Plant–Environment Response Pathway Regulation Uncovered by Investigating Non-Typical Legume Symbiosis and Nodulation
Abstract
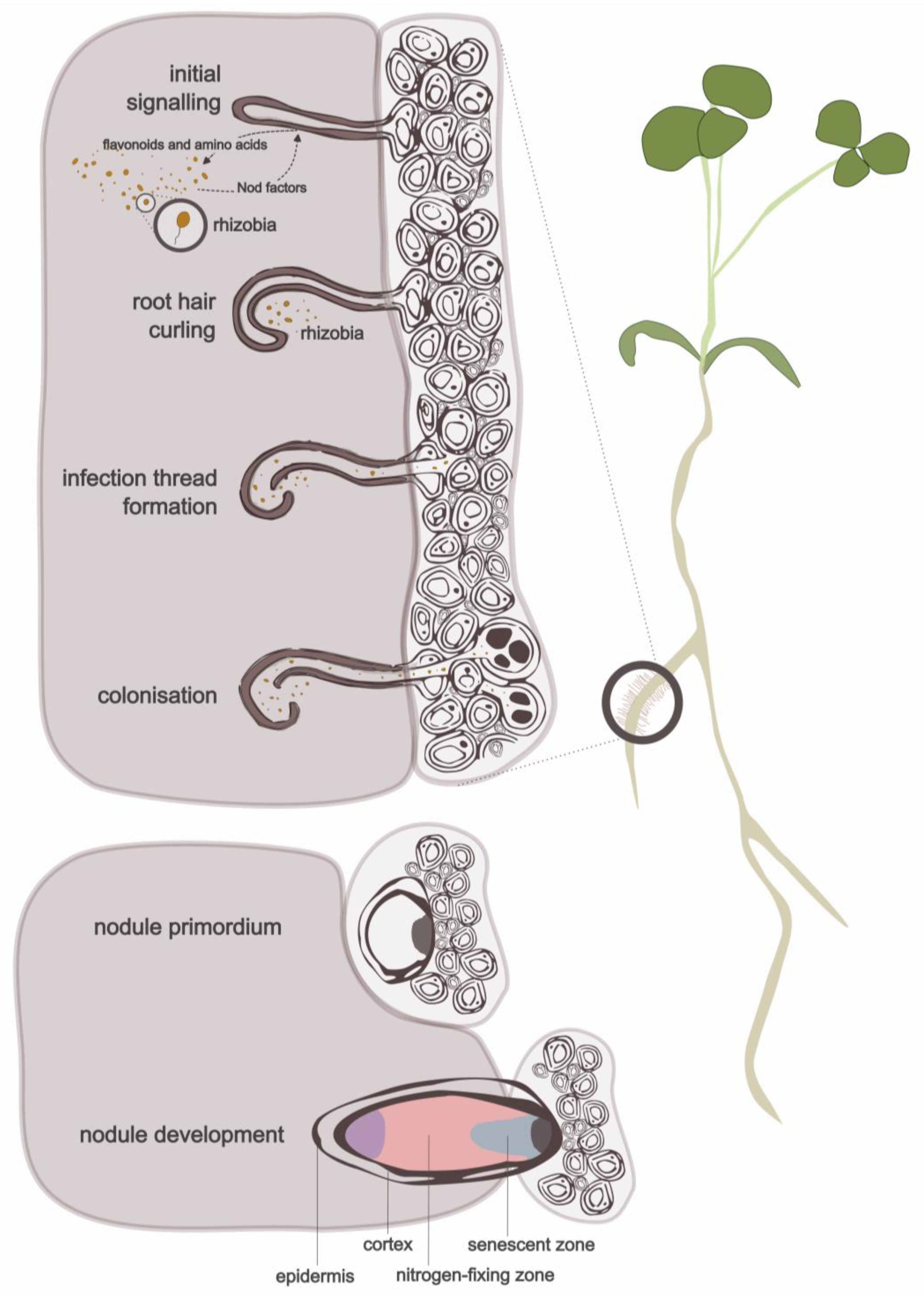
:1. Introduction
2. The (Relatively) Well-Known Rhizobial–Legume Symbiosis of Nodulation
3. Symbiotic Partnerships Outside of Legumes, and Outside of Rhizobia
3.1. Nodulation beyond Legumes: The Example of Parasponia
3.2. Nod-Factor-Independent Nodulation in Legumes
3.3. Symbiotic Partnerships in Non-Rhizobial Nodules
3.4. Plant–Microbe Symbiosis without Root Nodules
4. Extra Bonuses of Hosting Microbes
4.1. Defence Priming
4.2. Abiotic Stress Resistance
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef]
- Morere-Le Paven, M.-C.; Viau, L.; Hamon, A.; Vandecasteele, C.; Pellizzaro, A.; Bourdin, C.; Laffont, C.; Lapied, B.; Lepetit, M.; Frugier, F. Characterization of a dual-affinity nitrate transporter MtNRT1. 3 in the model legume Medicago truncatula. J. Exp. Bot. 2011, 62, 5595–5605. [Google Scholar] [CrossRef] [PubMed]
- Simon-Rosin, U.; Wood, C.; Udvardi, M.K. Molecular and cellular characterisation of LjAMT2; 1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol. Biol. 2003, 51, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Houassine, D.; Latati, M.; Rebouh, N.Y.; Gérard, F. Phosphorus acquisition processes in the field: Study of faba bean cultivated on calcareous soils in Algeria. Arch. Agron. Soil Sci. 2019, 66, 168–181. [Google Scholar] [CrossRef]
- le Roux, M.M.; Miller, J.T.; Waller, J.; Döring, M.; Bruneau, A. An expert curated global legume checklist improves the accuracy of occurrence, biodiversity and taxonomic data. Sci. Data 2022, 9, 708. [Google Scholar] [CrossRef]
- Xiao, T.T.; Schilderink, S.; Moling, S.; Deinum, E.E.; Kondorosi, E.; Franssen, H.; Kulikova, O.; Niebel, A.; Bisseling, T. Fate map of Medicago truncatula root nodules. Development 2014, 141, 3517–3528. [Google Scholar] [CrossRef]
- Kohlen, W.; Ng, J.L.P.; Deinum, E.E.; Mathesius, U. Auxin transport, metabolism, and signalling during nodule initiation: Indeterminate and determinate nodules. J. Exp. Bot. 2018, 69, 229–244. [Google Scholar] [CrossRef]
- Lopez-Gomez, M.; Sandal, N.; Stougaard, J.; Boller, T. Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. J. Exp. Bot. 2012, 63, 393–401. [Google Scholar] [CrossRef]
- Dong, W.; Song, Y. The significance of flavonoids in the process of biological nitrogen fixation. Int. J. Mol. Sci. 2020, 21, 5926. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, S.; Li, J.; Pei, R.; Tian, L.; Qi, J.; Azam, M.; Agyenim-Boateng, K.G.; Shaibu, A.S.; Liu, Y. Dual-function C2H2-type zinc-finger transcription factor GmZFP7 contributes to isoflavone accumulation in soybean. New Phytol. 2023, 237, 1794–1809. [Google Scholar] [CrossRef]
- Compton, K.K.; Hildreth, S.B.; Helm, R.F.; Scharf, B.E. An updated perspective on Sinorhizobium meliloti chemotaxis to alfalfa flavonoids. Front. Microbiol. 2020, 11, 581482. [Google Scholar] [CrossRef] [PubMed]
- Cullimore, J.; Fliegmann, J.; Gasciolli, V.; Gibelin-Viala, C.; Carles, N.; Luu, T.-B.; Girardin, A.; Cumener, M.; Maillet, F.; Pradeau, S. Evolution of lipochitooligosaccharide binding to a LysM-RLK for nodulation in Medicago truncatula. Plant Cell Physiol. 2023, pcad033. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, L.-Y.; Liu, W.; Tian, Y.; Xiong, J.-S.; Wang, Y.-H.; Li, R.-J.; Li, H.