GASA Proteins: Review of Their Functions in Plant Environmental Stress Tolerance
Abstract
1. Introduction
2. GASA Proteins Identified in Plants
3. GASA Protein’s Structure
4. Subcellular Localization of GASA Proteins
5. Tissue and Organs Specific Expression Patterns of GASA Genes
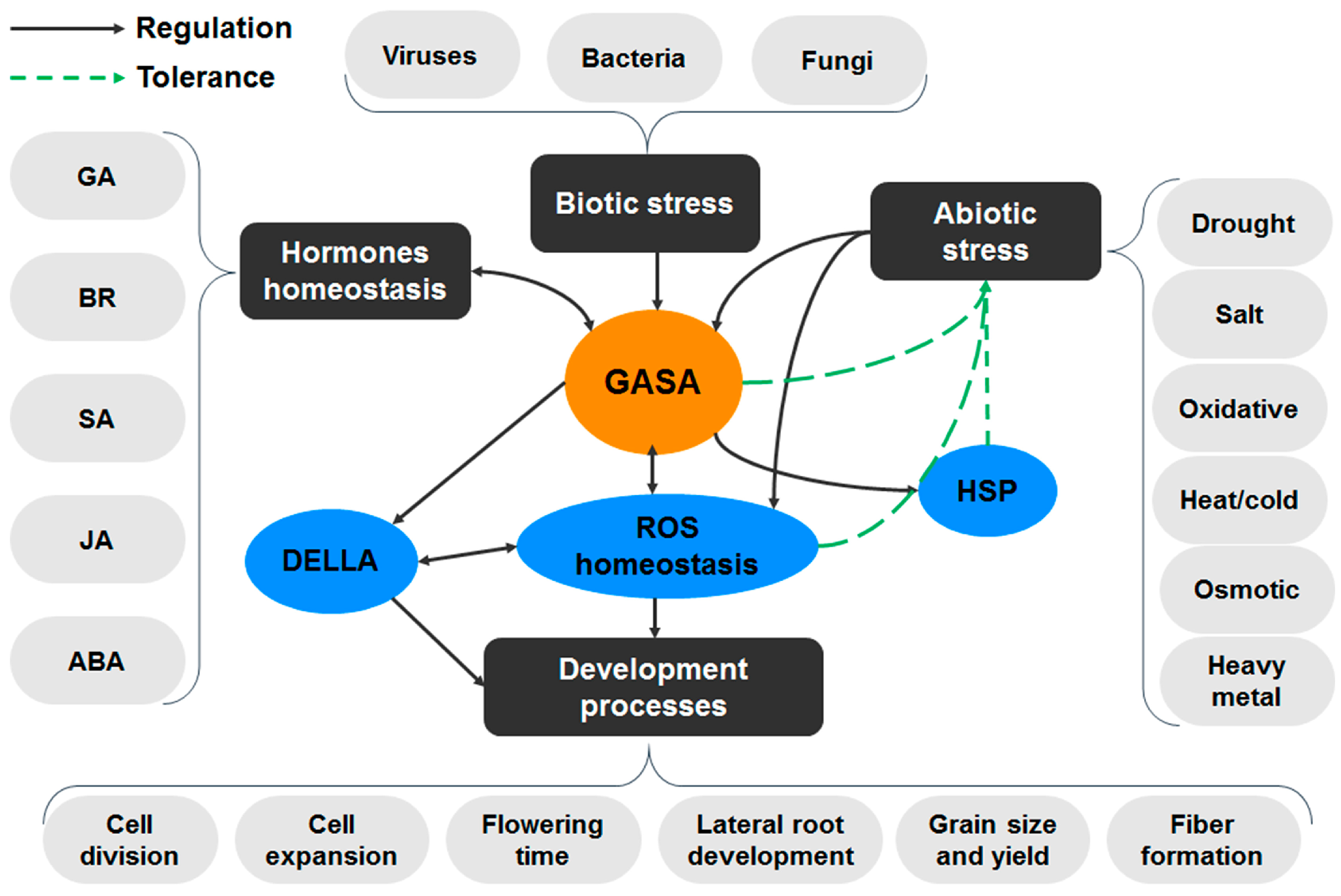
6. Involvement of GASA Genes in Plant Growth and Development
7. Phytohormones and GASA Proteins
8. Involvement of GASA in Abiotic Stress Tolerance and Redox Status Homeostasis
9. Involvement of GASA Proteins in Biotic Stress
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herzog, M.; Dorne, A.M.; Grellet, F. GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol. Biol. 1995, 27, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Aubert, D.; Chevillard, M.; Dorne, A.M.; Arlaud, G.; Herzog, M. Expression patterns of GASA genes in Arabidopsis thaliana: The GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol. Biol. 1998, 36, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Wigoda, N.; Ben-Nissan, G.; Granot, D.; Schwartz, A.; Weiss, D. The gibberellin-induced, cysteine-rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity. Plant J. 2006, 48, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, X. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Sci. Bull. 2008, 53, 3839–3846. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Z.; Xu, Y.; Joo, S.H.; Kim, S.K.; Xue, Z.; Xu, Z.; Wang, Z.; Chong, K. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 2009, 57, 498–510. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 2013, 64, 1637–1647. [Google Scholar] [CrossRef]
- Srivastava, R.; Liu, J.X.; Guo, H.; Yin, Y.; Howell, S.H. Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 2009, 59, 930–939. [Google Scholar] [CrossRef]
- Yu, F.; Qian, L.; Nibau, C.; Duan, Q.; Kita, D.; Levasseur, K.; Li, X.; Lu, C.; Li, H.; Hou, C.; et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 14693–14698. [Google Scholar] [CrossRef]
- Yamada, M.; Sawa, S. The roles of peptide hormones during plant root development. Curr. Opin. Plant Biol. 2013, 16, 56–61. [Google Scholar] [CrossRef]
- Miyakawa, T.; Hatano, K.; Miyauchi, Y.; Suwa, Y.; Sawano, Y.; Tanokura, M. A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol. 2014, 166, 766–778. [Google Scholar] [CrossRef]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; van der Does, D.; Ladwig, F.; Sticht, C.; Kolbeck, A.; Schurholz, A.K.; Augustin, S.; Keinath, N.; Rausch, T.; Greiner, S.; et al. A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 15261–15266. [Google Scholar] [CrossRef] [PubMed]
- Stes, E.; Gevaert, K.; De Smet, I. Phosphoproteomics-based peptide ligand-receptor kinase pairing. Commentary on: “A peptide hormone and its receptor protein kinase regulate plant cell expansion”. Front. Plant Sci. 2015, 6, 224. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, S.; Xu, D.; Liu, X.; Li, X.; Xiao, W.; Cao, J.; Jiang, H.; Min, X.; Wang, J.; et al. Identification and analysis of the GASR gene family in common wheat (Triticum aestivum L.) and characterization of TaGASR34, a gene associated with seed dormancy and germination. Front. Genet. 2019, 10, 980. [Google Scholar] [CrossRef]
- Zhang, K.; Hu, Y.; Yang, D.; Yan, C.; Li, N.; Li, Z.; Njogu, M.K.; Wang, X.; Jia, L. Genome-wide identification of GASA gene family in ten Cucurbitaceae species and expression analysis in Cucumber. Agronomy 2022, 12, 1978. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Z.; Jian, S. Genome-wide identification and functional analysis of the GASA gene family responding to multiple stressors in Canavalia rosea. Genes 2022, 13, 1988. [Google Scholar] [CrossRef]
