Transcriptomic and Physiological Analyses of Two Rice Restorer Lines under Different Nitrogen Supplies Provide Novel Insights into Hybrid Rice Breeding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rice Materials and Growth Conditions
2.2. RNA Isolation and Transcriptomic Analysis
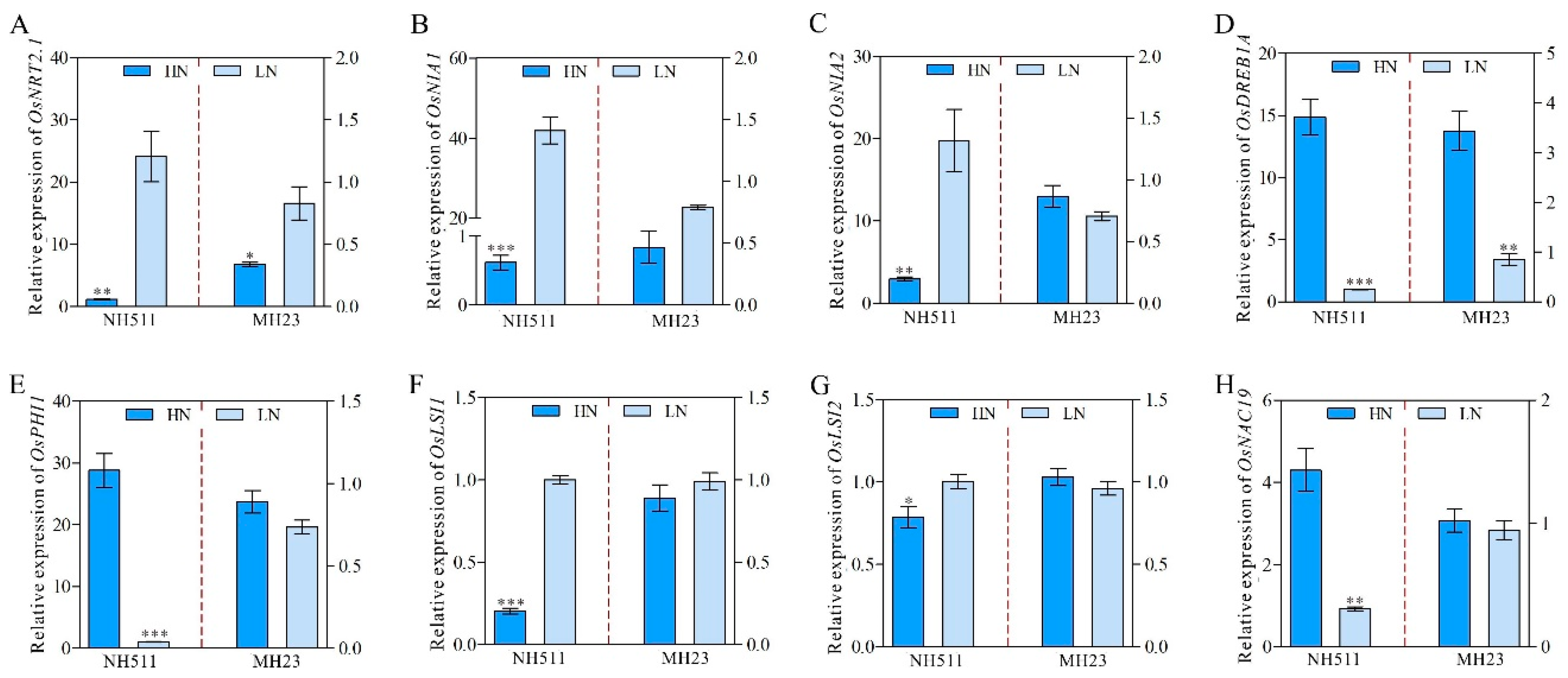
2.3. Quantitative RT-PCR
2.4. Enzyme Activity, N Content, and NUE Analysis
2.5. Chlorate Sensitivity Assay
2.6. Field Trial of Rice Cultivars
2.7. Agronomic Trait Analyses
3. Results
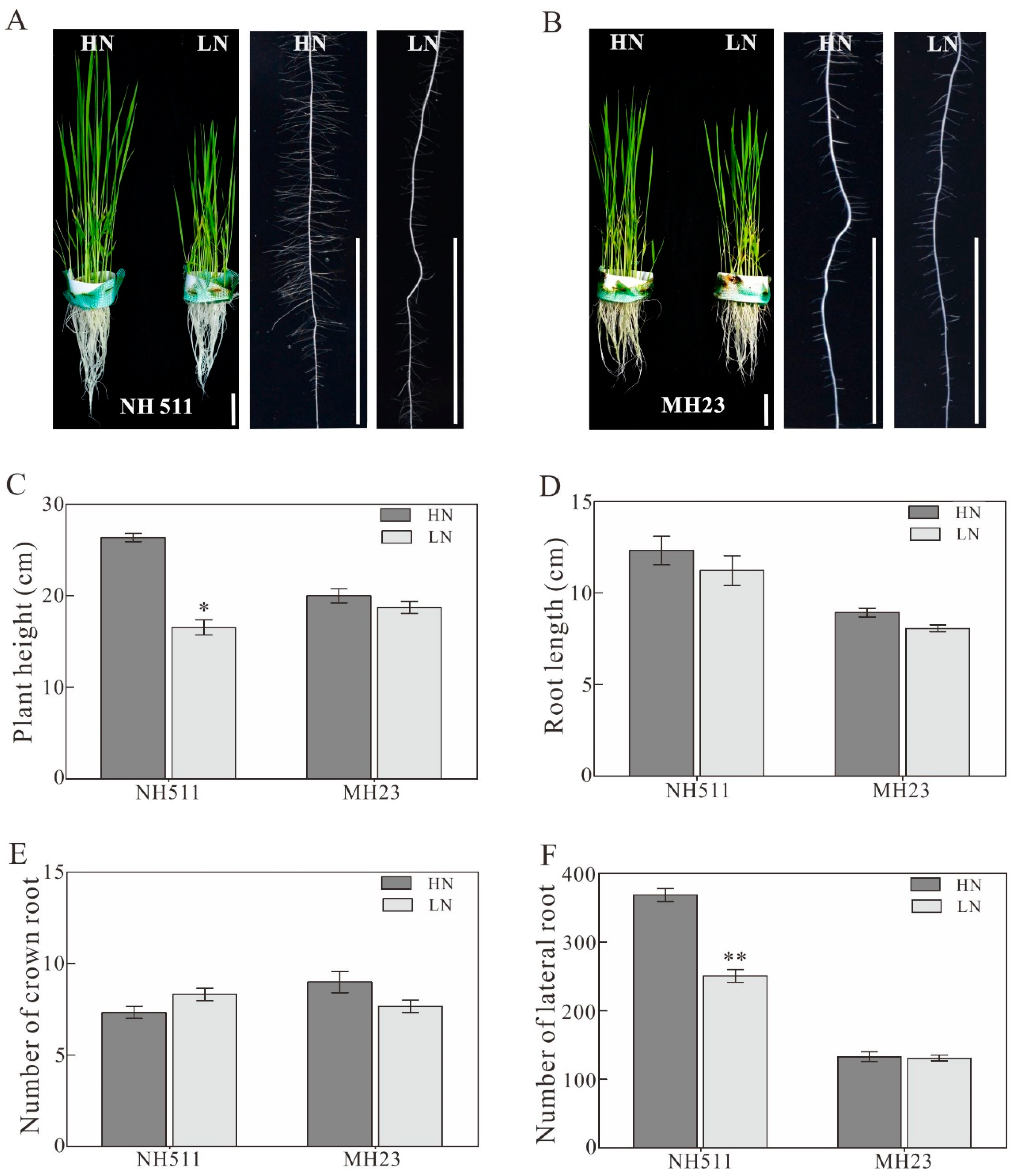
3.1. Morphological Variations in NH511 and MH23 Grown under HN and LN Conditions
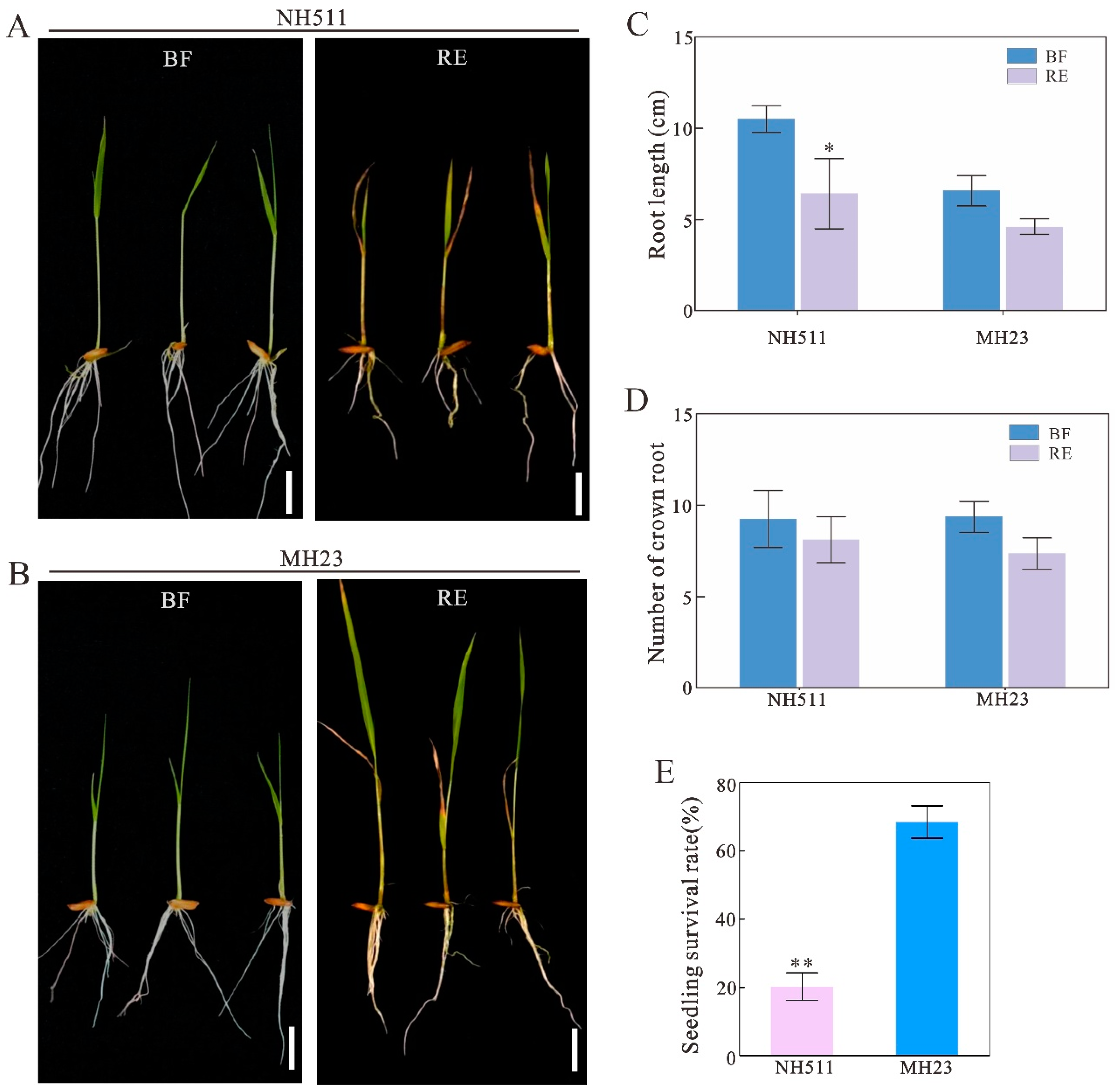
3.2. NH511 Enhances NUE Based on Chlorate Assays
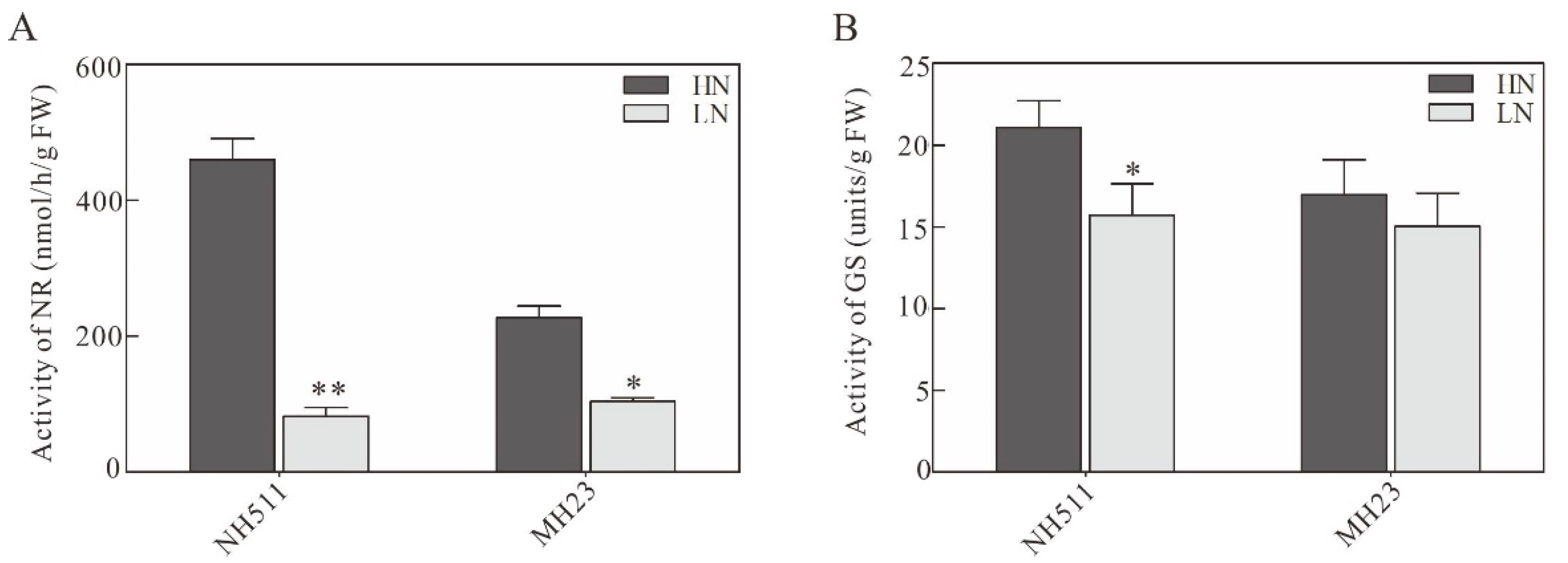
3.3. Effect of Different N Supplies on N-Metabolizing Enzymes
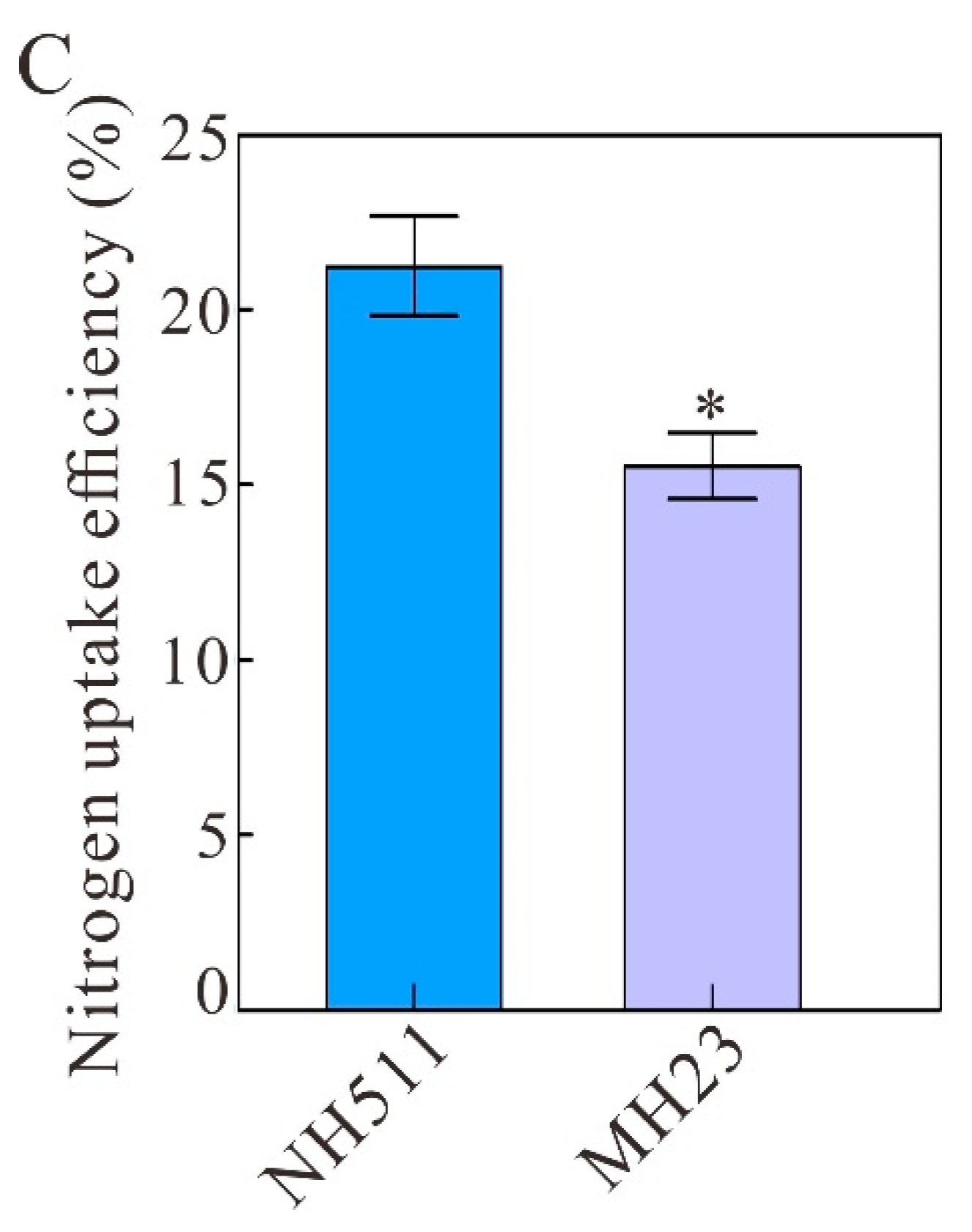
3.4. N Content and NUE under Different N-Supply Concentrations
3.5. NH511 Improves Grain Yield by Increasing the Number of Tillers under High-N Supply
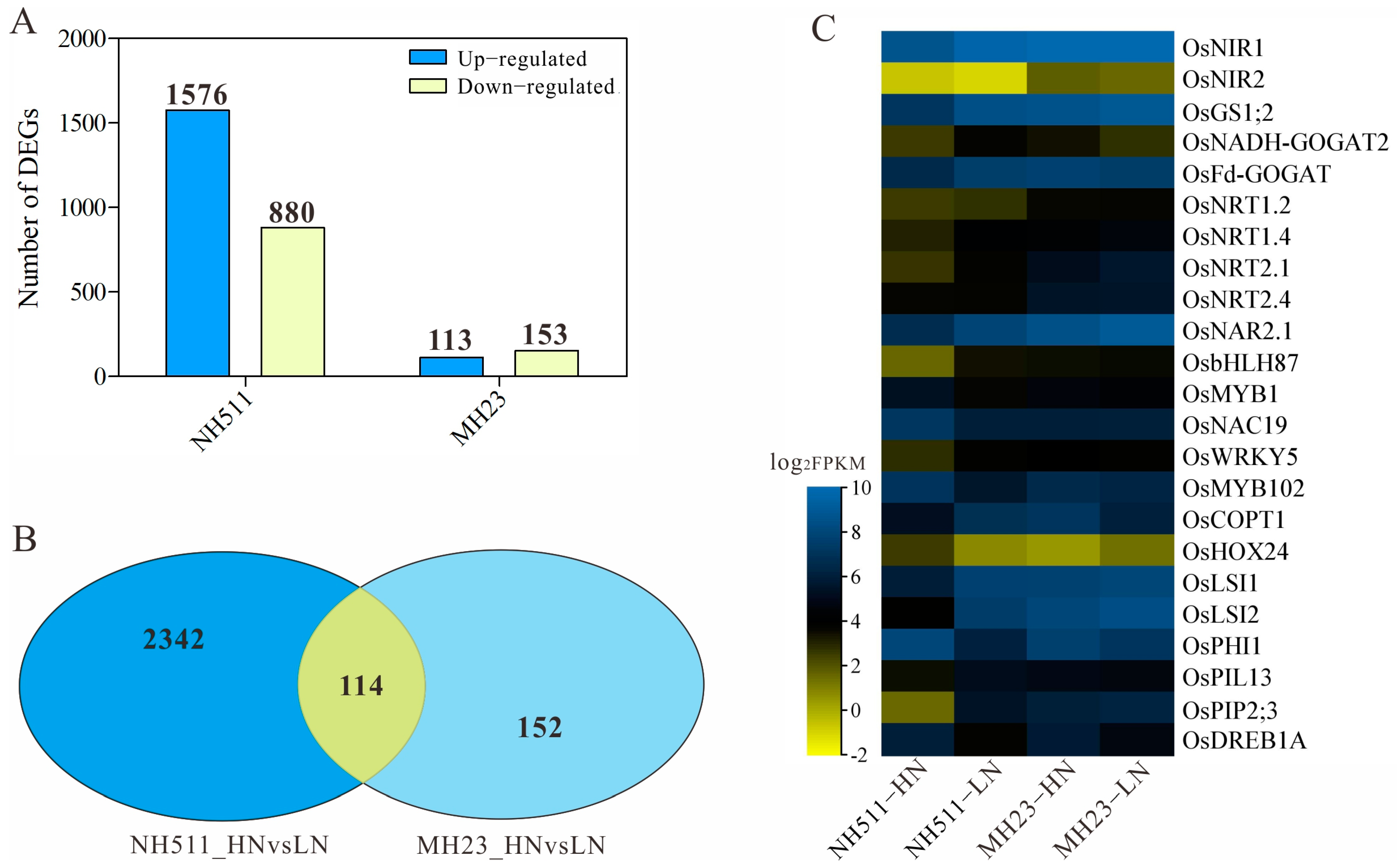
3.6. Differential Transcriptome Signature Underlies the Variation in N Metabolism between NH511 and MH23
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beukert, U.; Li, Z.; Liu, G.; Zhao, Y.; Ramachandra, N.; Mirdita, V.; Pita, F.; Pillen, K.; Reif, J.C. Genome-Based Identification of Heterotic Patterns in Rice. Rice 2017, 10, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Li, Z.; Liu, G.; Jiang, Y.; Maurer, H.P.; Wurschum, T.; Mock, H.P.; Matros, A.; Ebmeyer, E.; Schachschneider, R.; et al. Genome-based establishment of a high-yielding heterotic pattern for hybrid wheat breeding. Proc. Natl. Acad. Sci. USA 2015, 