LeGRXS14 Reduces Salt Stress Tolerance in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
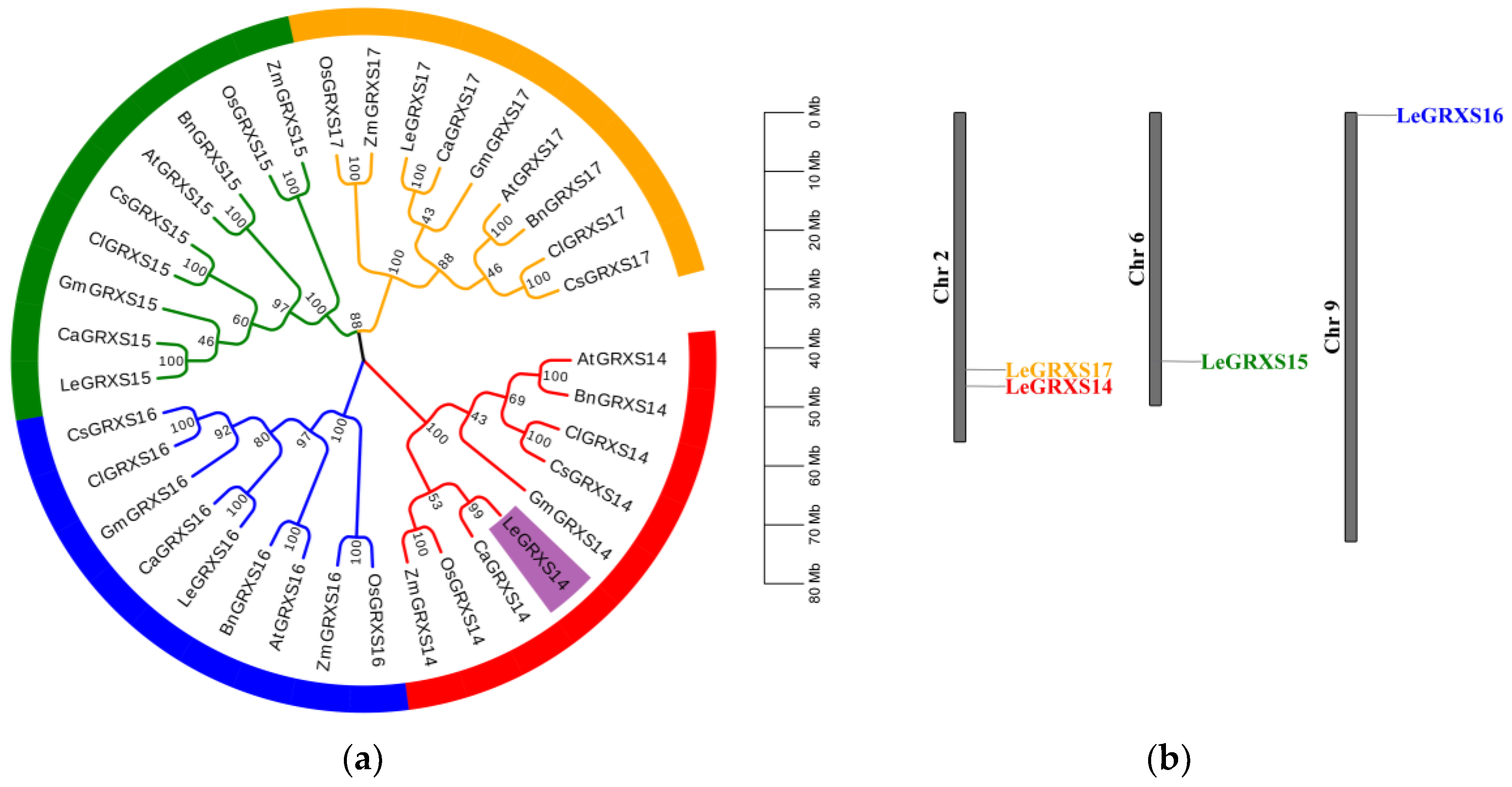
2.1. Phylogenetic Analysis and Chromosomal Distribution of CGFS-Type GRX Proteins
2.2. Multiple Sequence Alignment and Motif Analysis of GRXS14 in Tomatoes
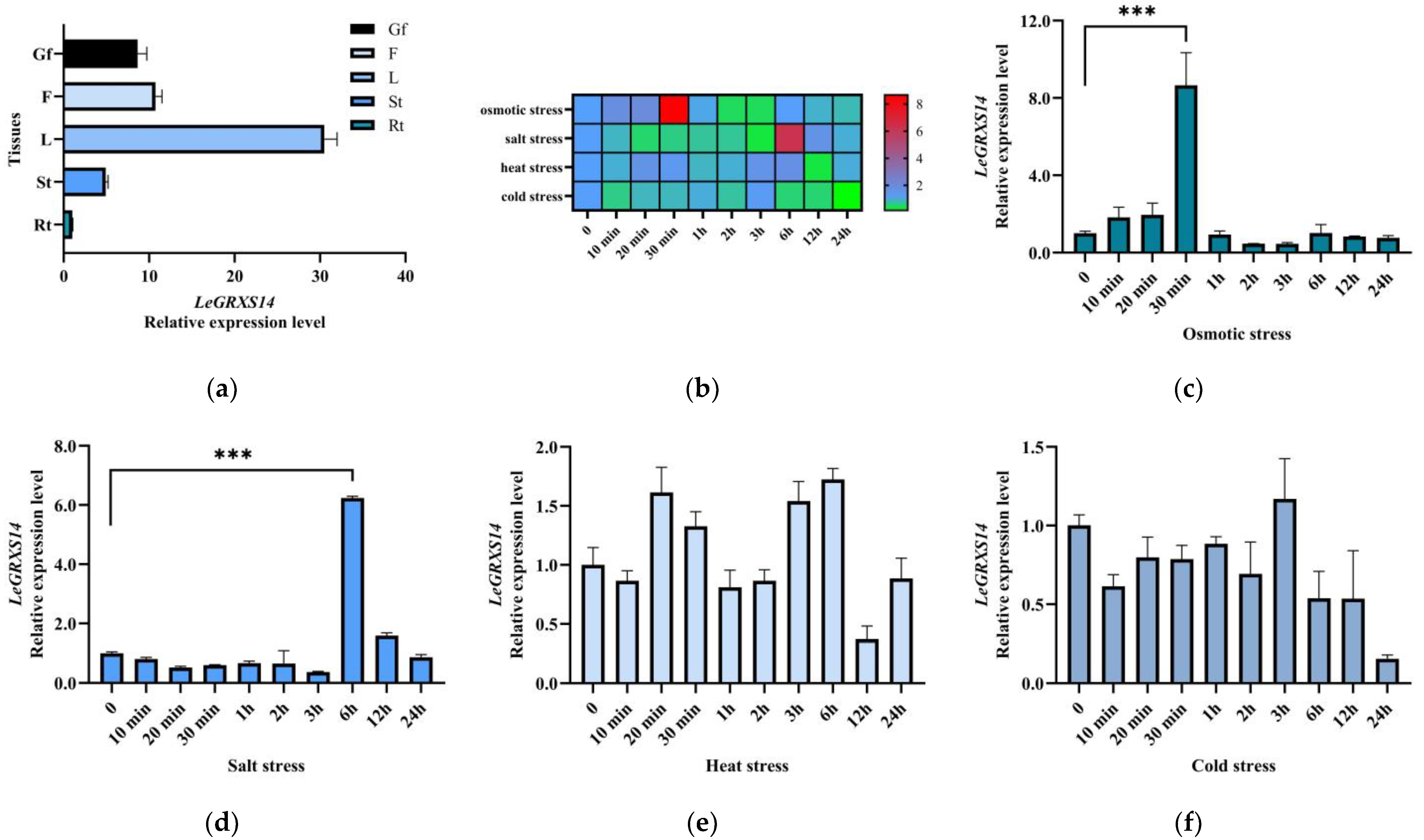
2.3. Expression Patterns of LeGRXS14 Genes in Tomato under Abiotic Stress
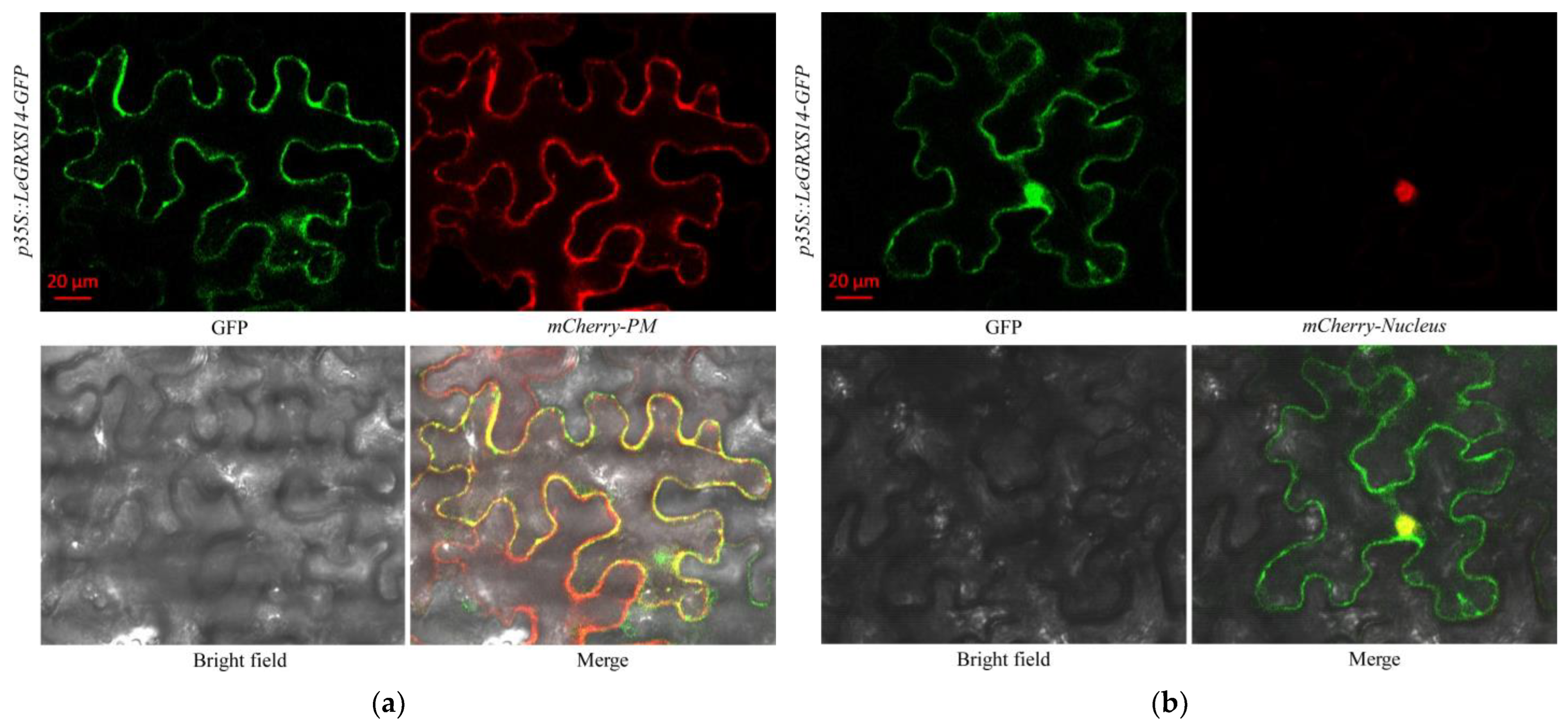
2.4. Subcellular Localization of LeGRXS14-GFP
2.5. LeGRXS14 Overexpression in Response to Salt Stress in A. thaliana
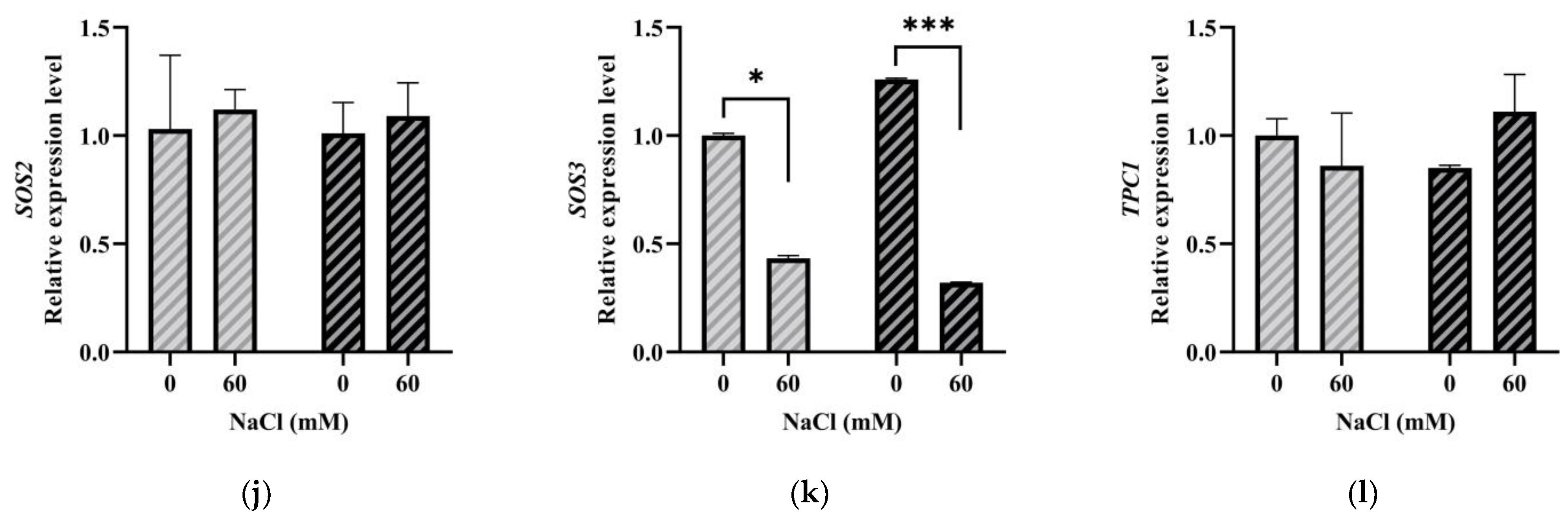
2.6. LeGRXS14 Is Related to Plant Resistance to Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. DNA Constructs and Transgenic Lines
4.3. PCR Primers and Vectors
4.4. RNA Extraction and qRT-PCR
4.5. GFP Subcellular Localization Analysis
4.6. Growth Responses to Salt Stress
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Najla, S.; Vercambre, G.; Pagès, L.; Grasselly, D.; Gautier, H.; Genard, M. Effect of Salinity on Tomato Plant Architecture. Acta Hortic. 2008, 801, 1183–1190. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Lennicke, C.; Cochemé, H.M. Redox metabolism: ROS as specific molecular regulators of cell signaling and function. Mol. Cell 2021, 81, 3691–3707. [Google Scholar] [CrossRef]
- Dvořák, P.; Krasylenko, Y.; Zeiner, A.; Šamaj, J.; Takáč, T. Signaling Toward Reactive Oxygen Species-Scavenging Enzymes in Plants. Front. Plant Sci. 2020, 11, 618835. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N. Plant glutaredoxins: Pivotal players in redox biology and iron-sulphur centre assembly. New Phytol. 2010, 186, 365–372. [Google Scholar] [CrossRef]
- Sharma, R.; Priya, P.; Jain, M. Modified expression of an auxin-responsive rice CC-type glutaredoxin gene affects multiple abiotic stress responses. Planta 2013, 238, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Holmgren, A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. USA 1976, 73, 2275–2279. [Google Scholar] [CrossRef]
- Ogata, F.T.; Branco, V.; Vale, F.F.; Coppo, L. Glutaredoxin: Discovery, redox defense and much more. Redox Biol. 2021, 43, 101975. [Google Scholar] [CrossRef]
- Sturlese, M.; Manta, B.; Bertarello, A.; Bonilla, M.; Lelli, M.; Zambelli, B.; Grunberg, K.; Mammi, S.; Comini, M.A.; Bellanda, M. The lineage-specific, intrinsically disordered N-terminal extension of monothiol glutaredoxin 1 from trypanosomes contains a regulatory region. Sci. Rep. 2018, 8, 13716. [Google Scholar] [CrossRef]
- Rouhier, N.; Lemaire, S.D.; Jacquot, J.P. The role of glutathione in photosynthetic organisms: Emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 2008, 59, 143–166. [Google Scholar] [CrossRef]
- Meyer, Y.; Buchanan, B.B.; Vignols, F.; Reichheld, J.P. Thioredoxins and Glutaredoxins: Unifying Elements in Redox Biology. Annu. Rev. Genet. 2009, 43, 335–367. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Zandalinas, S.I. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Rouhier, N.; Villarejo, A.; Srivastava, M.; Gelhaye, E.; Keech, O.; Droux, M.; Finkemeier, I.; Samuelsson, G.; Dietz, K.J.; Jacquot, J.P.; et al. Identification of plant glutaredoxin targets. Antioxid. Redox Signal. 2005, 7, 919–929. [Google Scholar] [CrossRef]
- Rouhier, N.; Gelhaye, E.; Jacquot, J.P. Plant glutaredoxins: Still mysterious reducing systems. Cell. Mol. Life Sci. 2004, 61, 1266–1277. [Google Scholar] [CrossRef]
