum L.). Plant Physiol. 1992, 100, 733–739. [Google Scholar]
- Blatt, M.R.; Jezek, M.; Lew, V.L.; Hills, A. What can mechanistic models tell us about guard cells, photosynthesis, and water use efficiency? Trends Plant Sci. 2022, 27, 166–179. [Google Scholar] [CrossRef]
- Singh, A.K.; McIntyre, L.M.; Sherman, L.A. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2003, 132, 1825–1839. [Google Scholar] [CrossRef]
- Strzepek, R.F.; Boyd, P.W.; Sunda, W.G. Photosynthetic adaptation to low iron, light, and temperature in Southern Ocean phytoplankton. Proc. Natl. Acad. Sci. USA 2019, 116, 4388–4393. [Google Scholar] [CrossRef]
- Larbi, A.; Abadia, A.; Morales, F.; Abadia, J. Fe Resupply to Fe-deficient Sugar Beet Plants Leads to Rapid Changes in the Violaxanthin Cycle and other Photosynthetic Characteristics without Significant de novo Chlorophyll Synthesis. Photosynth. Res. 2004, 79, 59–69. [Google Scholar] [CrossRef]
- Chen, W.; Zhao, L.; Liu, L.; Li, X.; Li, Y.; Liang, G.; Wang, H.; Yu, D. Iron deficiency-induced transcription factors bHLH38/100/101 negatively modulate flowering time in Arabidopsis thaliana. Plant Sci. 2021, 308, 110929. [Google Scholar] [CrossRef]
- Li, X.; Zhang, H.; Ai, Q.; Liang, G.; Yu, D. Two bHLH Transcription Factors, bHLH34 and bHLH104, Regulate Iron Homeostasis in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2478–2493. [Google Scholar] [CrossRef]
- Liang, G.; Zhang, H.; Li, X.; Ai, Q.; Yu, D. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 1743–1755. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wheeler, T.; Li, Z.; Kenerley, C.M.; He, P.; Shan, L. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 2011, 66, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Guerinot, M.L. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 1996, 10, 835–844. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids, the pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Qin, Z.; Bai, Y.; Muhammad, S.; Wu, X.; Deng, P.; Wu, J.; An, H.; Wu, L. Divergent roles of FT-like 9 in flowering transition under different day lengths in Brachypodium distachyon. Nat. Commun. 2019, 10, 812. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, C.; Cui, M.; Zhou, W.; Wu, H.; Ling, H. Four IVa bHLH Transcription Factors Are Novel Interactors of FIT and Mediate JA Inhibition of Iron Uptake in Arabidopsis. Molecular Plant. 2018, 11, 1166–1183. [Google Scholar] [CrossRef]
