Enhancing Crop Resilience to Drought Stress through CRISPR-Cas9 Genome Editing
Abstract
:1. Introduction
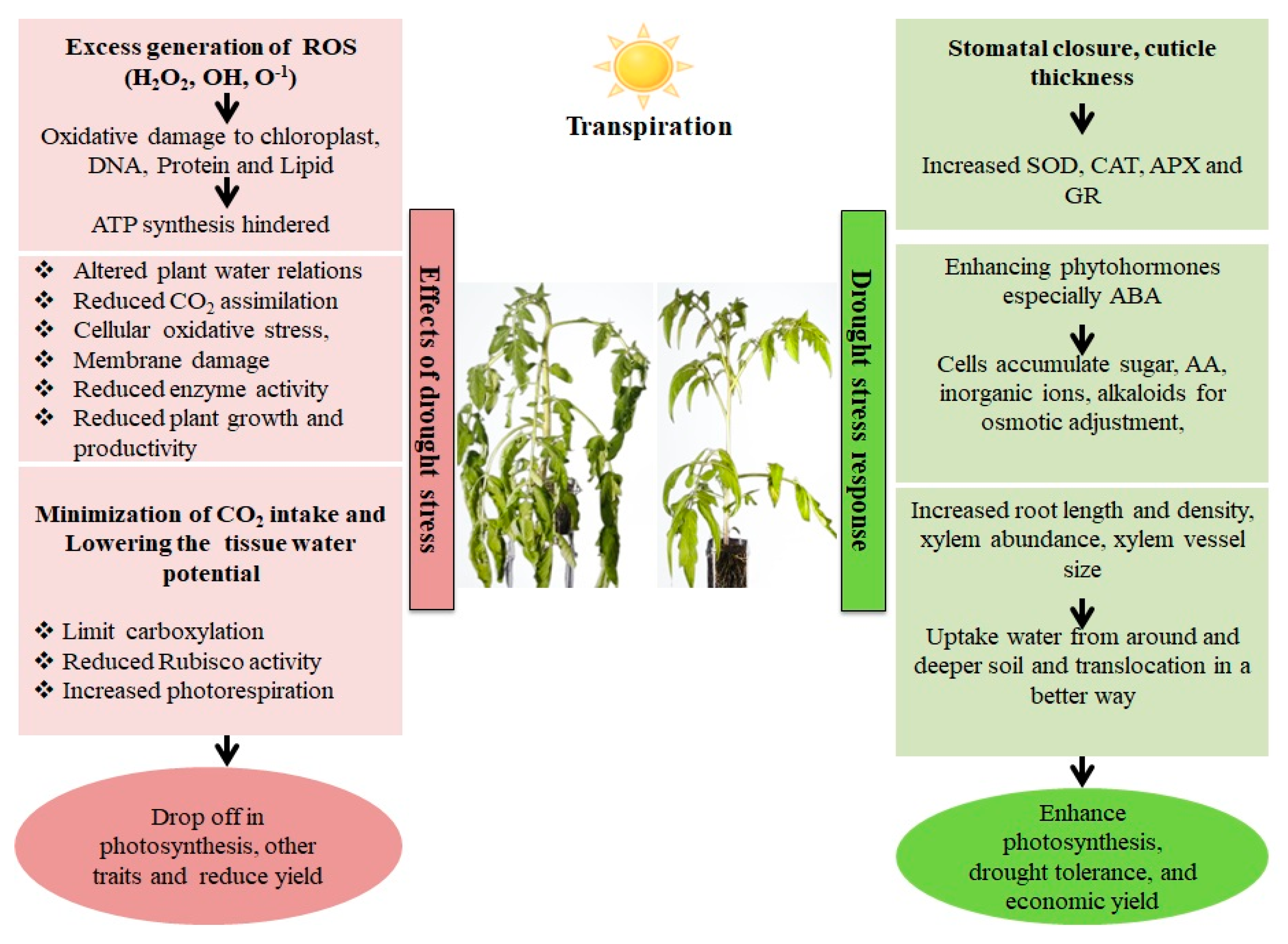
2. Detrimental Consequences of Drought Conditions on Plant
3. Molecular Aspects Underlying Resilience of Crops against Drought
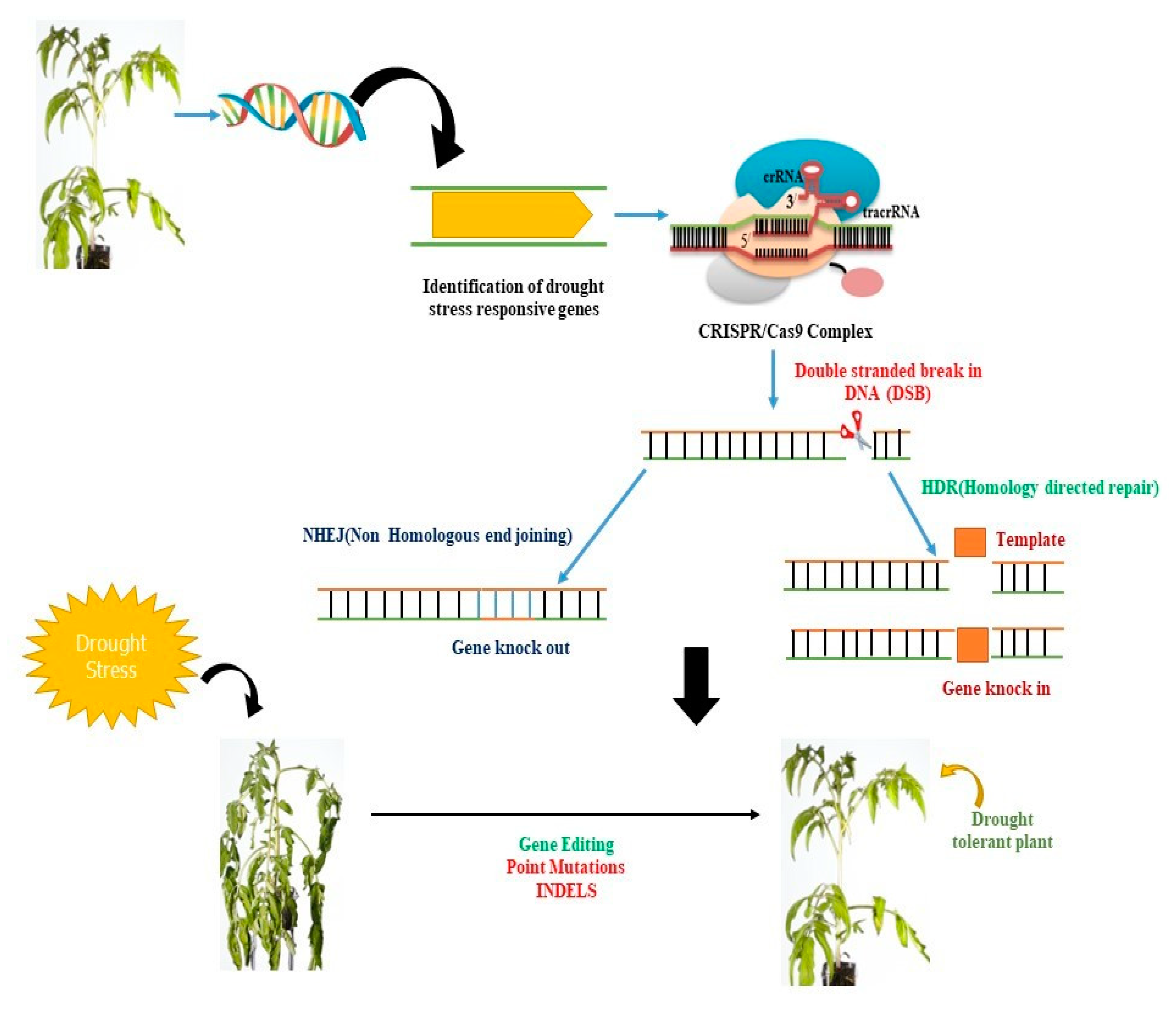
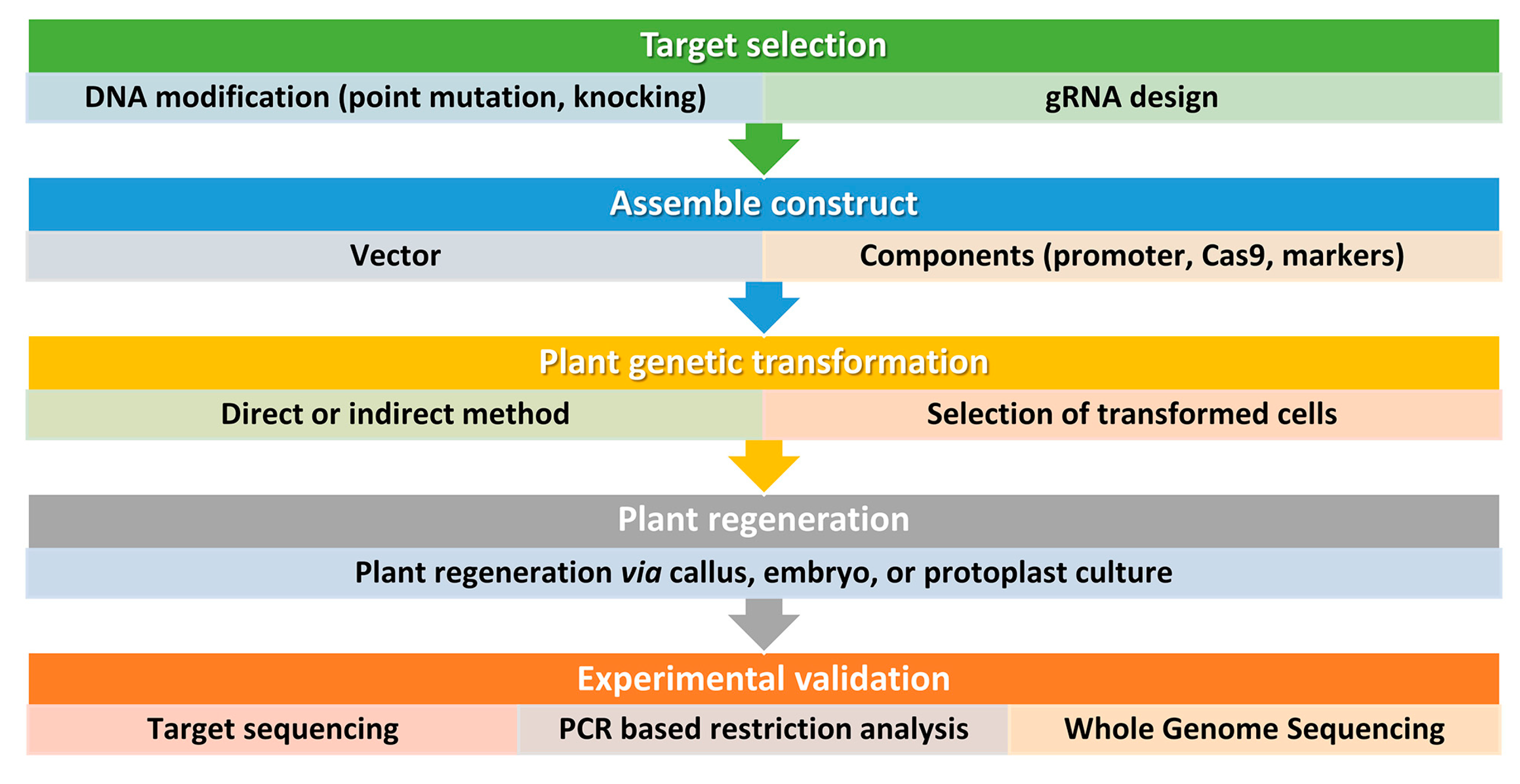
4. CRISPR/Cas9-Based Precise Genome Editing for Crop Drought Resilience
5. CRISPR and Crop Productivity in Drought Resilient Crops
6. Implications of CRISPR-Cas9 Promoting Drought Stress Tolerance by Modulating Ethylene Responsive Factors (ERFs)
7. Examples of CRISPR/Cas9 Modified Crops for Tolerance against Different Abiotic Stresses
8. Conclusions and Outlook
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liang, X.Z.; Wu, Y.; Chambers, R.G.; Schmoldt, D.L.; Gao, W.; Liu, C.; Liu, Y.A.; Sun, C.; Kennedy, J.A. Determining climate effects on US total agricultural productivity. Proc. Nat. Acad. Sci. USA 2017, 114, 2285–2292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, W.A.; Atri, N.; Ahmad, J.; Qureshi, M.I.; Singh, B.; Kumar, R.; Rai, V.; Pandey, S. Drought mediated physiological and molecular changes in muskmelon (Cucumis melo L.). PloS ONE 2019, 14, e0222647. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Ramegowda, V.; Senthil-Kumar, M. Shared and unique responses of plants to multiple individual stresses and stress combinations: Physiological and molecular mechanisms. Front. Plant Sci. 2015, 6, 723. [Google Scholar] [CrossRef] [Green Version]
- Troy, T.J.; Kipgen, C.; Pal, I. The impact of climate extremes and irrigation on US crop yields. Environ. Res. Lett. 2015, 10, 054013. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S.; Ansari, W.A.; Atri, N.; Singh, B.; Gupta, S.; Bhat, K.V. Standardization of screening technique and evaluation of muskmelon genotypes for drought tolerance. Plant Gen. Resour. 2018, 16, 1–8. [Google Scholar] [CrossRef]
- Zipper, S.C.; Qiu, J.; Kucharik, C.J. Drought effects on US maize and soybean production: Spatiotemporal patterns and historical changes. Environ. Res. Lett. 2016, 11, 094021. [Google Scholar] [CrossRef]
- Mushtaq, M.; Ahmad Dar, A.; Skalicky, M.; Tyagi, A.; Bhagat, N.; Basu, U.; Bhat, B.A.; Zaid, A.; Ali, S.; Dar, T.U.H.; et al. CRISPR-based genome editing tools: Insights into technological breakthroughs and future challenges. Genes 2021, 12, 797. [Google Scholar] [CrossRef]
- Mushtaq, M.; Dar, A.A.; Basu, U.; Bhat, B.A.; Mir, R.A.; Vats, S.; Dar, M.S.; Tyagi, A.; Ali, S.; Bansal, M.; et al. Integrating CRISPR-Cas and next generation sequencing in plant virology. Front. Genet. 2021, 12. [Google Scholar] [CrossRef]
