Role of NPR1 in Systemic Acquired Stomatal Immunity
Abstract
:1. Introduction
2. Results
2.1. Bacterial Colonization of SAR Mutants
2.2. Stomatal Density in the Course of Vegetative Development of Wild Type and npr1-1 Mutant
2.3. Stomatal Movement of Wild Type and npr1-1 in Response to Pst DC3000
2.4. Bacterial Colonization of Wild Type and npr1-1
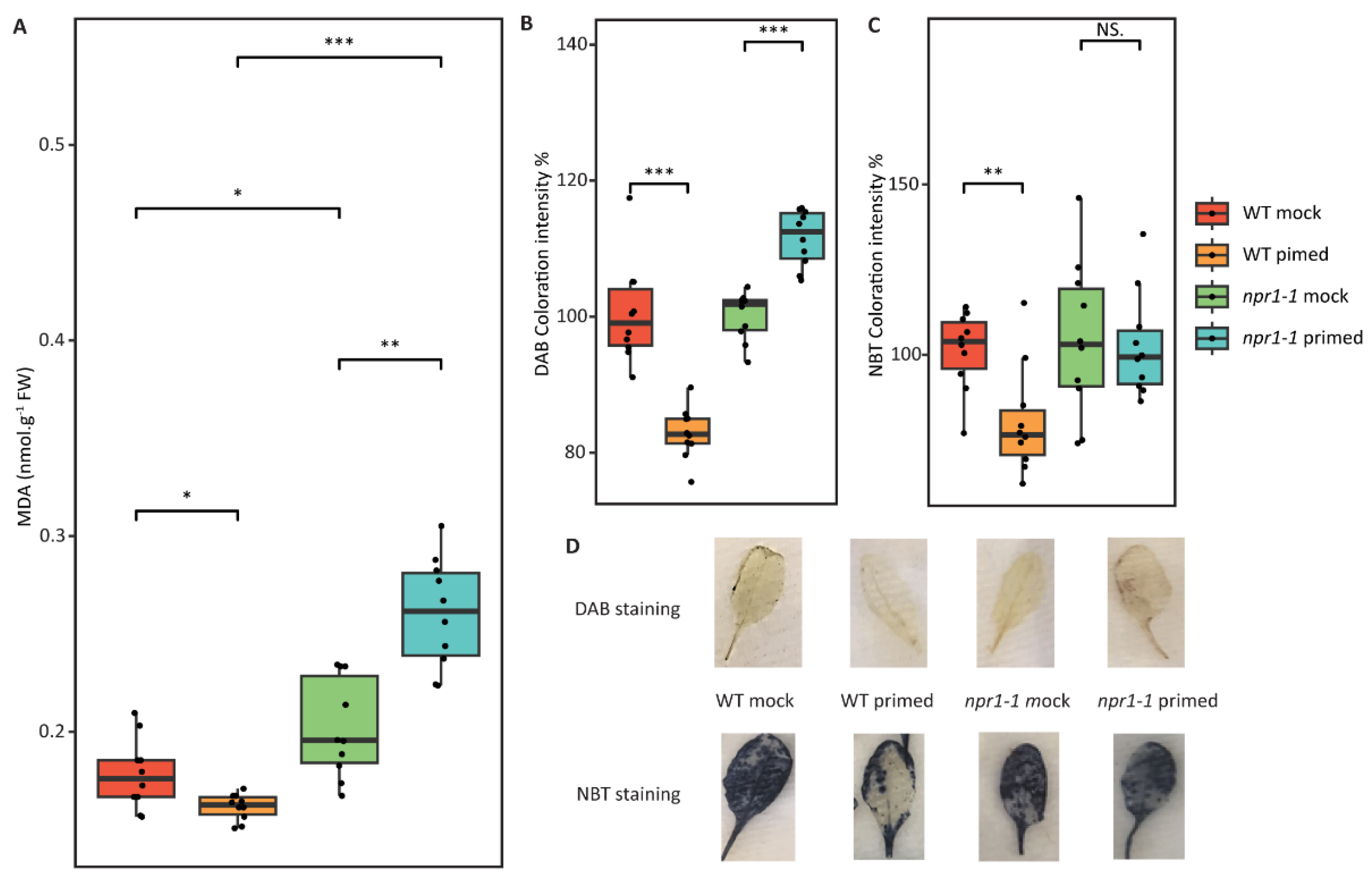
2.5. Redox Assays Revealed Decreased ROS and Lipid Peroxidation in Wild-Type Primed Leaves
2.6. Proteomic Differences of npr1-1 and Wild Type Arabidopsis
2.7. Proteome Changes in npr1-1 and Wild Type after Pst DC3000 Infection
2.8. Glutathione Metabolism of npr1-1 and Wild Type Arabidopsis after Pst DC3000 Infection
3. Discussion
3.1. NPR1 Does Not Regulate Stomatal Density
3.2. Stomatal Reopening by SCREW-NUT Function Is NPR1-Independent
3.3. Translation Regulation Is an Alternative Priming Effect of npr1-1 Knockout Arabidopsis
4. Materials and Methods
4.1. Plant Growth
4.2. Bacterial Growth
4.3. Stomata Aperture Measurements
4.4. Pst entry and Growth Assays
4.5. Stomata Density Assay
4.6. DAB, NBT, and MDA Analyses
4.7. Protein Extraction, Digestion, and LC-MS/MS
4.8. Proteomics Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Harmon, A.C.; Chen, S. Plant immune responses—From guard cells and local responses to systemic defense against bacterial pathogens. Plant Signal. Behav. 2019, 14, e1588667. [Google Scholar] [CrossRef] [PubMed]
- Withers, J.; Dong, X. Posttranslational Modifications of NPR1: A Single Protein Playing Multiple Roles in Plant Immunity and Physiology. PLoS Pathog. 2016, 12, e1005707. [Google Scholar] [CrossRef]
- Rufián, J.S.; Rueda-Blanco, J.; Beuzón, C.R.; Ruiz-Albert, J. Protocol: An improved method to quantify activation of systemic acquired resistance (SAR). Plant Methods 2019, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Naidoo, S.; van den Berg, N. The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) and Related Family: Mechanistic Insights in Plant Disease Resistance. Front. Plant Sci. 2019, 10, 102. [Google Scholar] [CrossRef]
- Melotto, M.; Zhang, L.; Oblessuc, P.R.; He, S.Y. Stomatal Defense a Decade Later. Plant Physiol. 2017, 174, 561–571. [Google Scholar] [CrossRef]
- Kinkema, M.; Fan, W.; Dong, X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 2000, 12, 2339–2350. [Google Scholar] [CrossRef]
- Lee, H.J.; Park, Y.J.; Seo, P.J.; Kim, J.H.; Sim, H.J.; Kim, S.G.; Park, C.M. Systemic Immunity Requires SnRK2.8-Mediated Nuclear Import of NPR1 in Arabidopsis. Plant Cell 2015, 27, 3425–3438. [Google Scholar] [CrossRef]