- Tissot, N.; Robe, K.; Gao, F.; Grant-Grant, S.; Boucherez, J.; Bellegarde, F.; Maghiaoui, A.; Marcelin, R.; Izquierdo, E.; Benhamed, M.; et al. Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 2019, 223, 1433–1446. [Google Scholar] [CrossRef]




| Genes | TRV2:GhbHLH121 +Fe | TRV2:00 -Fe | TRV2:GhbHLH121 -Fe |
|---|---|---|---|
| Subgroup bHLH IVc genes | |||
| GhbHLH104-A10 | 1.07 ± 0.24 | 2.46 ± 0.34 | 2.54 ± 0.32 |
| GhbHLH104-D10 | 0.84 ± 0.19 | 2.29 ± 0.51 | 2.18 ± 0.30 |
| GhbHLH115 A01 | 1.01 ± 0.21 | 2.36 ± 0.33 | 2.34 ± 0.31 |
| GhbHLH115 D01 | 0.95 ± 0.15 | 1.25 ± 0.12 | 1.12 ± 0.16 |
| GhbHLH115 A01-2 | 0.82 ± 0.11 | 0.78 ± 0.11 | 0.83 ± 0.10 |
| GhbHLH115 D01-2 | 0.97 ± 0.11 | 0.90 ± 0.13 | 0.85 ± 0.12 |
| GhbHLH115 A05 | 1.18 ± 0.17 | 1.28 ± 0.15 | 1.33 ± 0.17 |
| GhbHLH115 D05 | 0.93 ± 0.14 | 1.10 ± 0.12 | 1.06 ± 0.14 |
| GhbHLH115 A11 | 0.87 ± 0.12 | 0.84 ± 0.11 | 0.94 ± 0.11 |
| GhbHLH115 D11 | 0.99 ± 0.13 | 0.94 ± 0.13 | 0.98 ± 0.12 |
| GhbHLH115 A13 | 1.14 ± 0.12 | 1.01 ± 0.15 | 0.94 ± 0.13 |
| GhbHLH115 D13 | 0.93 ± 0.12 | 0.97 ± 0.12 | 0.95 ± 0.13 |
| Subgroup bHLH Ib genes | |||
| GhbHLH101 A11 | 0.94 ± 0.12 | 1.01 ± 0.12 | 0.92 ± 0.13 |
| GhbHLH101 D11 | 0.91 ± 0.13 | 1.07 ± 0.12 | 1.01 ± 0.14 |
| GhbHLH101 D11-2 | 0.90 ± 0.16 | 1.15 ± 0.12 | 1.23 ± 0.15 |
| GhbHLH38 A05 | 0.89 ± 0.23 | 58.80 ± 9.11 | 33.86 ± 7.88 ** |
| GhbHLH38 D05 | 0.80 ± 0.07 ** | 133.96 ± 17.10 | 53.94 ± 11.95 ** |
| GhbHLH38 A09 | 0.85 ± 0.20 | 57.27 ± 6.11 | 32.40 ± 5.67 ** |
| GhbHLH38 D09 | 0.92 ± 0.14 | 72.90 ± 8.12 | 65.44 ± 5.76 * |
| Fe uptake genes | |||
| GhFIT A05 | 0.48 ± 0.03 ** | 5.61 ± 0.06 | 0.87 ± 0.15 ** |
| GhFIT D05 | 0.31 ± 0.01 ** | 15.61 ± 0.14 | 7.58 ± 1.09 ** |
| GhFIT A09 | 0.63 ± 0.03 ** | 1.75 ± 0.18 | 1.03 ± 0.13 ** |
| GhFIT D09 | 0.71 ± 0.10 ** | 1.97 ± 0.15 | 1.11 ± 0.20 * |
| GhFRO2 | 0.95 ± 0.14 | 8.68 ± 1.22 | 4.08 ± 0.56 ** |
| GhIRT3 | 1.07 ± 0.01 | 5.60 ± 1.14 | 2.34 ± 0.35 ** |
| GhAHA2 | 1.03 ± 0.04 | 4.69 ± 1.13 | 3.06 ± 0.62 ** |
| Fe translocation genes | |||
| GhPYE | 1.05 ± 0.02 | 102.42 ± 12.07 | 73.34 ± 13.72 * |
| GhYSL1 | 1.14 ± 0.09 | 0.42 ± 0.11 | 0.31 ± 0.05 * |
| GhNAS4 | 0.88 ± 0.17 | 15.80 ± 0.21 | 5.00 ± 1.11 ** |
| GhAtZIF1 | 0.93 ± 0.08 | 4.15 ± 0.17 | 4.34 ± 0.55 |
| GhFRD3 | 1.00 ± 0.29 | 2.14 ± 0.43 | 2.22 ± 0.28 |
| Fe homeostasis regulation genes | |||
| GhBTS A04 | 0.55 ± 0.12 ** | 2.62 ± 0.27 | 0.91 ± 0.15 ** |
| GhBTS A11 | 0.39 ± 0.04 ** | 1.81 ± 0.35 | 1.09 ± 0.14 ** |