- Bouteraa, M.T.; Ben Romdhane, W.; Ben Hsouna, A.; Amor, F.; Ebel, C.; Ben Saad, R. Genome-wide characterization and expression profiling of GASA gene family in Triticum turgidum ssp. durum (desf.) husn. (Durum wheat) unveils its involvement in environmental stress responses. Phytochemistry 2023, 206, 113544. [Google Scholar] [CrossRef]
- Nahirnak, V.; Almasia, N.I.; Hopp, H.E.; Vazquez-Rovere, C. Snakin/GASA proteins: Involvement in hormone crosstalk and redox homeostasis. Plant Signal. Behav. 2012, 7, 1004–1008. [Google Scholar] [CrossRef]
- Shi, L.; Gast, R.T.; Gopalraj, M.; Olszewski, N.E. Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J. 1992, 2, 153–159. [Google Scholar]
- Nahirnak, V.; Almasia, N.I.; Fernandez, P.V.; Hopp, H.E.; Estevez, J.M.; Carrari, F.; Vazquez-Rovere, C. Potato snakin-1 gene silencing affects cell division, primary metabolism, and cell wall composition. Plant Physiol. 2012, 158, 252–263. [Google Scholar] [CrossRef]
- Nahirñak, V.; Rivarola, M.; Gonzalez de Urreta, M.; Paniego, N.; Hopp, H.E.; Almasia, N.I.; Vazquez-Rovere, C. Genome-wide analysis of the Snakin/GASA gene family in Solanum tuberosum cv. Kennebec. Am. J. Potato Res. 2016, 93, 172–188. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, D.; Zhang, L.; Gao, C.; Xin, M.; Tahir, M.M.; Li, Y.; Ma, J.; Han, M. Comprehensive analysis of GASA family members in the Malus domestica genome: Identification, characterization, and their expressions in response to apple flower induction. BMC Genom. 2017, 18, 827. [Google Scholar] [CrossRef] [PubMed]
- Rubinovich, L.; Ruthstein, S.; Weiss, D. The Arabidopsis cysteine-rich GASA5 is a redox-active metalloprotein that suppresses gibberellin responses. Mol. Plant 2014, 7, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, K.A.; Moskal, W.A., Jr.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef]
- Garcia, A.N.; Ayub, N.D.; Fox, A.R.; Gomez, M.C.; Dieguez, M.J.; Pagano, E.M.; Berini, C.A.; Muschietti, J.P.; Soto, G. Alfalfa snakin-1 prevents fungal colonization and probably coevolved with rhizobia. BMC Plant Biol. 2014, 14, 248. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, A.; Kumar, P.; Sarkar, A.K. Giberellic acid-stimulated transcript proteins evolved through successive conjugation of novel motifs and their Subfunctionalization. Plant Physiol. 2019, 180, 998–1012. [Google Scholar] [CrossRef]
- Zimmermann, R.; Sakai, H.; Hochholdinger, F. The gibberellic acid stimulated-like gene family in maize and its role in lateral root development. Plant Physiol. 2010, 152, 356–365. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Sana, A.; Jamil, A.; Nasir, J.A.; Ahmed, S.; Hameed, M.U.; Abdullah. A genome-wide approach to the comprehensive analysis of GASA gene family in Glycine max. Plant Mol. Biol. 2019, 100, 607–620. [Google Scholar] [CrossRef]
- Muhammad, I.; Li, W.Q.; Jing, X.Q.; Zhou, M.R.; Shalmani, A.; Ali, M.; Wei, X.Y.; Sharif, R.; Liu, W.T.; Chen, K.M. A systematic in silico prediction of gibberellic acid stimulated GASA family members: A novel small peptide contributes to floral architecture and transcriptomic changes induced by external stimuli in rice. J. Plant Physiol. 2019, 234–235, 117–132. [Google Scholar] [CrossRef]
- Han, S.; Jiao, Z.; Niu, M.X.; Yu, X.; Huang, M.; Liu, C.; Wang, H.L.; Zhou, Y.; Mao, W.; Wang, X.; et al. Genome-wide comprehensive analysis of the GASA gene family in Populus. Int. J. Mol. Sci. 2021, 22, 12336. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, Z.; Qi, F.; Zhao, M.; Dong, W.; Huang, B.; Zheng, Z.; Zhang, X. Comprehensive analysis of GASA family members in the Peanut genome: Identification, characterization, and their expressions in response to pod development. Agronomy 2022, 12, 3067. [Google Scholar] [CrossRef]