-M.; Wen, J.; Mysore, K.S. Role of the Nod factor hydrolase MtNFH1 in regulating Nod factor levels during rhizobial infection and in mature nodules of Medicago truncatula. Plant Cell 2018, 30, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Moura, F.T.; Ribeiro, R.A.; Helene, L.C.F.; Nogueira, M.A.; Hungria, M. So many rhizobial partners, so little nitrogen fixed: The intriguing symbiotic promiscuity of common bean (Phaseolus vulgaris L.). Symbiosis 2022, 86, 169–185. [Google Scholar] [CrossRef]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Rutten, L.; Limpens, E.; van der Molen, T.; van Velzen, R.; Chen, R.; Chen, Y.; Geurts, R.; Kohlen, W.; Kulikova, O. A remote cis-regulatory region is required for NIN expression in the pericycle to initiate nodule primordium formation in Medicago truncatula. Plant Cell 2019, 31, 68–83. [Google Scholar] [CrossRef]
- Peleg-Grossman, S.; Volpin, H.; Levine, A. Root hair curling and Rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. J. Exp. Bot. 2007, 58, 1637–1649. [Google Scholar] [CrossRef]
- Bhattacharjee, O.; Raul, B.; Ghosh, A.; Bhardwaj, A.; Bandyopadhyay, K.; Sinharoy, S. Nodule INception-independent epidermal events lead to bacterial entry during nodule development in peanut (Arachis hypogaea). New Phytol. 2022, 236, 2265–2281. [Google Scholar] [CrossRef]
- Ibáñez, F.; Wall, L.; Fabra, A. Starting points in plant-bacteria nitrogen-fixing symbioses: Intercellular invasion of the roots. J. Exp. Bot. 2017, 68, 1905–1918. [Google Scholar] [CrossRef]
- Cárdenas, L.; Martínez, A.; Sánchez, F.; Quinto, C. Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs). Plant J. 2008, 56, 802–813. [Google Scholar] [CrossRef]
- Fournier, J.; Teillet, A.; Chabaud, M.; Ivanov, S.; Genre, A.; Limpens, E.; de Carvalho-Niebel, F.; Barker, D.G. Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 2015, 167, 1233–1242. [Google Scholar] [CrossRef]
- Kitaeva, A.B.; Demchenko, K.N.; Tikhonovich, I.A.; Timmers, A.C.; Tsyganov, V.E. Comparative analysis of the tubulin cytoskeleton organization in nodules of Medicago truncatula and Pisum sativum: Bacterial release and bacteroid positioning correlate with characteristic microtubule rearrangements. New Phytol. 2016, 210, 168–183. [Google Scholar] [CrossRef]
- Mergaert, P.; Uchiumi, T.; Alunni, B.; Evanno, G.; Cheron, A.; Catrice, O.; Mausset, A.-E.; Barloy-Hubler, F.; Galibert, F.; Kondorosi, A. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 5230–5235. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.; Xiao, H.; Peng, Y.; Wang, J.; Lv, Q.; Wang, X. Phosphoenolpyruvate reallocation links nitrogen fixation rates to root nodule energy state. Science 2022, 378, 971–977. [Google Scholar] [CrossRef] [PubMed]
- van de Velde, W.; Guerra, J.C.P.; Keyser, A.D.; de Rycke, R.; Rombauts, S.; Maunoury, N.; Mergaert, P.; Kondorosi, E.; Holsters, M.; Goormachtig, S. Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol. 2006, 141, 711–720. [Google Scholar] [CrossRef]
- Dhanushkodi, R.; Matthew, C.; McManus, M.T.; Dijkwel, P.P. Drought-induced senescence of Medicago truncatula nodules involves serpin and ferritin to control proteolytic activity and iron levels. New Phytol. 2018, 220, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Serova, T.A.; Tikhonovich, I.A.; Tsyganov, V.E. Analysis of nodule senescence in pea (Pisum sativum L.) using laser microdissection, real-time PCR, and ACC immunolocalization. J. Plant Physiol. 2017, 212, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Griesmann, M.; Chang, Y.; Liu, X.; Song, Y.; Haberer, G.; Crook, M.B.; Billault-Penneteau, B.; Lauressergues, D.; Keller, J.; Imanishi, L. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 2018, 361, eaat1743. [Google Scholar] [CrossRef]