112, 15624–15629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, J.; Zhang, G.; Datta, K.; Xu, C.; He, Y.; Zhang, Q.; Khush, G.S.; Datta, S.K. Field performance of transgenic elite commercial hybrid rice expressing bacillus thuringiensis delta-endotoxin. Nat. Biotechnol. 2000, 18, 1101–1104. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.H.; Zhuang, J.Y.; Fan, Y.Y.; Du, J.H.; Cao, L.Y. Progress in research and development on hybrid rice: A super-domesticate in China. Ann. Bot. 2007, 100, 959–966. [Google Scholar] [CrossRef]
- Kim, Y.J.; Zhang, D. Molecular Control of Male Fertility for Crop Hybrid Breeding. Trends Plant. Sci. 2018, 23, 53–65. [Google Scholar] [CrossRef]
- Hussain, I.; Ali, S.; Liu, W.; Awais, M.; Li, J.; Liao, Y.; Zhu, M.; Fu, C.; Liu, D.; Wang, F. Identification of Heterotic Groups and Patterns Based on Genotypic and Phenotypic Characteristics among Rice Accessions of Diverse Origins. Front. Genet. 2022, 13, 811124. [Google Scholar] [CrossRef]
- Ouyang, Y.; Li, X.; Zhang, Q. Understanding the genetic and molecular constitutions of heterosis for developing hybrid rice. J. Genet. Genomics 2022, 49, 385–393. [Google Scholar] [CrossRef]
- Duan, D.; Zhang, H. A single SNP in NRT1.1B has a major impact on nitrogen use efficiency in rice. Sci. China Life Sci. 2015, 58, 827–828. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Wang, W.; Chen, J.; Liu, Y.; Chu, C. Genetic improvement toward nitrogen-use efficiency in rice: Lessons and perspectives. Mol. Plant. 2023, 16, 64–74. [Google Scholar] [CrossRef]