- Lindermayr, C.; Sell, S.; Müller, B.; Leister, D.; Durner, J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 2010, 22, 2894–2907. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L.; Jones, J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef]
- Daudi, A.; Cheng, Z.; O’Brien, J.A.; Mammarella, N.; Khan, S.; Ausubel, F.M.; Bolwell, G.P. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 2012, 24, 275–287. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Upadhyay, S.K. Calcium Channels, OST1 and Stomatal Defence: Current Status and Beyond. Cells 2023, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Leon-Reyes, A.; Van der Does, D.; De Lange, E.S.; Delker, C.; Wasternack, C.; Van Wees, S.C.; Ritsema, T.; Pieterse, C.M. Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 2010, 232, 1423–1432. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; He, S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hou, S.; Rodrigues, O.; Wang, P.; Luo, D.; Munemasa, S.; Lei, J.; Liu, J.; Ortiz-Morea, F.A.; Wang, X.; et al. Phytocytokine signalling reopens stomata in plant immunity and water loss. Nature 2022, 605, 332–339. [Google Scholar] [CrossRef]
- Toum, L.; Torres, P.S.; Gallego, S.M.; Benavídes, M.P.; Vojnov, A.A.; Gudesblat, G.E. Coronatine Inhibits Stomatal Closure through Guard Cell-Specific Inhibition of NADPH Oxidase-Dependent ROS Production. Front. Plant Sci. 2016, 7, 1851. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Gui, Y.; Li, Z.; Jiang, C.; Guo, J.; Niu, D. Induced Systemic Resistance for Improving Plant Immunity by Beneficial Microbes. Plants 2022, 11, 386. [Google Scholar] [CrossRef]
- David, L.; Kang, J.; Chen, S. Targeted Metabolomics of Plant Hormones and Redox Metabolites in Stomatal Immunity. Methods Mol. Biol. 2020, 2085, 79–92. [Google Scholar] [CrossRef]
- David, L.; Kang, J.; Dufresne, D.; Zhu, D.; Chen, S. Multi-Omics Revealed Molecular Mechanisms Underlying Guard Cell Systemic Acquired Resistance. Int. J. Mol. Sci. 2020, 22, 191. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Kang, J.; Chen, S. Untargeted Metabolomics of Arabidopsis Stomatal Immunity. Methods Mol. Biol. 2021, 2200, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Dommel, M.R.; Wang, C.; Li, Q.; Zhao, Q.; Zhang, X.; Dai, S.; Mou, Z. Differential Quantitative Requirements for NPR1 Between Basal Immunity and Systemic Acquired Resistance in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 570422. [Google Scholar] [CrossRef]
- Feys, B.J.; Moisan, L.J.; Newman, M.A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef]