- van Velzen, R.; Holmer, R.; Bu, F.; Rutten, L.; van Zeijl, A.; Liu, W.; Santuari, L.; Cao, Q.; Sharma, T.; Shen, D. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. USA 2018, 115, e4700–e4709. [Google Scholar] [CrossRef]
- van Zeijl, A.; Wardhani, T.A.; Seifi Kalhor, M.; Rutten, L.; Bu, F.; Hartog, M.; Linders, S.; Fedorova, E.E.; Bisseling, T.; Kohlen, W. CRISPR/Cas9-mediated mutagenesis of four putative symbiosis genes of the tropical tree Parasponia andersonii reveals novel phenotypes. Front. Plant Sci. 2018, 9, 284. [Google Scholar] [CrossRef]
- Wardhani, T.A.; Roswanjaya, Y.P.; Dupin, S.; Li, H.; Linders, S.; Hartog, M.; Geurts, R.; van Zeijl, A. Transforming, Genome Editing and Phenotyping the Nitrogen-fixing Tropical Cannabaceae Tree Parasponia andersonii. J. Vis. Exp. 2019. [Google Scholar] [CrossRef]
- Op den Camp, R.H.; Polone, E.; Fedorova, E.; Roelofsen, W.; Squartini, A.; Op den Camp, H.J.; Bisseling, T.; Geurts, R. Nonlegume Parasponia andersonii deploys a broad rhizobium host range strategy resulting in largely variable symbiotic effectiveness. Mol. Plant-Microbe Interact. 2012, 25, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Keet, J.-H.; Ellis, A.G.; Hui, C.; Le Roux, J.J. Legume–rhizobium symbiotic promiscuity and effectiveness do not affect plant invasiveness. Ann. Bot. 2017, 119, 1319–1331. [Google Scholar] [CrossRef] [PubMed]
- Ehinger, M.; Mohr, T.J.; Starcevich, J.B.; Sachs, J.L.; Porter, S.S.; Simms, E.L. Specialization-generalization trade-off in a Bradyrhizobium symbiosis with wild legume hosts. BMC Ecol. 2014, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- Harrison, T.L.; Simonsen, A.K.; Stinchcombe, J.R.; Frederickson, M.E. More partners, more ranges: Generalist legumes spread more easily around the globe. Biol. Lett. 2018, 14, 20180616. [Google Scholar] [CrossRef]
- Zarrabian, M.; Montiel, J.; Sandal, N.; Ferguson, S.; Jin, H.; Lin, Y.-Y.; Klingl, V.; Marín, M.; James, E.K.; Parniske, M. A promiscuity locus confers Lotus burttii nodulation with rhizobia from five different genera. Mol. Plant-Microbe Interact. 2022, 35, 1006–1017. [Google Scholar] [CrossRef]
- Dupin, S.E.; Geurts, R.; Kiers, E.T. The non-legume Parasponia andersonii mediates the fitness of nitrogen-fixing rhizobial symbionts under high nitrogen conditions. Front. Plant Sci. 2020, 10, 1779. [Google Scholar] [CrossRef]
- Rutten, L.; Miyata, K.; Roswanjaya, Y.P.; Huisman, R.; Bu, F.; Hartog, M.; Linders, S.; van Velzen, R.; van Zeijl, A.; Bisseling, T. Duplication of symbiotic lysin motif receptors predates the evolution of nitrogen-fixing nodule symbiosis. Plant Physiol. 2020, 184, 1004–1023. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, Z.; Dong, X.; Tang, Z.; Ju, B.; Du, Z.; Wang, E.; Xie, Z. Bradyrhizobium aeschynomenes sp. nov., a root and stem nodule microsymbiont of Aeschynomene indica. Syst. Appl. Microbiol. 2022, 45, 126337. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Pan, X.; Shao, S.; Liu, W.; Wang, E.-T.; Xie, Z. Aeschynomene indica-nodulating rhizobia lacking Nod factor synthesis genes: Diversity and evolution in Shandong Peninsula, China. Appl. Environ. Microbiol. 2019, 85, e00782-19. [Google Scholar] [CrossRef]
- Kaló, P.; Gleason, C.; Edwards, A.; Marsh, J.; Mitra, R.M.; Hirsch, S.; Jakab, J.; Sims, S.; Long, S.R.; Rogers, J. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 2005, 308, 1786–1789. [Google Scholar] [CrossRef] [PubMed]
- Quilbé, J.; Nouwen, N.; Pervent, M.; Guyonnet, R.; Cullimore, J.; Gressent, F.; Araújo, N.H.; Gully, D.; Klopp, C.; Giraud, E. A mutant-based analysis of the establishment of Nod-independent symbiosis in the legume Aeschynomene evenia. Plant Physiol. 2022, 190, 1400–1417. [Google Scholar] [CrossRef] [PubMed]