- Ahrens, T.D.; Lobell, D.B.; Ortiz-Monasterio, J.I.; Li, Y.; Matson, P.A. Narrowing the agronomic yield gap with improved nitrogen use efficiency: A modeling approach. Ecol. Appl. 2010, 20, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Dobermann, A.; Cassman, K.G. Cereal area and nitrogen use efficiency are drivers of future nitrogen fertilizer consumption. Sci. China C Life Sci. 2005, 48, 745–758. [Google Scholar] [PubMed]
- Zhu, S.; Liu, L.; Xu, Y.; Yang, Y.; Shi, R. Application of controlled release urea improved grain yield and nitrogen use efficiency: A meta-analysis. PLoS ONE 2020, 15, e0241481. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Yu, M.; Li, Z.; Ai, Z.; Chen, J. Molecular Regulatory Networks for Improving Nitrogen Use Efficiency in Rice. Int. J. Mol. Sci. 2021, 22, 9040. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, M.; Cao, G.; Han, L. Effects of genetic background on expression of QTL for nitrogen efficiency in irrigated rice and upland rice. Sci. Agric. Sin. 2010, 43, 4331–4340. [Google Scholar]
- Wei, D.; Cui, K.; Ye, G.; Pan, J.; Xiang, J.; Huang, J.; Nie, L. QTL mapping for nitrogen-use efficiency and nitrogen-deficiency tolerance traits in rice. Plant. Soil. 2012, 359, 281–295. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, Y.; Tang, D.; Wang, J.; Zhong, J.; Wang, Y.; Yuan, Q.; Yu, X.; Zhang, Y.; Wang, Y.; et al. Identification of QTL associated with nitrogen uptake and nitrogen use efficiency using high throughput genotyped CSSLs in rice (Oryza sativa L.). Front. Plant. Sci. 2017, 8, 1166. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Wang, W.; Ou, S.; Tang, J.; Li, H.; Che, R.; Zhang, Z.; Chai, X.; Wang, H.; Wang, Y.; et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015, 47, 834–838. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Jiang, Z.; Wang, W.; Xu, R.; Wang, Q.; Zhang, Z.; Li, A.; Liang, Y.; Ou, S.; et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 2021, 590, 600–605. [Google Scholar] [CrossRef]