- Li, Z.; Gao, J.; Wang, G.; Wang, S.; Chen, K.; Pu, W.; Wang, Y.; Xia, Q.; Fan, X. Genome-wide identification and characterization of GASA gene family in Nicotiana tabacum. Front. Genet. 2021, 12, 768942. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhong, Y.; Chen, M.; Wu, B.; Wang, T.; Jiang, B.; Zhong, G. Analysis of CcGASA family members in Citrus clementina (Hort. ex Tan.) by a genome-wide approach. BMC Plant Biol. 2021, 21, 565. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Yao, J.; Zhang, S.; Li, X.; Zhang, X.; Yadav, V.; Wang, X. Genome-wide characterization and expression profiling of GASA genes during different stages of seed development in grapevine (Vitis vinifera L.) predict their involvement in seed development. Int. J. Mol. Sci. 2020, 21, 1088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Geng, X.; Zhang, H.; Zhou, C.; Zhao, A.; Wang, F.; Zhao, Y.; Tian, X.; Hu, Z.; Xin, M.; et al. Isolation and characterization of heat-responsive gene TaGASR1 from wheat (Triticum aestivum L.). J. Plant Biol. 2017, 60, 57–65. [Google Scholar] [CrossRef]
- Mao, Z.; Zheng, J.; Wang, Y.; Chen, G.; Yang, Y.; Feng, D.; Xie, B. The new CaSn gene belonging to the snakin family induces resistance against root-knot nematode infection in pepper. Phytoparasitica 2011, 39, 151–164. [Google Scholar] [CrossRef]
- Alonso-Ramirez, A.; Rodriguez, D.; Reyes, D.; Jimenez, J.A.; Nicolas, G.; Lopez-Climent, M.; Gomez-Cadenas, A.; Nicolas, C. Cross-talk between gibberellins and salicylic acid in early stress responses in Arabidopsis thaliana seeds. Plant Signal. Behav. 2009, 4, 750–751. [Google Scholar] [CrossRef]
- de la Fuente, J.I.; Amaya, I.; Castillejo, C.; Sanchez-Sevilla, J.F.; Quesada, M.A.; Botella, M.A.; Valpuesta, V. The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J. Exp. Bot. 2006, 57, 2401–2411. [Google Scholar] [CrossRef]
- Moyano-Canete, E.; Bellido, M.L.; Garcia-Caparros, N.; Medina-Puche, L.; Amil-Ruiz, F.; Gonzalez-Reyes, J.A.; Caballero, J.L.; Munoz-Blanco, J.; Blanco-Portales, R. FaGAST2, a strawberry ripening-related gene, acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 2013, 54, 218–236. [Google Scholar] [CrossRef]
- Kotilainen, M.; Helariutta, Y.; Mehto, M.; Pollanen, E.; Albert, V.A.; Elomaa, P.; Teeri, T.H. GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 1999, 11, 1093–1104. [Google Scholar] [CrossRef]
- Peng, J.; Lai, L.; Wang, X. PRGL: A cell wall proline-rich protein containning GASA domain in Gerbera hybrida. Sci. China C Life Sci. 2008, 51, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Li, K.L.; Bai, X.; Li, Y.; Cai, H.; Ji, W.; Tang, L.L.; Wen, Y.D.; Zhu, Y.M. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J. Plant Physiol. 2011, 168, 2153–2160. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Ma, C.; Lv, J.; Zhang, C.; Ma, Q.; Fan, S. Identification, characterization, and expression profiles of the GASA genes in cotton. J. Cotton Res. 2021, 4, 7. [Google Scholar] [CrossRef]
- An, B.; Wang, Q.; Zhang, X.; Zhang, B.; Luo, H.; He, C. Comprehensive transcriptional and functional analyses of HbGASA genes reveal their roles in fungal pathogen resistance in Hevea brasiliensis. Tree Genet. Genomes 2018, 14, 41. [Google Scholar] [CrossRef]
- Rodriguez-Decuadro, S.; da Rosa, G.; Radio, S.; Barraco-Vega, M.; Benko-Iseppon, A.M.; Dans, P.D.; Smircich, P.; Cecchetto, G. Antimicrobial peptides in the seedling transcriptome of the tree legume Peltophorum dubium. Biochimie 2021, 180, 229–242. [Google Scholar] [CrossRef]
- Ben-Nissan, G.; Lee, J.Y.; Borohov, A.; Weiss, D. GIP, a Petunia hybrida GA-induced cysteine-rich protein: A possible role in shoot elongation and transition to flowering. Plant J. 2004, 37, 229–238. [Google Scholar] [CrossRef]