- Meyer, Y.; Belin, C.; Delorme-Hinoux, V.; Reichheld, J.P.; Riondet, C. Thioredoxin and glutaredoxin systems in plants: Molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 2012, 17, 1124–1160. [Google Scholar] [CrossRef]
- Lillig, C.H.; Berndt, C. Glutaredoxins in Thiol/Disulfide Exchange. Antioxid. Redox Signal. 2013, 18, 1654–1665. [Google Scholar] [CrossRef] [PubMed]
- Gutsche, N.; Thurow, C.; Zachgo, S.; Gatz, C. Plant-specific CC-type glutaredoxins: Functions in developmental processes and stress responses. Biol. Chem. 2015, 396, 495–509. [Google Scholar] [CrossRef]
- Kakeshpour, T.; Tamang, T.M.; Motolai, G.; Fleming, Z.W.; Park, J.-E.; Wu, Q.; Park, S. CGFS-type glutaredoxin mutations reduce tolerance to multiple abiotic stresses in tomato. Physiol. Plant 2021, 173, 1263–1279. [Google Scholar] [CrossRef] [PubMed]
- Picciocchi, A.; Saguez, C.; Boussac, A.; Cassier-Chauvat, C.; Chauvat, F. CGFS-Type Monothiol Glutaredoxins from the Cyanobacterium Synechocystis PCC6803 and Other Evolutionary Distant Model Organisms Possess a Glutathione-Ligated [2Fe-2S] Cluster. Biochemistry 2007, 46, 15018–15026. [Google Scholar] [CrossRef]