- Tang, W.; Ye, J.; Yao, X.; Zhao, P.; Xuan, W.; Tian, Y.; Zhang, Y.; Xu, S.; An, H.; Chen, G.; et al. Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 2019, 10, 5279. [Google Scholar] [CrossRef] [Green Version]
- Guo, N.; Hu, J.; Yan, M.; Qu, H.; Luo, L.; Tegeder, M.; Xu, G. Oryza sativa Lysine-Histidine-type Transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. Plant. J. 2020, 103, 395–411. [Google Scholar] [CrossRef]
- Wang, Q.; Nian, J.; Xie, X.; Yu, H.; Zhang, J.; Bai, J.; Dong, G.; Hu, J.; Bai, B.; Chen, L.; et al. Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice. Nat. Commun. 2018, 9, 735. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Ali, J.; Zhou, S.; Ren, G.; Xie, H.; Xu, J.; Yu, X.; Zhou, F.; Peng, S.; Ma, L.; et al. From Green Super Rice to green agriculture: Reaping the promise of functional genomics research. Mol. Plant. 2022, 15, 9–26. [Google Scholar] [CrossRef]
- Zhang, Q. Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. USA 2007, 104, 16402–16409. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.Y.; Jeong, J.S.; Lim, J.Y.; Kim, T.; Park, J.H.; Kim, J.K.; Shin, C. Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genomics 2018, 19, 532. [Google Scholar] [CrossRef] [PubMed]
- Mandal, V.K.; Jangam, A.P.; Chakraborty, N.; Raghuram, N.; Jagadhesan, B.; Sathee, L.; Meena, H.S.; Jha, S.K.; Chinnusamy, V.; Kumar, A.; et al. Nitrate-responsive transcriptome analysis reveals additional genes/processes and associated traits viz. height, tillering, heading date, stomatal density and yield in japonica rice. Planta 2022, 255, 42. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.M.; Kant, S.; Clarke, J.; Gidda, S.; Ming, F.; Xu, J.; Rochon, A.; Shelp, B.J.; Hao, L.; Zhao, R.; et al. Increased nitrogen-use efficiency in transgenic rice plants over-expressing a nitrogen-responsive early nodulin gene identified from rice expression profiling. Plant. Cell. Environ. 2009, 32, 1749–1760. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Lu, Y.; Xie, W.; Zhu, T.; Lian, X. Transcriptome response to nitrogen starvation in rice. J. Biosci. 2012, 37, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Sevanthi, V.A.; Chaudhary, S.; Tyagi, P.; Venkadesan, S.; Rani, M.; Mandal, P.K. Transcriptome Analysis of Two Rice Varieties Contrasting for Nitrogen Use Efficiency under Chronic N Starvation Reveals Differences in Chloroplast and Starch Metabolism-Related Genes. Genes 2018, 9, 206. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Yoon, J.; Choi, H.; Fan, Y.; Chen, R.; An, G. Transcriptome analysis of nitrogen-starvation-responsive genes in rice. BMC Plant. Biol. 2015, 15, 31. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Yan, H.H.; Yang, Y.Q.; Liang, Y.S.; Nan, W.B.; Zhang, H.M.; Qin, X.J. Screening and research of different rice (Oryza sativa) varieties based on nitrate absorption and utilization in seedlings. Plant. Physiol. J. 2016, 52, 1941–1949. [Google Scholar] [CrossRef]