- Guha, S.; Molla, F.; Sarkar, M.; Ibañez, F.; Fabra, A.; DasGupta, M. Nod factor-independent ‘crack-entry’symbiosis in dalbergoid legume Arachis hypogaea. Environ. Microbiol. 2022, 24, 2732–2746. [Google Scholar] [CrossRef]
- Ratu, S.T.N.; Teulet, A.; Miwa, H.; Masuda, S.; Nguyen, H.P.; Yasuda, M.; Sato, S.; Kaneko, T.; Hayashi, M.; Giraud, E. Rhizobia use a pathogenic-like effector to hijack leguminous nodulation signalling. Sci. Rep. 2021, 11, 2034. [Google Scholar] [CrossRef] [PubMed]
- Teulet, A.; Busset, N.; Fardoux, J.; Gully, D.; Chaintreuil, C.; Cartieaux, F.; Jauneau, A.; Comorge, V.; Okazaki, S.; Kaneko, T. The rhizobial type III effector ErnA confers the ability to form nodules in legumes. Proc. Natl. Acad. Sci. USA 2019, 116, 21758–21768. [Google Scholar] [CrossRef] [PubMed]
- Diagne, N.; Arumugam, K.; Ngom, M.; Nambiar-Veetil, M.; Franche, C.; Narayanan, K.K.; Laplaze, L. Use of Frankia and actinorhizal plants for degraded lands reclamation. BioMed Res. Int. 2013, 2013, 948258. [Google Scholar] [CrossRef] [PubMed]
- Dawson, J. Ecology of actinorhizal plants. In Nitrogen-Fixing Actinorhizal Symbioses; Springer: Dordrecht, The Netherlands, 2007; pp. 199–234. [Google Scholar]
- Bélanger, P.-A.; Beaudin, J.; Roy, S. High-throughput screening of microbial adaptation to environmental stress. J. Microbiol. Methods 2011, 85, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Chen, H.; Pan, Y.; Feng, H.; Fang, D.; Yang, J.; Wang, Y.; Yang, J.; Sahu, S.K.; Liu, J. Genome of Hippophae rhamnoides provides insights into a conserved molecular mechanism in actinorhizal and rhizobial symbioses. New Phytol. 2022, 235, 276–291. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Y.; Meng, J.; Wang, Y.; Nie, S.; Zhang, Z.; Wang, H.; Yang, Y.; Gao, Y.; Wu, J. Chromosome-scale de novo genome assembly and annotation of three representative Casuarina species: C. equisetifolia, C. glauca, and C. cunninghamiana. Plant J. 2023. [Google Scholar] [CrossRef]
- Echbab, H.; Arahou, M.; Ducousso, M.; Nourissier-Mountou, S.; Duponnois, R.; Lahlou, H.; Prin, Y. Successful nodulation of Casuarina by Frankia in axenic conditions. J. Appl. Microbiol. 2007, 103, 1728–1737. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Wibberg, D.; Vigil-Stenman, T.; Berckx, F.; Battenberg, K.; Demchenko, K.N.; Blom, J.; Fernandez, M.P.; Yamanaka, T.; Berry, A.M. Frankia-enriched metagenomes from the earliest diverging symbiotic Frankia cluster: They come in teams. Genome Biol. Evol. 2019, 11, 2273–2291. [Google Scholar] [CrossRef] [PubMed]
- Nouioui, I.; Cortés-Albayay, C.; Carro, L.; Castro, J.F.; Gtari, M.; Ghodhbane-Gtari, F.; Klenk, H.-P.; Tisa, L.S.; Sangal, V.; Goodfellow, M. Genomic insights into plant-growth-promoting potentialities of the genus Frankia. Front. Microbiol. 2019, 10, 1457. [Google Scholar] [CrossRef] [PubMed]
- Gasser, M.; Alloisio, N.; Fournier, P.; Balmand, S.; Kharrat, O.; Tulumello, J.; Carro, L.; Heddi, A.; Da Silva, P.; Normand, P. A Nonspecific Lipid Transfer Protein with Potential Functions in Infection and Nodulation. Mol. Plant-Microbe Interact. 2022, 35, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Meesters, T.M.; van Vliet, W.M.; Akkermans, A.D. Nitrogenase is restricted to the vesicles in Frankia strain EAN1pec. Physiol. Plant. 1987, 70, 267–271. [Google Scholar] [CrossRef]
- Parsons, R.; Silvester, W.B.; Harris, S.; Gruijters, W.; Bullivant, S. Frankia vesicles provide inducible and absolute oxygen protection for nitrogenase. Plant Physiol. 1987, 83, 728–731. [Google Scholar] [CrossRef]