- Ansari, W.A.; Atri, N.; Singh, B.; Pandey, S. Changes in antioxidant enzyme activities and gene expression in two muskmelon genotypes under progressive water stress. Biologia Plantarum. 2017, 61, 333–341. [Google Scholar] [CrossRef]
- Rai, G.K.; Parveen, A.; Jamwal, G.; Basu, U.; Kumar, R.R.; Rai, P.K.; Sharma, J.P.; Alalawy, A.I.; Al-Duais, M.A.; Hossain, M.A.; et al. Leaf Proteome Response to Drought Stress and Antioxidant Potential in Tomato (Solanum lycopersicum L.). Atmosphere 2021, 12, 1021. [Google Scholar] [CrossRef]
- Zlatev, Z.; Lidon, F.C. An overview on drought induced changes in plant growth, water relations and photosynthesis. Emirates J. Food Agric. 2012, 57–72. [Google Scholar]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.S.P. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, W.A.; Atri, N.; Singh, B.; Kumar, P.; Pandey, S. Morpho-physiological and biochemical responses of muskmelon genotypes to different degree of water deficit. Photosynthetica 2018, 56, 1019–1030. [Google Scholar] [CrossRef]
- Sharma, A.; Zheng, B. Melatonin mediated regulation of drought stress: Physiological and molecular aspects. Plants 2019, 8, 190. [Google Scholar] [CrossRef] [Green Version]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, R.M.; Rao, N.S. Influence of pod load on response of okra to water stress. Indian J. Plant Physiol. 2005, 10, 54. [Google Scholar]
- Razmjoo, K.; Heydarizadeh, P.; Sabzalian, M.R. Effect of salinity and drought stresses on growth parameters and essential oil content of Matricaria chamomile. Int. J. Agric. Biol. 2008, 10, 451–454. [Google Scholar]
- Hoang, T.M.L.; Tran, T.N.; Nguyen, T.K.T.; Williams, B.; Wurm, P.; Bellairs, S.; Mundree, S. Improvement of salinity stress tolerance in rice: Challenges and opportunities. Agronomy 2016, 6, 54. [Google Scholar] [CrossRef] [Green Version]
- Ansari, W.A.; Chandanshive, S.U.; Bhatt, V.; Nadaf, A.B.; Vats, S.; Katara, J.L.; Sonah, H.; Deshmukh, R. Genome editing in cereals: Approaches, applications and challenges. Int. J. Mol. Sci. 2020, 21, 4040. [Google Scholar] [CrossRef] [PubMed]
- Ansari, W.A.; Atri, N.; Pandey, M.; Singh, A.K.; Singh, B.; Pandey, S. Influence of drought stress on morphological, physiological and biochemical attributes of plants: A review. Biosci. Biotechnol. Res. Asia. 2019, 16, 697–709. [Google Scholar] [CrossRef]