- Büyük, İ.; Okay, A.; Gorska, M.; İLhan, E.; Aras, E.S. Identification and characterization of the Pvul-GASA gene family in the Phaseolus vulgaris and expression patterns under salt stress. Turk. J. Bot. 2021, 45, 655–670. [Google Scholar] [CrossRef]
- Hou, D.; Bai, Q.; Li, J.; Xie, L.; Li, X.; Cheng, Z.; Gao, J. The gibberellic acid-stimulated transcript gene family in Moso Bamboo: A Genome-wide survey and expression profiling during development and abiotic stresses. J. Plant Growth Regul. 2018, 37, 1135–1147. [Google Scholar] [CrossRef]
- Wang, H.; Wei, T.; Wang, X.; Zhang, L.; Yang, M.; Chen, L.; Song, W.; Wang, C.; Chen, C. Transcriptome analyses from mutant Salvia miltiorrhiza reveals Important roles for SmGASA4 during plant development. Int. J. Mol. Sci. 2018, 19, 2088. [Google Scholar] [CrossRef]
- Faccio, P.; Vazquez-Rovere, C.; Hopp, E.; González, G.; Décima-Oneto, C.; Favret, E.; Díaz Paleo, A.; Franzone, P. Increased tolerance to wheat powdery mildew by heterologous constitutive expression of the Solanum chacoense Snakin-1 gene. Czech J. Genet. Plant Breed 2011, 47, S135–S141. [Google Scholar] [CrossRef]
- Su, D.; Liu, K.; Yu, Z.; Li, Y.; Zhang, Y.; Zhu, Y.; Wu, Y.; He, H.; Zeng, X.; Chen, H.; et al. Genome-wide characterization of the tomato GASA family identifies SlGASA1 as a repressor of fruit ripening. Hortic. Res. 2023, 10, uhac222. [Google Scholar] [CrossRef] [PubMed]
- Filiz, E.; Kurt, F. Antimicrobial peptides Snakin/GASA gene family in sorghum (Sorghum bicolor): Genome-wide identification and bioinformatics analyses. Gene Rep. 2020, 20, 100766. [Google Scholar] [CrossRef]
- Abdullah; Faraji, S.; Mehmood, F.; Malik, H.M.T.; Ahmed, I.; Heidari, P.; Poczai, P. The GASA gene family in Cacao (Theobroma cacao, Malvaceae): Genome wide identification and expression analysis. Agronomy 2021, 11, 1425. [Google Scholar] [CrossRef]
- Oliveira-Lima, M.; Benko-Iseppon, A.M.; Neto, J.; Rodriguez-Decuadro, S.; Kido, E.A.; Crovella, S.; Pandolfi, V. Snakin: Structure, roles and applications of a plant antimicrobial peptide. Curr. Protein Pept. Sci. 2017, 18, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Almasia, N.I.; Bazzini, A.A.; Hopp, H.E.; Vazquez-Rovere, C. Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol. Plant Pathol. 2008, 9, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Almasia, N.I.; Molinari, M.P.; Maroniche, G.A.; Nahirnak, V.; Barrios Baron, M.P.; Taboga, O.A.; Vazquez Rovere, C. Successful production of the potato antimicrobial peptide Snakin-1 in baculovirus-infected insect cells and development of specific antibodies. BMC Biotechnol. 2017, 17, 75. [Google Scholar] [CrossRef] [PubMed]
- Berrocal-Lobo, M.; Segura, A.; Moreno, M.; Lopez, G.; Garcia-Olmedo, F.; Molina, A. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002, 128, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Kovalskaya, N.; Hammond, R.W. Expression and functional characterization of the plant antimicrobial snakin-1 and defensin recombinant proteins. Protein Expr. Purif. 2009, 63, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Kuddus, M.R.; Rumi, F.; Tsutsumi, M.; Takahashi, R.; Yamano, M.; Kamiya, M.; Kikukawa, T.; Demura, M.; Aizawa, T. Expression, purification and characterization of the recombinant cysteine-rich antimicrobial peptide snakin-1 in Pichia pastoris. Protein Expr. Purif. 2016, 122, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Qi, L.; Wang, J.; Du, L.; Xu, H.; Wang, A.; Zhang, Z. Expression of a potato antimicrobial peptide SN1 increases resistance to take-all pathogen Gaeumannomyces graminis var. tritici in transgenic wheat. Funct. Integr. Genom. 2013, 13, 403–409. [Google Scholar] [CrossRef]