- Wall, L.G.; Valverde, C.; Huss-Danell, K. Regulation of nodulation in the absence of N2 is different in actinorhizal plants with different infection pathways. J. Exp. Bot. 2003, 54, 1253–1258. [Google Scholar] [CrossRef]
- Chabaud, M.; Gherbi, H.; Pirolles, E.; Vaissayre, V.; Fournier, J.; Moukouanga, D.; Franche, C.; Bogusz, D.; Tisa, L.S.; Barker, D.G. Chitinase-resistant hydrophilic symbiotic factors secreted by Frankia activate both Ca2+ spiking and NIN gene expression in the actinorhizal plant Casuarina glauca. New Phytol. 2016, 209, 86–93. [Google Scholar] [CrossRef]
- Gueddou, A.; Sarker, I.; Sen, A.; Ghodhbane-Gtari, F.; Benson, D.R.; Armengaud, J.; Gtari, M. Effect of actinorhizal root exudates on the proteomes of Frankia soli NRRL B-16219, a strain colonizing the root tissues of its actinorhizal host via intercellular pathway. Res. Microbiol. 2022, 173, 103900. [Google Scholar] [CrossRef]
- Cissoko, M.; Hocher, V.; Gherbi, H.; Gully, D.; Carré-Mlouka, A.; Sane, S.; Pignoly, S.; Champion, A.; Ngom, M.; Pujic, P. Actinorhizal signaling molecules: Frankia root hair deforming factor shares properties with NIN inducing factor. Front. Plant Sci. 2018, 9, 1494. [Google Scholar] [CrossRef]
- Tsurugi-Sakurada, A.; Kaneko, T.; Takemoto, K.; Yoneda, Y.; Yamanaka, T.; Kawai, S. Cyclic diarylheptanoids as potential signal compounds during actinorhizal symbiosis between Alnus sieboldiana and Frankia. Fitoterapia 2022, 162, 105284. [Google Scholar] [CrossRef]
- Iwanycki Ahlstrand, N.; Stevenson, D.W. Retracing origins of exceptional cycads in botanical collections to increase conservation value. Plants People Planet 2021, 3, 94–98. [Google Scholar] [CrossRef]
- Kanesaki, Y.; Hirose, M.; Hirose, Y.; Fujisawa, T.; Nakamura, Y.; Watanabe, S.; Matsunaga, S.; Uchida, H.; Murakami, A. Draft genome sequence of the nitrogen-fixing and hormogonia-inducing cyanobacterium Nostoc cycadae strain WK-1, isolated from the coralloid roots of Cycas revoluta. Genome Announc. 2018, 6, e00021-18. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.C.G.; Chen, T.; Li, N.; Duan, J. Perspectives on endosymbiosis in coralloid roots: Association of cycads and cyanobacteria. Front. Microbiol. 2019, 10, 1888. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-García, K.; Bustos-Díaz, E.D.; Corona-Gómez, J.A.; Ramos-Aboites, H.E.; Sélem-Mojica, N.; Cruz-Morales, P.; Pérez-Farrera, M.A.; Barona-Gómez, F.; Cibrián-Jaramillo, A. Cycad coralloid roots contain bacterial communities including cyanobacteria and Caulobacter spp. that encode niche-specific biosynthetic gene clusters. Genome Biol. Evol. 2019, 11, 319–334. [Google Scholar] [CrossRef]
- Hashidoko, Y.; Nishizuka, H.; Tanaka, M.; Murata, K.; Murai, Y.; Hashimoto, M. Isolation and characterization of 1-palmitoyl-2-linoleoyl-sn-glycerol as a hormogonium-inducing factor (HIF) from the coralloid roots of Cycas revoluta (Cycadaceae). Sci. Rep. 2019, 9, 4751. [Google Scholar] [CrossRef]
- Burnat, M.; Herrero, A.; Flores, E. Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium. Proc. Natl. Acad. Sci. USA 2014, 111, 3823–3828. [Google Scholar] [CrossRef]
- Compaoré, J.; Stal, L.J. Oxygen and the light–dark cycle of nitrogenase activity in two unicellular cyanobacteria. Environ. Microbiol. 2010, 12, 54–62. [Google Scholar] [CrossRef]
- Mohr, W.; Lehnen, N.; Ahmerkamp, S.; Marchant, H.K.; Graf, J.S.; Tschitschko, B.; Yilmaz, P.; Littmann, S.; Gruber-Vodicka, H.; Leisch, N. Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature 2021, 600, 105–109. [Google Scholar] [CrossRef] [PubMed]