| GhMYB72 A01 | 0.89 ± 0.13 | 3.52 ± 0.42 | 3.24 ± 0.47 |
| GhMYB72 A08 | 0.99 ± 0.18 | 7.34 ± 0.33 | 5.84 ± 0.98 * |
| Genes | GhbHLH121OE +Fe | WT -Fe | GhbHLH121OE -Fe |
|---|---|---|---|
| Subgroup bHLH IVc genes | |||
| AtbHLH34 | 1.34 ± 0.13 | 1.23 ± 0.17 | 1.53 ± 0.15 |
| AtbHLH104 | 0.87 ± 0.12 | 0.89 ± 0.11 | 1.01 ± 0.11 |
| AtbHLH105 | 1.02 ± 0.06 | 0.88 ± 0.12 | 1.29 ± 0.11 |
| AtbHLH115 | 1.08 ± 0.15 | 2.03 ± 0.13 | 2.01 ± 0.25 |
| Subgroup bHLH Ib genes | |||
| AtbHLH38 | 4.46 ± 0.76 ** | 490.66 ± 36.56 | 1980.02 ± 62.31 ** |
| AtbHLH39 | 0.80 ± 0.13 | 15.13 ± 3.10 | 40.96 ± 3.92 ** |
| AtbHLH100 | 4.40 ± 0.52 ** | 295.60 ± 34.55 | 2234.81 ± 37.54 ** |
| AtbHLH101 | 2.81 ± 0.32 ** | 19.59 ± 2.35 | 57.68 ± 8.48 ** |
| Fe uptake genes | |||
| AtFIT | 2.01 ± 0.37 ** | 5.62 ± 0.25 | 10.84 ± 0.71 ** |
| AtFRO2 | 2.70 ± 0.05 ** | 27.32 ± 0.34 | 39.78 ± 3.47 ** |
| AtIRTl | 4.44 ± 0.30 ** | 133.25 ± 11.56 | 230.75 ± 16.92 ** |
| AtIAHA2 | 1.03 ± 0.17 | 2.68 ± 0.13 | 6.08 ± 0.34 ** |
| Fe translocation genes | |||
| AtPYE | 1.64 ± 0.23 | 3.02 ± 0.20 | 9.69 ± 0.38 ** |
| AtYSL1 | 0.96 ± 0.04 | 0.37 ± 0.12 | 0.37 ± 0.04 |
| AtNAS4 | 2.34 ± 0.04 ** | 7.72 ± 0.29 | 14.56 ± 0.98 ** |
| AtZIF1 | 1.53 ± 0.17 * | 1.82 ± 0.19 | 2.92 ± 0.23 ** |
| AtFRD3 | 0.81 ± 0.13 | 0.62 ± 0.10 | 1.05 ± 0.07** |
| Fe homeostasis regulation genes | |||
| AtBTS | 1.72 ± 0.15 ** | 1.54 ± 0.21 | 5.96 ± 0.19 ** |
| AtMYB10 | 0.98 ± 0.19 | 6.73 ± 0.12 | 36.19 ± 0.85 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Nie, K.; Wang, L.; Zhao, Y.; Qu, M.; Yang, D.; Guan, X. The Molecular Mechanism of GhbHLH121 in Response to Iron Deficiency in Cotton Seedlings. Plants 2023, 12, 1955. https://doi.org/10.3390/plants12101955
Li J, Nie K, Wang L, Zhao Y, Qu M, Yang D, Guan X. The Molecular Mechanism of GhbHLH121 in Response to Iron Deficiency in Cotton Seedlings. Plants. 2023; 12(10):1955. https://doi.org/10.3390/plants12101955
Chicago/Turabian StyleLi, Jie, Ke Nie, Luyao Wang, Yongyan Zhao, Mingnan Qu, Donglei Yang, and Xueying Guan. 2023. "The Molecular Mechanism of GhbHLH121 in Response to Iron Deficiency in Cotton Seedlings" Plants 12, no. 10: 1955. https://doi.org/10.3390/plants12101955
APA StyleLi, J., Nie, K., Wang, L., Zhao, Y., Qu, M., Yang, D., & Guan, X. (2023). The Molecular Mechanism of GhbHLH121 in Response to Iron Deficiency in Cotton Seedlings. Plants, 12(10), 1955. https://doi.org/10.3390/plants12101955