- Sourour, A.; Afef, O.; Mounir, R.; Mongi, B.Y. A review: Morphological, physiological, biochemical and molecular plant responses to water deficit stress. Int. J. Eng. Sci. 2017, 6, 1–4. [Google Scholar] [CrossRef]
- Queiroz, M.S.; Oliveira, C.E.; Steiner, F.; Zuffo, A.M.; Zoz, T.; Vendruscolo, E.P.; Silva, M.V.; Mello, B.F.F.R.; Cabra, R.C.; Menis, F.T. Drought stresses on seed germination and early growth of maize and sorghum. J. Agric. Sci. 2019, 11, 310–318. [Google Scholar] [CrossRef]
- Khan, A.; Pan, X.; Najeeb, U.; Tan, D.K.Y.; Fahad, S.; Zahoor, R.; Luo, H. Coping with drought: Stress and adaptive mechanisms, and management through cultural and molecular alternatives in cotton as vital constituents for plant stress resilience and fitness. Biol. Res. 2018, 51. [Google Scholar] [CrossRef] [Green Version]
- Shinwari, Z.K.; Jan, S.A.; Nakashima, K.; Yamaguchi-Shinozaki, K. Genetic engineering approaches to understanding drought tolerance in plants. Plant Biotechnol. Rep. 2020, 14, 151–162. [Google Scholar] [CrossRef]
- Haque, E.; Taniguchi, H.; Hassan, M.M.; Bhowmik, P.; Karim, M.R.; Śmiech, M.; Zhao, K.; Rahman, M.; Islam, T. Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: Recent progress, prospects, and challenges. Front. Plant Sci. 2018, 9, 617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brokowski, C.; Adli, M. CRISPR ethics: Moral considerations for applications of a powerful tool. J. Mol. Biol. 2019, 431, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Charagh, S.; Razzaq, A.; Javed, R.; Khan RS, A.; Hasanuzzaman, M. Brassicaceae plants response and tolerance to drought stress: Physiological and molecular interventions. In The Plant Family Brassicaceae: Biology and Physiological Responses to Environmental Stresses; Springer: Berlin/Heidelberg, Germany, 2020; pp. 229–261. [Google Scholar]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. The Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Li, X.Y.; Liu, X.; Yao, Y.; Li, Y.H.; Liu, S.; He, C.Y.; Li, J.M.; Lin, Y.Y.; Li, L. Overexpression of Arachis hypogaea AREB1 gene enhances drought tolerance by modulating ROS scavenging and maintaining endogenous ABA content. Int. J. Mol. Sci. 2013, 14, 12827–12842. [Google Scholar] [CrossRef] [Green Version]
- Ansari, W.A.; Atri, N.; Yang, L.; Singh, B.; Pandey, S. Genetic diversity in muskmelon based on SSR markers and morphological traits under well-watered and water-deficit condition. Biocat. Agric. Biotechnol. 2020, 26, 101630. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nature Biotechnol. 2014, 31, 686–688. [Google Scholar] [CrossRef]