- García-Márquez, M.G.; Rodríguez-Castañeda, J.C.; Agawin, N.S. Sunscreen exposure interferes with physiological processes while inducing oxidative stress in seagrass Posidonia oceanica (L.) Delile. Mar. Pollut. Bull. 2023, 187, 114507. [Google Scholar] [CrossRef]
- Zotz, G.; Winkler, U. Aerial roots of epiphytic orchids: The velamen radicum and its role in water and nutrient uptake. Oecologia 2013, 171, 733–741. [Google Scholar] [CrossRef]
- Sma-Air, S.; Ritchie, R.J. Photosynthesis in a Vanda sp. orchid with Photosynthetic Roots. J. Plant Physiol. 2020, 251, 153187. [Google Scholar] [CrossRef] [PubMed]
- Tsavkelova, E.A.; Glukhareva, I.D.; Volynchikova, E.A.; Egorova, M.A.; Leontieva, M.R.; Malakhova, D.V.; Kolomeitseva, G.L.; Netrusov, A.I. Cyanobacterial Root Associations of Leafless Epiphytic Orchids. Microorganisms 2022, 10, 1006. [Google Scholar] [CrossRef] [PubMed]
- van Deynze, A.; Zamora, P.; Delaux, P.-M.; Heitmann, C.; Jayaraman, D.; Rajasekar, S.; Graham, D.; Maeda, J.; Gibson, D.; Schwartz, K.D. Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol. 2018, 16, e2006352. [Google Scholar] [CrossRef] [PubMed]
- Bhar, A.; Chatterjee, M.; Gupta, S.; Das, S. Salicylic acid regulates systemic defense signaling in chickpea during Fusarium oxysporum f. sp. ciceri race 1 infection. Plant Mol. Biol. Rep. 2018, 36, 162–175. [Google Scholar] [CrossRef]
- Díaz-Valle, A.; López-Calleja, A.C.; Alvarez-Venegas, R. Enhancement of Pathogen Resistance in Common Bean Plants by Inoculation With Rhizobium etli. Front. Plant Sci. 2019, 10, 1317. [Google Scholar] [CrossRef]
- Smigielski, L.; Laubach, E.-M.; Pesch, L.; Glock, J.M.L.; Albrecht, F.; Slusarenko, A.; Panstruga, R.; Kuhn, H. Nodulation induces systemic resistance of Medicago truncatula and Pisum sativum against Erysiphe pisi and primes for powdery mildew-triggered salicylic acid accumulation. Mol. Plant-Microbe Interact. 2019, 32, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Carlier, A.L.; Omasits, U.; Ahrens, C.H.; Eberl, L. Proteomics analysis of Psychotria leaf nodule symbiosis: Improved genome annotation and metabolic predictions. Mol. Plant-Microbe Interact. 2013, 26, 1325–1333. [Google Scholar] [CrossRef]
- Lemaire, B.; Smets, E.; Dessein, S. Bacterial leaf symbiosis in Ardisia (Myrsinoideae, Primulaceae): Molecular evidence for host specificity. Res. Microbiol. 2011, 162, 528–534. [Google Scholar] [CrossRef]
- Carlier, A.; Fehr, L.; Pinto-Carbó, M.; Schäberle, T.; Reher, R.; Dessein, S.; König, G.; Eberl, L. The genome analysis of Candidatus Burkholderia crenata reveals that secondary metabolism may be a key function of the Ardisia crenata leaf nodule symbiosis. Environ. Microbiol. 2016, 18, 2507–2522. [Google Scholar] [CrossRef]
- Pinto-Carbó, M.; Gademann, K.; Eberl, L.; Carlier, A. Leaf nodule symbiosis: Function and transmission of obligate bacterial endophytes. Curr. Opin. Plant Biol. 2018, 44, 23–31. [Google Scholar] [CrossRef]
- Danneels, B.; Blignaut, M.; Marti, G.; Sieber, S.; Vandamme, P.; Meyer, M.; Carlier, A. Cyclitol metabolism is a central feature of Burkholderia leaf symbionts. Environ. Microbiol. 2023, 25, 454–472. [Google Scholar] [CrossRef] [PubMed]