- Segura, A.; Moreno, M.; Madueno, F.; Molina, A.; Garcia-Olmedo, F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol. Plant Microbe Interact. 1999, 12, 16–23. [Google Scholar] [CrossRef]
- Harris, P.W.; Yang, S.H.; Molina, A.; Lopez, G.; Middleditch, M.; Brimble, M.A. Plant antimicrobial peptides snakin-1 and snakin-2: Chemical synthesis and insights into the disulfide connectivity. Chemistry 2014, 20, 5102–5110. [Google Scholar] [CrossRef]
- Lopez-Solanilla, E.; Gonzalez-Zorn, B.; Novella, S.; Vazquez-Boland, J.A.; Rodriguez-Palenzuela, P. Susceptibility of Listeria monocytogenes to antimicrobial peptides. FEMS Microbiol. Lett. 2003, 226, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Sakaguchi, N.; Shimada, H. Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet. Syst. 2006, 81, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Rezaee, S.; Ahmadizadeh, M.; Heidari, P. Genome-wide characterization, expression profiling, and post-transcriptional study of GASA gene family. Gene Rep. 2020, 20, 100795. [Google Scholar] [CrossRef]
- Yeung, H.; Squire, C.J.; Yosaatmadja, Y.; Panjikar, S.; Lopez, G.; Molina, A.; Baker, E.N.; Harris, P.W.; Brimble, M.A. Radiation damage and racemic protein crystallography reveal the unique structure of the GASA/Snakin protein superfamily. Angew. Chem. Int. Ed. Engl. 2016, 55, 7930–7933. [Google Scholar] [CrossRef]
- Su, T.; Han, M.; Cao, D.; Xu, M. Molecular and Biological Properties of Snakins: The foremost cysteine-rich plant host defense peptides. J. Fungi 2020, 6, 220. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Franco, O.L. Theoretical structural insights into the snakin/GASA family. Peptides 2013, 44, 163–167. [Google Scholar] [CrossRef]
- Broekaert, W.F.; Cammue, B.; De Bolle, M.; Thevissen, K.; De Samblanx, G.W.; Osborn, R.W.; Nielson, K. Antimicrobial peptides from plants. Crit. Rev. Plant Sci. 1997, 16, 297–323. [Google Scholar] [CrossRef]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial Peptides from Plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef]
- Herbel, V.; Schafer, H.; Wink, M. Recombinant Production of Snakin-2 (an antimicrobial peptide from tomato) in E. coli and analysis of its bioactivity. Molecules 2015, 20, 14889–14901. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Decuadro, S.; Barraco-Vega, M.; Dans, P.D.; Pandolfi, V.; Benko-Iseppon, A.M.; Cecchetto, G. Antimicrobial and structural insights of a new snakin-like peptide isolated from Peltophorum dubium (Fabaceae). Amino Acids 2018, 50, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Cheng, C.; Zhong, Y.; Lv, Y.; Ma, Y.; Zhong, G. Molecular characterization of the gibberellin-stimulated transcript of GASA4 in Citrus. Plant Growth Regul. 2020, 91, 89–99. [Google Scholar] [CrossRef]
- Li, X.; Shi, S.; Tao, Q.; Tao, Y.; Miao, J.; Peng, X.; Li, C.; Yang, Z.; Zhou, Y.; Liang, G. OsGASR9 positively regulates grain size and yield in rice (Oryza sativa). Plant Sci. 2019, 286, 17–27. [Google Scholar] [CrossRef]
- Nahirnak, V.; Rivarola, M.; Almasia, N.I.; Barrios Baron, M.P.; Hopp, H.E.; Vile, D.; Paniego, N.; Vazquez Rovere, C. Snakin-1 affects reactive oxygen species and ascorbic acid levels and hormone balance in potato. PLoS ONE 2019, 14, e0214165. [Google Scholar] [CrossRef]
- Almasia, N.I.; Narhirñak, V.; Hopp, H.E.; Vazquez-Rovere, C. Isolation and characterization of the tissue and development-specific potato snakin-1 promoter inducible by temperature and wounding. Electron. J. Biotechnol. 2010, 13, 8–9. [Google Scholar] [CrossRef]