- Dixon, D.P.; Edwards, R. Glutathione transferases. In Arabidopsis Book; American Society of Plant Biologist: Rockville, MD, USA, 2010; Volume 8, p. e0131. [Google Scholar] [CrossRef]
- Cheng, N.H.; Hirschi, K.D. Cloning and characterization of CXIP1, a novel PICOT domain-containing Arabidopsis protein that associates with CAX1. J. Biol. Chem. 2003, 278, 6503–6509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jiang, L.; Huang, J.; Ding, Y.; Liu, Z. Loss of proton/calcium exchange 1 results in the activation of plant defense and accelerated senescence in Arabidopsis. Plant Sci. 2020, 296, 110472. [Google Scholar] [CrossRef]
- Guerra, T.; Schilling, S.; Hake, K.; Gorzolka, K.; Sylvester, F.P.; Conrads, B.; Westermann, B.; Romeis, T. Calcium-dependent protein kinase 5 links calcium signaling with N-hydroxy-l-pipecolic acid- and SARD1-dependent immune memory in systemic acquired resistance. New Phytol. 2020, 225, 310–325. [Google Scholar] [CrossRef]
- Ota, M.; Imada, K.; Sasaki, K.; Kajihara, H.; Sakai, S.; Ito, S. MgO-induced defence against bacterial wilt disease in Arabidopsis thaliana. Plant Pathol. 2019, 68, 323–333. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Treder, K.; Király, L. Glutathione Contribution in Interactions between Turnip mosaic virus and Arabidopsis thaliana Mutants Lacking Respiratory Burst Oxidase Homologs D and F. Int. J. Mol. Sci. 2023, 24, 7128. [Google Scholar] [CrossRef]
- Uddin, S.; Bae, D.; Cha, J.Y.; Ahn, G.; Kim, W.Y.; Kim, M.G. Coronatine Induces Stomatal Reopening by Inhibiting Hormone Signaling Pathways. J. Plant Biol. 2022, 65, 403–411. [Google Scholar] [CrossRef]
- Talaat, N.B. Role of reactive oxygen species signaling in plant growth and development. In Reactive Oxygen, Nitrogen and Sulfur Species in Plants; John Wiley & Sons: Hoboken, NJ, USA, 2019; Volume 10, pp. 225–266. [Google Scholar] [CrossRef]
- Mindrebo, J.T.; Nartey, C.M.; Seto, Y.; Burkart, M.D.; Noel, J.P. Unveiling the functional diversity of the alpha/beta hydrolase superfamily in the plant kingdom. Curr. Opin. Struct. Biol. 2016, 41, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Gorlach, J. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef]
- Xin, X.F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar] [CrossRef]
- Lawrence, S.; Pang, Q.; Kong, W.; Chen, S. Stomata Tape-Peel: An Improved Method for Guard Cell Sample Preparation. J. Vis. Exp. 2018, 137, 57422. [Google Scholar] [CrossRef]