- de Meyer, F.; Danneels, B.; Acar, T.; Rasolomampianina, R.; Rajaonah, M.T.; Jeannoda, V.; Carlier, A. Adaptations and evolution of a heritable leaf nodule symbiosis between Dioscorea sansibarensis and Orrella dioscoreae. ISME J. 2019, 13, 1831–1844. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.M.; Mescher, M.C.; de Moraes, C.M. Plant dependence on rhizobia for nitrogen influences induced plant defenses and herbivore performance. Int. J. Mol. Sci. 2014, 15, 1466–1480. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Singh, A.; Dwivedi, D.H.; Arora, N.K. Zinc and phosphate solubilizing Rhizobium radiobacter (LB2) for enhancing quality and yield of loose leaf lettuce in saline soil. Environ. Sustain. 2020, 3, 209–218. [Google Scholar] [CrossRef]
- Rani, L.; Thapa, K.; Kanojia, N.; Sharma, N.; Singh, S.; Grewal, A.S.; Srivastav, A.L.; Kaushal, J. An extensive review on the consequences of chemical pesticides on human health and environment. J. Clean. Prod. 2021, 283, 124657. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Hussain, S.; Zhang, J.-H.; Zhong, C.; Zhu, L.-F.; Cao, X.-C.; Yu, S.-M.; Bohr, J.A.; Hu, J.-J.; Jin, Q.-Y. Effects of salt stress on rice growth, development characteristics, and the regulating ways: A review. J. Integr. Agric. 2017, 16, 2357–2374. [Google Scholar] [CrossRef]
- Shirzaei, M.; Khoshmanesh, M.; Ojha, C.; Werth, S.; Kerner, H.; Carlson, G.; Sherpa, S.F.; Zhai, G.; Lee, J.-C. Persistent impact of spring floods on crop loss in US Midwest. Weather. Clim. Extrem. 2021, 34, 100392. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, Z.; Shi, H. Rhizobium Symbiosis Leads to Increased Drought Tolerance in Chinese Milk Vetch (Astragalus sinicus L.). Agronomy 2022, 12, 725. [Google Scholar] [CrossRef]
- Staudinger, C.; Mehmeti-Tershani, V.; Gil-Quintana, E.; Gonzalez, E.M.; Hofhansl, F.; Bachmann, G.; Wienkoop, S. Evidence for a rhizobia-induced drought stress response strategy in Medicago truncatula. J. Proteom. 2016, 136, 202–213. [Google Scholar] [CrossRef]
- Marinković, J.; Bjelić, D.; Đorđević, V.; Balešević-Tubić, S.; Jošić, D.; Vucelić-Radović, B. Performance of different Bradyrhizobium strains in root nodule symbiosis under drought stress. Acta Physiol. Plant. 2019, 41, 37. [Google Scholar] [CrossRef]
- Mnasri, B.; Aouani, M.E.; Mhamdi, R. Nodulation and growth of common bean (Phaseolus vulgaris) under water deficiency. Soil Biol. Biochem. 2007, 39, 1744–1750. [Google Scholar] [CrossRef]
- Benmoussa, S.; Nouairi, I.; Rajhi, I.; Rezgui, S.; Manai, K.; Taamali, W.; Abbes, Z.; Zribi, K.; Brouquisse, R.; Mhadhbi, H. Growth Performance and Nitrogen Fixing Efficiency of Faba Bean (Vicia faba L.) Genotypes in Symbiosis with Rhizobia under Combined Salinity and Hypoxia Stresses. Agronomy 2022, 12, 606. [Google Scholar] [CrossRef]
- Bertrand, A.; Bipfubusa, M.; Dhont, C.; Chalifour, F.-P.; Drouin, P.; Beauchamp, C.J. Rhizobial strains exert a major effect on the amino acid composition of alfalfa nodules under NaCl stress. Plant Physiol. Biochem. 2016, 108, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, A.; Dhont, C.; Bipfubusa, M.; Chalifour, F.-P.; Drouin, P.; Beauchamp, C.J. Improving salt stress responses of the symbiosis in alfalfa using salt-tolerant cultivar and rhizobial strain. Appl. Soil Ecol. 2015, 87, 108–117. [Google Scholar] [CrossRef]
- Mangal, V.; Lal, M.K.; Tiwari, R.K.; Altaf, M.A.; Sood, S.; Kumar, D.; Bharadwaj, V.; Singh, B.; Singh, R.K.; Aftab, T. Molecular insights into the role of reactive oxygen, nitrogen and sulphur species in conferring salinity stress tolerance in plants. J. Plant Growth Regul. 2022, 42, 554–574. [Google Scholar] [CrossRef]
- Kalloniati, C.; Krompas, P.; Karalias, G.; Udvardi, M.K.; Rennenberg, H.; Herschbach, C.; Flemetakis, E. Nitrogen-fixing nodules are an important source of reduced sulfur, which triggers global changes in sulfur metabolism in Lotus japonicus. Plant Cell 2015, 27, 2384–2400. [Google Scholar] [CrossRef]