- Fang, Y.; Mei, H.; Zhou, B.; Xiao, X.; Yang, M.; Huang, Y.; Long, X.; Hu, S.; Tang, C. De novo transcriptome analysis reveals distinct defense mechanisms by young and mature leaves of Hevea brasiliensis (Para Rubber Tree). Sci. Rep. 2016, 6, 33151. [Google Scholar] [CrossRef]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.; Opsahl-Sorteberg, H.G. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, C.; Peng, J.; Sun, S.; Wang, X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol. Biol. 2009, 69, 745–759. [Google Scholar] [CrossRef]
- Ben-Nissan, G.; Weiss, D. The petunia homologue of tomato gast1: Transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol. Biol. 1996, 32, 1067–1074. [Google Scholar] [CrossRef]
- Qu, J.; Kang, S.G.; Hah, C.; Jang, J.C. Molecular and cellular characterization of GA-stimulated transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016, 246, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Lai, L.; Wang, X. Temporal and spatial expression analysis of PRGL in Gerbera hybrida. Mol. Biol. Rep. 2010, 37, 3311–3317. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Wang, F.; Liu, T.; Dong, Z.; Li, A.; Jing, R.; Mao, L.; Li, Y.; Liu, X.; Zhang, K. Natural variation of TaGASR7-A1 affects grain length in common wheat under multiple cultivation conditions. Mol. Breed. 2014, 34, 937–947. [Google Scholar] [CrossRef]
- Lee, S.C.; Kim, S.J.; Han, S.K.; An, G.; Kim, S.R. A gibberellin-stimulated transcript, OsGASR1, controls seedling growth and alpha-amylase expression in rice. J. Plant Physiol. 2017, 214, 116–122. [Google Scholar] [CrossRef]
- Ke, S.; Liu, X.; Luan, X.; Yang, W.; Zhu, H.; Liu, G.; Zhang, G.; Wang, S. Genome-wide transcriptome profiling provides insights into panicle development of rice (Oryza sativa L.). Gene 2018, 675, 285–300. [Google Scholar] [CrossRef]
- Dong, B.; Deng, Y.; Wang, H.; Gao, R.; Stephen, G.K.; Chen, S.; Jiang, J.; Chen, F. Gibberellic acid signaling Is required to induce flowering of Chrysanthemums grown under both short and long days. Int. J. Mol. Sci. 2017, 18, 1259. [Google Scholar] [CrossRef]
- Miceli, A.; Moncada, A.; Sabatino, L.; Vetrano, F. Effect of gibberellic acid on growth, yield, and quality of leaf lettuce and rocket grown in a floating system. Agronomy 2019, 9, 382. [Google Scholar] [CrossRef]
- Nagai, K.; Mori, Y.; Ishikawa, S.; Furuta, T.; Gamuyao, R.; Niimi, Y.; Hobo, T.; Fukuda, M.; Kojima, M.; Takebayashi, Y.; et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 2020, 584, 109–114. [Google Scholar] [CrossRef]
- Rubinovich, L.; Weiss, D. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 2010, 64, 1018–1027. [Google Scholar] [CrossRef]
- Zhong, C.; Xu, H.; Ye, S.; Wang, S.; Li, L.; Zhang, S.; Wang, X. Gibberellic acid-stimulated Arabidopsis serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis. Plant Physiol. 2015, 169, 2288–2303. [Google Scholar]
- Lee, S.C.; Han, S.; Kim, S.J. Salt-and ABA-inducible OsGASR1 is involved in salt tolerance. J. Plant Biol. 2015, 58, 96–101. [Google Scholar] [CrossRef]
- Ko, C.B.; Woo, Y.M.; Lee, D.J.; Lee, M.C.; Kim, C.S. Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol. Biochem. 2007, 45, 722–728. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X. Overexpression of GASA5 increases the sensitivity of Arabidopsis to heat stress. J. Plant Physiol. 2011, 168, 2093–2101. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Ramirez, A.; Rodriguez, D.; Reyes, D.; Jimenez, J.A.; Nicolas, G.; Lopez-Climent, M.; Gomez-Cadenas, A.; Nicolas, C. Evidence for a role of gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol. 2009, 150, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Mo, H.; Sun, W.; Guo, Y.; Li, J. Systematic isolation and characterization of Cadmium tolerant genes in tobacco: A cDNA library construction and screening approach. PLoS ONE 2016, 11, e0161147. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Meiyalaghan, S.; Thomson, S.J.; Fiers, M.; Barrell, P.J.; Latimer, J.M.; Mohan, S.; Jones, E.E.; Conner, A.J.; Jacobs, J.M. Structure and expression of GSL1 and GSL2 genes encoding gibberellin stimulated-like proteins in diploid and highly heterozygous tetraploid potato reveals their highly conserved and essential status. BMC Genom. 2014, 15, 2. [Google Scholar] [CrossRef]