- Ferrario-Mery, S.; Valadier, M.H.; Foyer, C.H. Overexpression of nitrate reductase in tobacco delays drought-induced decreases in nitrate reductase activity and mRNA. Plant. Physiol. 1998, 117, 293–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, C.; Chen, K.; Cui, Y.; Chen, J.; Mi, X.; Zhu, S.; Zhu, Y.; Ali, J.; Ye, G.; Li, Z.; et al. QTL Mapping and Favorable Allele Mining of Nitrogen Deficiency Tolerance Using an Interconnected Breeding Population in Rice. Front. Genet. 2021, 12, 616428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.S.; Xia, J.Q.; Alfatih, A.; Song, Y.; Huang, Y.J.; Sun, L.Q.; Wan, G.Y.; Wang, S.M.; Wang, Y.P.; Hu, B.H.; et al. Rice NIN-LIKE PROTEIN 3 modulates nitrogen use efficiency and grain yield under nitrate-sufficient conditions. Plant. Cell. Environ. 2022, 45, 1520–1536. [Google Scholar] [CrossRef]
- Hakeem, K.R.; Ahmad, A.; Iqbal, M.; Gucel, S.; Ozturk, M.; Neeraja, C.N.; Barbadikar, K.M.; Krishnakanth, T.; Bej, S.; Rao, I.S.; et al. Nitrogen-efficient rice cultivars can reduce nitrate pollution Down regulation of transcripts involved in selective metabolic pathways as an acclimation strategy in nitrogen use efficient genotypes of rice under low nitrogen. Environ. Sci. Pollut. Res. Int. 2011, 18, 1184–1193. [Google Scholar] [CrossRef]
- Kabange, N.R.; Park, S.Y.; Lee, J.Y.; Shin, D.; Lee, S.M.; Kwon, Y.; Cha, J.K.; Cho, J.H.; Duyen, D.V.; Ko, J.M.; et al. New Insights into the Transcriptional Regulation of Genes Involved in the Nitrogen Use Efficiency under Potassium Chlorate in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2021, 22, 583785. [Google Scholar] [CrossRef]
- Wang, J.; Song, K.; Sun, L.; Qin, Q.; Sun, Y.; Pan, J.; Xue, Y. Morphological and Transcriptome Analysis of Wheat Seedlings Response to Low Nitrogen Stress. Plants 2019, 8, 369. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.L.; Han, W.X.; Zhang, W.F.; Christie, P.; Goulding, K.W.; Vitousek, P.M.; Zhang, F.S. Significant acidification in major Chinese croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Deng, Y.; Ding, Y.; Guo, J.; Qiu, J.; Wang, B.; Wang, C.; Xie, Y.; Zhang, Z.; Chen, J.; et al. Rice functional genomics: Decades’ efforts and roads ahead. Sci. China Life Sci. 2022, 65, 33–92. [Google Scholar] [CrossRef]
- Han, B.; Xue, Y.; Li, J.; Deng, X.W.; Zhang, Q. Rice functional genomics research in China. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 1009–1021. [Google Scholar] [CrossRef]
- Li, P.; Chen, Y.H.; Lu, J.; Zhang, C.Q.; Liu, Q.Q.; Li, Q.F. Genes and Their Molecular Functions Determining Seed Structure, Components, and Quality of Rice. Rice 2022, 15, 18. [Google Scholar] [CrossRef]
- Tian, X.H.; Tsutomu, M.; Li, S.H.; Lin, J.C. High temperature stress on rice anthesis: Research progress and prospects. Ying Yong Sheng Tai Xue Bao 2007, 18, 2632–2636. (In Chinese) [Google Scholar] [PubMed]
- ElShamey, E.A.Z.; Sakran, R.M.; ElSayed, M.A.A.; Aloufi, S.; Alharthi, B.; Alqurashi, M.; Mansour, E.; Abd El-Moneim, D. Heterosis and combining ability for floral and yield characters in rice using cytoplasmic male sterility system. Saudi J. Biol. Sci. 2022, 29, 3727–3738. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lu, Q.; Qiu, S.; Yu, H.; Wang, Z.; Yu, Z.; Lu, Y.; Wang, L.; Xia, F.; Wu, Y.; et al. Fujian cytoplasmic male sterility and the fertility restorer gene OsRf19 provide a promising breeding system for hybrid rice. Proc. Natl. Acad. Sci. USA 2022, 119, e2208759119. [Google Scholar] [CrossRef] [PubMed]