- Goormachtig, S.; Capoen, W.; James, E.K.; Holsters, M. Switch from intracellular to intercellular invasion during water stress-tolerant legume nodulation. Proc. Natl. Acad. Sci. USA 2004, 101, 6303–6308. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, X.; Dong, X.; Yan, J.; Xie, Z.; Luo, Y. The effect of Azorhizobium caulinodans ORS571 and γ-aminobutyric acid on salt tolerance of Sesbania rostrata. Front. Plant Sci. 2022, 13, 926850. [Google Scholar] [CrossRef]
- Capoen, W.; Oldroyd, G.; Goormachtig, S.; Holsters, M. Sesbania rostrata: A case study of natural variation in legume nodulation. New Phytol. 2010, 186, 340–345. [Google Scholar] [CrossRef]
- Liu, X.; Xie, Z.; Wang, Y.; Sun, Y.; Dang, X.; Sun, H. A dual role of amino acids from Sesbania rostrata seed exudates in the chemotaxis response of Azorhizobium caulinodans ORS571. Mol. Plant-Microbe Interact. 2019, 32, 1134–1147. [Google Scholar] [CrossRef]
- Parsons, R.; Raven, J.; Sprent, J. A simple open flow system used to measure acetylene reduction activity of Sesbania rostrata stem and root nodules. J. Exp. Bot. 1992, 43, 595–604. [Google Scholar] [CrossRef]
- Edmonds, D.A.; Caldwell, R.L.; Brondizio, E.S.; Siani, S.M. Coastal flooding will disproportionately impact people on river deltas. Nat. Commun. 2020, 11, 4741. [Google Scholar] [CrossRef]
- Pokhrel, Y.; Felfelani, F.; Satoh, Y.; Boulange, J.; Burek, P.; Gädeke, A.; Gerten, D.; Gosling, S.N.; Grillakis, M.; Gudmundsson, L. Global terrestrial water storage and drought severity under climate change. Nat. Clim. Chang. 2021, 11, 226–233. [Google Scholar] [CrossRef]
- Corwin, D.L. Climate change impacts on soil salinity in agricultural areas. Eur. J. Soil Sci. 2021, 72, 842–862. [Google Scholar] [CrossRef]
- Kazmierczak, T.; Yang, L.; Boncompagni, E.; Meilhoc, E.; Frugier, F.; Frendo, P.; Bruand, C.; Gruber, V.; Brouquisse, R. Legume nodule senescence: A coordinated death mechanism between bacteria and plant cells. Adv. Bot. Res. 2020, 94, 181–212. [Google Scholar]
- Behm, J.E.; Geurts, R.; Kiers, E.T. Parasponia: A novel system for studying mutualism stability. Trends Plant Sci. 2014, 19, 757–763. [Google Scholar] [CrossRef]
- Pawlowski, K.; Demchenko, K.N. The diversity of actinorhizal symbiosis. Protoplasma 2012, 249, 967–979. [Google Scholar] [CrossRef]
- Fleischman, D.; Kramer, D. Photosynthetic rhizobia. Biochim. Et Biophys. Acta (BBA) Bioenerg. 1998, 1364, 17–36. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilkinson, H.; Coppock, A.; Richmond, B.L.; Lagunas, B.; Gifford, M.L. Plant–Environment Response Pathway Regulation Uncovered by Investigating Non-Typical Legume Symbiosis and Nodulation. Plants 2023, 12, 1964. https://doi.org/10.3390/plants12101964
Wilkinson H, Coppock A, Richmond BL, Lagunas B, Gifford ML. Plant–Environment Response Pathway Regulation Uncovered by Investigating Non-Typical Legume Symbiosis and Nodulation. Plants. 2023; 12(10):1964. https://doi.org/10.3390/plants12101964
Chicago/Turabian StyleWilkinson, Helen, Alice Coppock, Bethany L. Richmond, Beatriz Lagunas, and Miriam L. Gifford. 2023. "Plant–Environment Response Pathway Regulation Uncovered by Investigating Non-Typical Legume Symbiosis and Nodulation" Plants 12, no. 10: 1964. https://doi.org/10.3390/plants12101964
APA StyleWilkinson, H., Coppock, A., Richmond, B. L., Lagunas, B., & Gifford, M. L. (2023). Plant–Environment Response Pathway Regulation Uncovered by Investigating Non-Typical Legume Symbiosis and Nodulation. Plants, 12(10), 1964. https://doi.org/10.3390/plants12101964