- Herbel, V.; Sieber-Frank, J.; Wink, M. The antimicrobial peptide snakin-2 is upregulated in the defense response of tomatoes (Solanum lycopersicum) as part of the jasmonate-dependent signaling pathway. J. Plant Physiol. 2017, 208, 1–6. [Google Scholar] [CrossRef]
- Balaji, V.; Smart, C.D. Over-expression of snakin-2 and extensin-like protein genes restricts pathogen invasiveness and enhances tolerance to Clavibacter michiganensis subsp. michiganensis in transgenic tomato (Solanum lycopersicum). Transgenic Res. 2012, 21, 23–37. [Google Scholar]
- Mohan, S.; Meiyalaghan, S.; Latimer, J.M.; Gatehouse, M.L.; Monaghan, K.S.; Vanga, B.R.; Pitman, A.R.; Jones, E.E.; Conner, A.J.; Jacobs, J.M. GSL2 over-expression confers resistance to Pectobacterium atrosepticum in potato. Appl. Genet. 2014, 127, 677–689. [Google Scholar] [CrossRef]
- Balaji, V.; Sessa, G.; Smart, C.D. Silencing of host basal defense response-related gene expression increases susceptibility of Nicotiana benthamiana to Clavibacter michiganensis subsp. michiganensis. Phytopathology 2011, 101, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Boonpa, K.; Tantong, S.; Weerawanich, K.; Panpetch, P.; Pringsulaka, O.; Yingchutrakul, Y.; Roytrakul, S.; Sirikantaramas, S. Heterologous expression and antimicrobial activity of OsGASR3 from rice (Oryza sativa L.). J. Plant Physiol. 2018, 224–225, 95–102. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Yang, X.; Xun, H.; Lou, X.; Li, S.; Zhang, Z.; Jiang, L.; Dong, Y.; Wang, S.; Pang, J.; et al. Over-expression of GmSN1 enhances virus resistance in Arabidopsis and soybean. Plant Cell. Rep. 2017, 36, 1441–1455. [Google Scholar] [CrossRef]
- Thevissen, K.; Kristensen, H.H.; Thomma, B.P.; Cammue, B.P.; Francois, I.E. Therapeutic potential of antifungal plant and insect defensins. Drug. Discov. Today 2007, 12, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Padovan, L.; Scocchi, M.; Tossi, A. Structural aspects of plant antimicrobial peptides. Curr. Protein Pept. Sci. 2010, 11, 210–219. [Google Scholar] [CrossRef]
- Silva, O.N.; Mulder, K.C.; Barbosa, A.E.; Otero-Gonzalez, A.J.; Lopez-Abarrategui, C.; Rezende, T.M.; Dias, S.C.; Franco, O.L. Exploring the pharmacological potential of promiscuous host-defense peptides: From natural screenings to biotechnological applications. Front. Microbiol. 2011, 2, 232. [Google Scholar]
- Shwaiki, L.N.; Sahin, A.W.; Arendt, E.K. Study on the inhibitory activity of a synthetic Defensin derived from barley endosperm against common food spoilage Yeast. Molecules 2020, 26, 165. [Google Scholar] [CrossRef]
| Species | Protein Family | Characterized Members | Protein Length (aa) | Ref. |
|---|---|---|---|---|
| Arabidopsis thaliana | AtGASA | 15 | 87–275 | [22] |
| Arachis duranensis | AdGASA | 20 | Ø | [31] |
| Arachis ipaensis | AiGASA | 22 | Ø | [31] |
| Arachis hypogaea | AhGASA | 40 | 60–202 | [31] |
| Benincasa hispida | ø | 9 | 80–232 | [15] |
| Canavalia rosea | CrGASA | 23 | 70–233 | [16] |
| Capsicum annuum | CaSn | 1 | 104 | [36] |
| Citrullus lanatus | ø | 9 | 88–216 | [15] |
| Citrus clementina | CcGASA | 18 | 70–206 | [33] |
| Cucumis melo | ø | 10 | 80–222 | [15] |
| Cucumis sativus | ø | 9 | 61–231 | [15] |
| Cucurbita moschata | ø | 10 | 85–516 | [15] |
| Fagus sylvatica | FsGASA | 1 | 107 | [37] |