- Bhat, J.A.; Ali, S.; Salgotra, R.K.; Mir, Z.A.; Dutta, S.; Jadon, V.; Tyagi, A.; Mushtaq, M.; Jain, N.; Singh, P.K.; et al. Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 2016, 7, 221. [Google Scholar] [CrossRef] [Green Version]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Bendaou, N.; Bouzayen, M.; Zsögön, A.; Smouni, A.; et al. Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef] [Green Version]
- Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Melo, B.P.D.; de Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 1–9. [Google Scholar]
- Bota, J.; Medrano, H.; Flexas, J. Is photosynthesis limited by decreased Rubisco activity and RuBP content under progressive water stress. New Phytol. 2004, 162, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Yoshida, R.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc. Nat. Acad. Sci. USA 2004, 101, 17306–17311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein kinases-key regulators of plant response to abiotic stresses. Omics A J. Integr. Biol. 2011, 15, 859–872. [Google Scholar] [CrossRef] [Green Version]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [Green Version]
- Hyun, T.K. CRISPR/Cas-based genome editing to improve abiotic stress tolerance in plants. Bot. Serbica 2020, 44, 121–127. [Google Scholar] [CrossRef]
- Martignago, D.; Rico-Medina, A.; Blasco-Escámez, D.; Fontanet-Manzaneque, J.B.; Caño-Delgado, A.I. Drought resistance by engineering plant tissue-specific responses. Front. Plant Sci. 2020, 10, 1676. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Al-Sadi, A.M.; Pour-Aboughadareh, A.; Burritt, D.J.; Tran, L.S.P. Genome editing using CRISPR/Cas9–targeted mutagenesis: An opportunity for yield improvements of crop plants grown under environmental stresses. Plant Physiol. Biochem. 2018, 131, 31–36. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Liu, C.; Zhao, R.; Wang, L.; Chen, L.; Yu, W.; Zhang, S.; Sheng, J.; Shen, L. CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019, 19, 38. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Guo, L.; Wang, Z.; Zhang, X.; Zhao, B.; Guo, Y.D.; Zhang, N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020, 301, 110683. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. CRISPR/ Cas9-Induced mutagenesis of semi-rolled leaf1, 2 confers curled leaf phenotype and drought tolerance by influencing protein expression patterns and ROS scavenging in rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef] [Green Version]