- Su, J.; Zhang, M.; Zhang, L.; Sun, T.; Liu, Y.; Lukowitz, W.; Xu, J.; Zhang, S. Regulation of Stomatal Immunity by Interdependent Functions of a Pathogen-Responsive MPK3/MPK6 Cascade and Abscisic Acid. Plant Cell 2017, 29, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Han, H.; Liu, A.; Guan, Q.; Kang, J.; David, L.; Dufresne, C.; Chen, S.; Tian, J. Combined ultraviolet and darkness regulation of medicinal metabolites in Mahonia bealei revealed by proteomics and metabolomics. J. Proteom. 2021, 233, 104081. [Google Scholar] [CrossRef] [PubMed]





| Locus ID | Protein Name | p Value | Log2FC |
|---|---|---|---|
| AT5G13650 | Elongation factor family protein | 0.04 | −5.30 |
| AT1G33140 | Ribosomal protein L6 family | 0.03 | 5.16 |
| AT3G47800 | Galactose mutarotase-like superfamily protein | 0.01 | −3.76 |
| AT5G10470 | Kinesin like for actin-based chloroplast movement 1 | 0.01 | −3.70 |
| AT1G09340 | Chloroplast RNA binding | 0.05 | −3.21 |
| AT5G14590 | Isocitrate/isopropylmalate dehydrogenase family protein | 0.05 | −3.18 |
| AT1G16720 | High chlorophyll fluorescence phenotype 173 | 0.03 | −3.01 |
| AT5G64050 | Glutamate tRNA synthetase | 0.01 | −3.01 |
| AT3G23810 | S-adenosyl-l-homocysteine hydrolase 2 | 0.00 | −2.96 |
| AT1G79720 | Eukaryotic aspartyl protease family protein | 0.03 | −2.94 |
| AT2G35040 | AICARFT/IMPCHase bienzyme family protein | 0.00 | −2.80 |
| AT1G71720 | Nucleic acid-binding proteins superfamily | 0.02 | −2.77 |
| AT3G46740 | Translocon at the outer envelope of chloroplasts 75-III | 0.04 | −2.71 |
| AT4G02930 | GTP binding elongation factor Tu family protein | 0.00 | −2.70 |
| AT3G13750 | Beta galactosidase 1 | 0.04 | −2.42 |
| AT3G18190 | TCP-1/cpn60 chaperonin family protein | 0.03 | −2.41 |
| AT3G07810 | RNA-binding (RRM/RBD/RNP motifs) family protein | 0.04 | 2.40 |
| AT1G30530 | UDP-glucosyl transferase 78D1 | 0.02 | −2.35 |
| AT5G60600 | 4-hydroxy-3-methylbut-2-enyl diphosphate synthase | 0.03 | −2.28 |
| AT1G67700 | Unknown protein | 0.04 | −2.22 |
| Locus ID | Protein Name | WT Mock | WT Primed | npr1-1 Mock | npr1-1 Primed |
|---|---|---|---|---|---|
| AT1G66250 | O-Glycosyl hydrolases family 17 protein | ↑ | ↑ | ↑ | ↑ |
| AT5G48540 | Receptor-like protein kinase-related | ↑ | ↑ | ↑ | ↑ |
| AT5G52310 | Low-temperature-responsive protein 78 | ↑ | ↑ | ↑ | ↑ |
| AT3G44860 | Farnesoic acid carboxyl-O-methyltransferase | ↑ | ↑ | ↑ | ↑ |
| AT5G03630 | pyridine nucleotide-disulphide oxidoreductase | ↑ | ↑ | ↑ | ↑ |
| AT5G37360 | Unknown protein | ↑ | ↑ | ↑ | ↑ |
| AT5G14040 | Phosphate transporter 3;1 | ↑ | ↑ | ↑ | ↑ |
| ATCG00020 | Photosystem II reaction center protein A | ↑ | ↑ | ↑ | ↑ |
| AT5G36700 | 2-phosphoglycolate phosphatase 1 | ↑ | ↑ | ↑ | ↑ |
| AT2G34930 | Disease resistance / LRR family protein | ↑ | ↑ | ↑ | ↑ |