| Fragaria × ananassa (strawberry) | FaGAST | 2 | 86–91 | [38,39] |
| Gerbera hybrida | GEG PRGL | 1 1 | 150 (PRGL) | [40,41] |
| Glycine soja | GsGASA | 1 | 97 | [42] |
| Glycine max | GmGASA | 37 | 66–198 | [28] |
| Gossypium arboreum | GmGASA | 17 | 69–213 | [43] |
| Gossypium barbadense | GbGASA | 33 | 73–870 | [43] |
| Gossypium herbaceum | GheGASA | 19 | 89–926 | [43] |
| Gossypium hirsutum | GhGASA | 38 | 76–264 | [43] |
| Gossypium raimondii | GrGASA | 25 | 72–297 | [43] |
| Hevea brasiliensis | HbGASA | 16 | 88–275 | [44] |
| Lagenaria siceraria | ø | 8 | 80–212 | [15] |
| Luffa cylindrica | ø | 9 | 85–221 | [15] |
| Malus domestica | MdGASA | 26 | 88–305 | [22] |
| Medicago sativa | MsSN1 | 1 | 108 | [25] |
| Momordica charantia | ø | 15 | 59–370 | [15] |
| Nicotiana tabacum | NtGASA | 18 | 61–147 | [32] |
| Oryza sativa | OsGASA | 10 | 92–152 | [29] |
| Peltophorum dubium | PdSN | 12 | 63–95 | [45] |
| Petunia hybrida | GIP | 4 | Ø | [46] |
| Phaseolus vulgaris | PvulGASA | 23 | 88–179 | [47] |
| Phyllostachys edulis | PheGAST | 8 | 81–113 | [48] |
| Populus euphratica | PeuGASA | 19 | 88–222 | [30] |
| Populus trichocarpa | PtrGASA | 21 | 88–191 | [30] |
| Salvia miltiorrhiza | SmGASA | 1 | 110 | [49] |
| Sechium edule | ø | 16 | 86–223 | [15] |
| Solanum chacoense | Snakin-1 | 1 | Ø | [50] |
| Solanum lycopersicum | SlGASA | 17 | 88–146 | [51] |
| Solanum tuberosum | Snakin | 18 | 88–143 | [21] |
| Sorghum bicolor | SbSN | 12 | 93–137 | [52] |
| Theobroma cacao | TcGASA | 17 | 88–320 | [53] |
| Trichosanthes anguina | ø | 18 | 80–234 | [15] |
| Triticum aestivum | TaGASR | 37 | 261–1172 | [14] |
| Triticum durum | TdGASA | 19 | 92–222 | [17] |
| Vitis vinifera | VvGASA | 14 | 74–298 | [34] |
| Zea mays | ZmGSL | 10 | 75–128 | [27] |
| GASA Protein | Subcellular Localization | Signal Peptide Length (aa) | Ref. |
|---|---|---|---|
| AtGASA14 | Plasma Membrane | Ø | [6] |
| CcGASA4 | Cell Membrane—Nucleus | Ø | [73] |
| GIP | Plasma Membrane—Endoplasmic Reticulum | 19 (GIP1) | [46] |
| GsGASA1 | Plasma Membrane—Cytoplasm—Nucleus | 27 | [42] |
| HbGASA5—HbGASA9 | Nucleus—Cytoplasm | Ø | [72] |
| OsGASR1—OsGASR9 | Apoplasm—Cell Wall | 29 | [64,74] |
| PRGL | Cell Wall | 19 | [41] |
| SlGASA1 | Cytoplasm—Nucleus | 18–29 | [51] |
| Snakin-1 | Plasma Membrane | 25 | [20,21] |
| TaGASR1 | Cell Membrane—Cytosol | Ø | [35] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouteraa, M.T.; Ben Romdhane, W.; Baazaoui, N.; Alfaifi, M.Y.; Chouaibi, Y.; Ben Akacha, B.; Ben Hsouna, A.; Kačániová, M.; Ćavar Zeljković, S.; Garzoli, S.; et al. GASA Proteins: Review of Their Functions in Plant Environmental Stress Tolerance. Plants 2023, 12, 2045. https://doi.org/10.3390/plants12102045
Bouteraa MT, Ben Romdhane W, Baazaoui N, Alfaifi MY, Chouaibi Y, Ben Akacha B, Ben Hsouna A, Kačániová M, Ćavar Zeljković S, Garzoli S, et al. GASA Proteins: Review of Their Functions in Plant Environmental Stress Tolerance. Plants. 2023; 12(10):2045. https://doi.org/10.3390/plants12102045
Chicago/Turabian StyleBouteraa, Mohamed Taieb, Walid Ben Romdhane, Narjes Baazaoui, Mohammad Y. Alfaifi, Yosra Chouaibi, Bouthaina Ben Akacha, Anis Ben Hsouna, Miroslava Kačániová, Sanja Ćavar Zeljković, Stefania Garzoli, and et al. 2023. "GASA Proteins: Review of Their Functions in Plant Environmental Stress Tolerance" Plants 12, no. 10: 2045. https://doi.org/10.3390/plants12102045
APA StyleBouteraa, M. T., Ben Romdhane, W., Baazaoui, N., Alfaifi, M. Y., Chouaibi, Y., Ben Akacha, B., Ben Hsouna, A., Kačániová, M., Ćavar Zeljković, S., Garzoli, S., & Ben Saad, R. (2023). GASA Proteins: Review of Their Functions in Plant Environmental Stress Tolerance. Plants, 12(10), 2045. https://doi.org/10.3390/plants12102045