- Usman, B.; Nawaz, G.; Zhao, N.; Liao, S.; Liu, Y.; Li, R. Precise editing of the OsPYL9 gene by RNA-guided Cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza sativa L.) by regulating circadian rhythm and abiotic stress responsive proteins. Int. J. Mol. Sci. 2020, 21, E7854. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogata, T.; Ishizaki, T.; Fujita, M.; Fujita, Y. CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS ONE 2020, 15. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Functional Integ. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Ma, J.; Zhang, X.; Yang, Y.; Zhou, D.; Yu, Q.; Que, Y.; Xu, L.; Guo, J. A novel non-specific lipid transfer protein gene from sugarcane (NsLTPs), obviously responded to abiotic stresses and signaling molecules of SA and MeJA. Sugar Tech. 2017, 19, 17–25. [Google Scholar] [CrossRef]
- Zhong, X.; Hong, W.; Shu, Y.; Li, J.; Liu, L.; Chen, X.; Islam, F.; Zhou, W.; Tang, G. CRISPR/Cas9 mediated gene-editing of GmHdz4 transcription factor enhances drought tolerance in soybean (Glycine max [L.] Merr.). Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, A.; Razzaq, A.; Ahmad, M.; Jenks, M.A. Water stress causes differential effects on germination indices, total soluble sugar and proline content in wheat (Triticum aestivum L.) genotypes. Afr. J. Biotechnol. 2011, 10, 14038–14045. [Google Scholar]
- Zia, R.; Nawaz, M.S.; Siddique, M.J.; Hakim, S.; Imran, A. Plant survival under drought stress: Implications, adaptive responses, and integrated rhizosphere management strategy for stress mitigation. Microbiol. Res. 2021, 242, 126626. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Kriz, A.J.; Sharp, P.A. Target specificity of the CRISPR-Cas9 system. Quant. Biol. 2014, 2, 59–70. [Google Scholar] [CrossRef] [Green Version]
- Shimatani, Z.; Fujikura, U.; Ishii, H.; Matsui, Y.; Suzuki, M.; Ueke, Y.; Taoka, K.I.; Terada, R.; Nishida, K.; Kondo, A. Inheritance of co-edited genes by CRISPR-based targeted nucleotide substitutions in rice. Plant Physiol. Biochem. 2018, 131, 78–83. [Google Scholar] [CrossRef]
- Pereira, A. Plant abiotic stress challenges from the changing environment. Front. Plant Sci. 2016, 7, 1123. [Google Scholar] [CrossRef] [Green Version]
- Sasano, Y.; Nagasawa, K.; Kaboli, S.; Sugiyama, M.; Harashima, S. CRISPR-PCS: A powerful new approach to inducing multiple chromosome splitting in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, 30278. [Google Scholar] [CrossRef] [Green Version]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Jain, M. Function genomics of abiotic stress tolerance in plants: A CRISPR approach. Front. Plant Sci. 2015, 6, 375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mushtaq, M.; Bhat, J.A.; Mir, Z.A.; Sakina, A.; Ali, S.; Singh, A.K.; Tyagi, A.; Salgotra, R.K.; Dar, A.A.; Bhat, R. CRISPR/Cas approach: A new way of looking at plant-abiotic interactions. J. Plant Physiol. 