| AT1G53280 | Class I glutamine amidotransferase-like | ↑ | ↑ | ↑ | ↑ |
| AT2G21170 | Triosephosphate isomerase | ↑ | ↑ | ↑ | ↑ |
| AT3G12780 | Phosphoglycerate kinase 1 | ↑ | ↑ | ↑ | ↑ |
| AT5G19760 | Mitochondrial substrate carrier | ↑ | ↑ | ↑ | ↑ |
| AT2G38270 | CAX-interacting protein 2 (CXIP2) | ↑ | ↑ | ↓ | ↓ |
| AT5G58250 | Unknown protein | ↓ | ↓ | ↓ | ↓ |
| AT1G15820 | Light harvesting complex PS II subunit 6 | ↓ | ↓ | ↓ | ↓ |
| AT4G25050 | Acyl carrier protein 4 | ↓ | ↓ | ↓ | ↓ |
| AT5G42980 | Thioredoxin 3 | ↓ | ↓ | ↓ | ↓ |
| AT1G08520 | ALBINA 1 | ↓ | ↓ | ↓ | ↓ |
| AT1G35680 | Ribosomal protein L21 | ↓ | ↓ | ↓ | ↓ |
| Locus ID | Protein Name | p-Value | Log2FC |
|---|---|---|---|
| AT4G12830 | Alpha/beta-Hydrolases (ABH) superfamily protein | 0.01 | −6.47 |
| AT3G09630 | Ribosomal protein L4/L1 family | 0.01 | 6.07 |
| AT5G54600 | Translation protein SH3-like family protein | 0.02 | 5.72 |
| AT2G39460 | Ribosomal protein L23AA | 0.03 | 5.08 |
| AT3G49010 | 60S ribosomal protein L13 | 0.02 | 5.00 |
| AT1G20450 | Dehydrin family protein | 0.04 | 4.57 |
| AT1G74970 | Ribosomal protein S9 | 0.03 | 4.50 |
| AT5G07090 | Ribosomal protein S4 (RPS4A) family protein | 0.03 | 4.28 |
| AT1G23290 | Ribosomal protein L18e/L15 superfamily protein | 0.01 | 4.24 |
| AT4G31700 | Ribosomal protein S6 | 0.03 | 4.02 |
| AT3G60770 | Ribosomal protein S13/S15 | 0.02 | 3.95 |
| AT4G35090 | Catalase 2 | 0.02 | 3.94 |
| AT2G40510 | Ribosomal protein S26e family protein | 0.01 | 3.80 |
| AT2G43030 | Ribosomal protein L3 family protein | 0.02 | 3.73 |
| AT3G02080 | Ribosomal protein S19e family protein | 0.01 | 3.60 |
| AT1G26910 | Ribosomal protein L16p/L10e family protein | 0.01 | 3.42 |
| AT5G15200 | Ribosomal protein S4 | 0.01 | 3.28 |
| AT4G18100 | Ribosomal protein L32e | 0.03 | 3.13 |
| AT3G58510 | DEA(D/H)-box RNA helicase family protein | 0.04 | −3.03 |
| AT3G04550 | Unknown protein | 0.01 | −2.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, Q.; David, L.; Moran, R.; Grela, I.; Ortega, A.; Scott, P.; Warnock, L.; Chen, S. Role of NPR1 in Systemic Acquired Stomatal Immunity. Plants 2023, 12, 2137. https://doi.org/10.3390/plants12112137
Guan Q, David L, Moran R, Grela I, Ortega A, Scott P, Warnock L, Chen S. Role of NPR1 in Systemic Acquired Stomatal Immunity. Plants. 2023; 12(11):2137. https://doi.org/10.3390/plants12112137
Chicago/Turabian StyleGuan, Qijie, Lisa David, Riley Moran, Ivan Grela, Angelica Ortega, Peter Scott, Lindsey Warnock, and Sixue Chen. 2023. "Role of NPR1 in Systemic Acquired Stomatal Immunity" Plants 12, no. 11: 2137. https://doi.org/10.3390/plants12112137