- Jie, R.S.; Liu, F.P.; Yang, C.H.; He, P.; Mao, J.X. Breeding of New Rice Restorer Line Nanhui 511 with High Combining Ability. Hybrid Rice 2005, 20, 15–16. [Google Scholar]
- Luo, L.G.; Xu, J.F.; Zhai, H.Q.; Wan, J.M. Analysis of photoperiod-sensitivity genes in Minghui63, an restorer line of indica rice (Oryza sativa L.). Yi Chuan Xue Bao 2003, 30, 804–810. (In Chinese) [Google Scholar] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, X.; Li, X.; Xiao, J.; Wu, Q.; Li, Y.; Li, C.; Jiang, D.; Tang, T.; Nan, W.; Liang, Y.; et al. Transcriptomic and Physiological Analyses of Two Rice Restorer Lines under Different Nitrogen Supplies Provide Novel Insights into Hybrid Rice Breeding. Plants 2023, 12, 2276. https://doi.org/10.3390/plants12122276
Qin X, Li X, Xiao J, Wu Q, Li Y, Li C, Jiang D, Tang T, Nan W, Liang Y, et al. Transcriptomic and Physiological Analyses of Two Rice Restorer Lines under Different Nitrogen Supplies Provide Novel Insights into Hybrid Rice Breeding. Plants. 2023; 12(12):2276. https://doi.org/10.3390/plants12122276
Chicago/Turabian StyleQin, Xiaojian, Xiaowei Li, Juan Xiao, Qian Wu, Yuntong Li, Cuiping Li, Dan Jiang, Tingting Tang, Wenbin Nan, Yongshu Liang, and et al. 2023. "Transcriptomic and Physiological Analyses of Two Rice Restorer Lines under Different Nitrogen Supplies Provide Novel Insights into Hybrid Rice Breeding" Plants 12, no. 12: 2276. https://doi.org/10.3390/plants12122276
APA StyleQin, X., Li, X., Xiao, J., Wu, Q., Li, Y., Li, C., Jiang, D., Tang, T., Nan, W., Liang, Y., & Zhang, H. (2023). Transcriptomic and Physiological Analyses of Two Rice Restorer Lines under Different Nitrogen Supplies Provide Novel Insights into Hybrid Rice Breeding. Plants, 12(12), 2276. https://doi.org/10.3390/plants12122276