2018, 224, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Debbarma, J.; Sarki, Y.N.; Saikia, B.; Boruah, H.P.D.; Singha, D.L.; Chikkaputtaiah, C. Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR-Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: A review. Mol. Biotechnol. 2019, 61, 153–172. [Google Scholar] [CrossRef]
- Curtin, S.J.; Xiong, Y.; Michno, J.M.; Campbell, B.W.; Stec, A.O.; Čermák, T.; Starker, C.; Voytas, D.F.; Eamens, A.L.; Stupar, R.M. CRISPR/Cas9 and TALENs Generate Heritable Mutations for Genes Involved in Small RNA Processing of Glycine max and Medicago truncatula. Plant Biotech. J. 2018, 16, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.; bin, X.; et al. Identification and Characterization of GmMYB118 Responses to Drought and Salt Stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.; Lin, J.; Liu, X.; Chu, W.; Li, J.; Gao, Y.; An, K.; Song, W.; Xin, M.; Yao, Y.; et al. Histone Acetyltransferase TaHAG1 Acts as a Crucial Regulator to Strengthen Salt Tolerance of Hexaploid Wheat. Plant Physiol. 2021, 186, 1951–1969. [Google Scholar] [CrossRef]
- Wu, J.; Yan, G.; Duan, Z.; Wang, Z.; Kang, C.; Guo, L.; Liu, K.; Tu, J.; Shen, J.; Yi, B.; et al. Roles of the Brassica napus DELLA protein BnaA6. RGA, in modulating drought tolerance by interacting with the ABA signaling component BnaA10. ABF2. Front. Plant Sci. 2020, 11, 577. [Google Scholar] [CrossRef]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted Mutagenesis, Precise Gene Editing, and Site-Specific Gene Insertion in Maize Using Cas9 and Guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Zhu, X.; Liu, X.; Wu, C.; Yu, C.; Hu, G.; Chen, L.; Chen, R.; Bouzayen, M.; Zouine, M.; et al. Knockout of Auxin Response Factor SlARF4 Improves Tomato Resistance to Water Deficit. Int. J. Mol. Sci. 2021, 22, 3347. [Google Scholar] [CrossRef]
- Tran, M.T.; Doan, D.T.H.; Kim, J.; Song, Y.J.; Sung, Y.W.; Das, S.; Kim, E.J.; Son, G.H.; Kim, S.H.; van Vu, T.; et al. CRISPR/Cas9-Based Precise Excision of SlHyPRP1 Domain(s) to Obtain Salt Stress-Tolerant Tomato. Plant Cell Rep. 2021, 40, 999–1011. [Google Scholar] [CrossRef]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 Transcription Factor Regulates Heat Stress Tolerance through FERONIA Receptor-like Kinase-Mediated Reactive Oxygen Species Signaling in Tomato. Plant Cell Physiol. 2018, 59, 2239–2245. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Reduction of Tomato-Plant Chilling Tolerance by CRISPRCas9-Mediated SlCBF1 Mutagenesis. J. Agric. Food Chem. 2018, 66, 9042–9051. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Kim, E.; Park, H.; Koo, Y. Selection of the Highly Efficient SgRNA for CRISPR-Cas9 to Edit Herbicide Related Genes, PDS, ALS, and EPSPS in Tomato. Appl. Biol. Chem. 2022, 65, 13. [Google Scholar] [CrossRef]
- Jogam, P.; Sandhya, D.; Alok, A.; Peddaboina, V.; Allini, V.R.; Zhang, B. A review on CRISPR/Cas-based epigenetic regulation plants. Int. J. Biol. Macromol. 2022, 219, 1261–1271. [Google Scholar] [CrossRef]
- Kumar, M.; Prusty, M.R.; Pandey, M.K.; Singh, P.K.; Guo, B.; Varshney, R.K. Application of CRISPR-Cas9-Mediated Gene Editing for Abiotic Stress Management in Crop Plants. Front. Plant Sci. 2023, 14, 1157678. [Google Scholar] [CrossRef] [PubMed]
- Shelake, R.M.; Pramanik, D.; Kim, J.Y. In Vivo rapid investigation of CRISPR-Based base editing components in Escherichia coli (IRI-CCE): A platform for evaluating base editing tools and their components. Int. J. Mol. Sci. 2022, 23, 1145. [Google Scholar] [CrossRef]
- Jiang, Y.Y.; Chai, Y.P.; Lu, M.H.; Han, X.L.; Lin, Q.; Zhang, Y.; Zhang, Q.; Zhou, Y.; Wang, X.C.; Gao, C.; et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 2020, 21, 1–10. [Google Scholar] [CrossRef]
- Lu, Y.; Tian, Y.; Shen, R.; Yao, Q.; Wang, M.; Chen, M.; Dong, J.; Zhang, T.; Li, F.; Lei, M.; et al. Targeted, efficient sequence insertion and replacement in rice. Nature Biotechnol. 2020, 38, 1402–1407. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Anand, U.; Pal, T.; Mandal, S.; Kumar, M.; Gopalakrishnan, A.V.; Lastra, J.M.P.D.L.; Dey, A. Exploring the potential of CRISPR/Cas genome editing for vegetable crop improvement: An overview of challenges and approaches. Biotechnol. Bioeng. 2023, 120, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
| Crop | Gene of Interest | Abbreviation Key | Gene Function | Trait | Reference |
|---|---|---|---|---|---|
| Maize | ARGOS8 | - | Transcription Factor | Enhanced grain yield in filed (under stress conditions) | [43] |
| Tomato | SlNPR1 | Nonexpresser of Pathogenesis- Related Genes 1 | Transcriptional coactivator | Reduced drought tolerance in the mutants | [44] |
| SlLBD40 | Lateral Organ Boundaries Domain | Transcription Factor | Drought tolerance in the knockout lines | [45] | |
| SlMAPK3 | Mitogen-Activated Protein Kinases | Signal transduction | Reduced drought tolerance in the mutants in greenhouse conditions | [46] | |
| Rice | SAPK2 | Stress-Activated Protein Kinase 2 | Signal transduction | Mutants more sensitive to drought stress | [39] |
| OsSRL1, OsSRL2 | Semi-Rolled Leaf | Transcription Factor | Drought tolerance (higher grain filling under stress) | [47] | |
| OsPYL9 | Pyrabactin Resistance-Like | Transcription Factor | Higher yield under normal and drought conditions (in growth chamber) | [48] | |
| OsDSL | Drought and Salt Tolerance | Transcription factor | High tolerance to NaCl moderate tolerance to osmotic stress at seedling stage | [49] | |
| OsRR22 | Response Regulator | Transcriptional regulator/Signaling | Salinity tolerance at seedling stage | [50] | |
| OsERA1 | Enhanced Response to ABA | Transcriptional regulator/Signaling | Enhanced response to drought stress | [51] | |
| Wheat | TaDREB2 | Dehydration Responsive Element Binding protein 2 | Transcription factor | Enhanced drought tolerance | [52] |
| TaERF3 | Ethylene Responsive Factor 3 | Transcription factor | Enhanced drought tolerance | [52] | |
| Sugar- cane | ScNLTP | Non-specific Lipid Transfer protein | Structural gene | Alteration of MeJA-induced pathways | [53] |
| Soybean | GmHdz4 | Homeodomain- Leucine Zipper | Transcription Factor | Higher drought tolerance | [54] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, G.K.; Khanday, D.M.; Kumar, P.; Magotra, I.; Choudhary, S.M.; Kosser, R.; Kalunke, R.; Giordano, M.; Corrado, G.; Rouphael, Y.; et al. Enhancing Crop Resilience to Drought Stress through CRISPR-Cas9 Genome Editing. Plants 2023, 12, 2306. https://doi.org/10.3390/plants12122306
Rai GK, Khanday DM, Kumar P, Magotra I, Choudhary SM, Kosser R, Kalunke R, Giordano M, Corrado G, Rouphael Y, et al. Enhancing Crop Resilience to Drought Stress through CRISPR-Cas9 Genome Editing. Plants. 2023; 12(12):2306. https://doi.org/10.3390/plants12122306
Chicago/Turabian StyleRai, Gyanendra Kumar, Danish Mushtaq Khanday, Pradeep Kumar, Isha Magotra, Sadiya M. Choudhary, Rafia Kosser, Raviraj Kalunke, Maria Giordano, Giandomenico Corrado, Youssef Rouphael, and et al. 2023. "Enhancing Crop Resilience to Drought Stress through CRISPR-Cas9 Genome Editing" Plants 12, no. 12: 2306. https://doi.org/10.3390/plants12122306
APA StyleRai, G. K., Khanday, D. M., Kumar, P., Magotra, I., Choudhary, S. M., Kosser, R., Kalunke, R., Giordano, M., Corrado, G., Rouphael, Y., & Pandey, S. (2023). Enhancing Crop Resilience to Drought Stress through CRISPR-Cas9 Genome Editing. Plants, 12(12), 2306. https://doi.org/10.3390/plants12122306