- Son, S.; Kim, H.; Lee, K.S.; Kim, S.; Park, S.R. Rice glutaredoxin GRXS15 confers broad-spectrum resistance to Xanthomonas oryzae pv. oryzae and Fusarium fujikuroi. Biochem. Biophys. Res. Commun. 2020, 533, 1385–1392. [Google Scholar] [CrossRef]
- Martins, L.; Knuesting, J.; Bariat, L.; Dard, A.; Freibert, S.A.; Marchand, C.H.; Young, D.; Dung, N.H.T.; Voth, W.; Debures, A.; et al. Redox Modification of the Iron-Sulfur Glutaredoxin GRXS17 Activates Holdase Activity and Protects Plants from Heat Stress. Plant Physiol. 2020, 184, 676–692. [Google Scholar] [CrossRef]
- Guo, Y.; Huang, C.; Xie, Y.; Song, F.; Zhou, X. A tomato glutaredoxin gene SlGRX1 regulates plant responses to oxidative, drought and salt stresses. Planta 2010, 232, 1499–1509. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Michelet, L.; Massot, V.; Trost, P.; Lemaire, S.D. Biochemical Characterization of Glutaredoxins from Chlamydomonas reinhardtii Reveals the Unique Properties of a Chloroplastic CGFS-type Glutaredoxin*. J. Biol. Chem. 2008, 283, 8868–8876. [Google Scholar] [CrossRef]
- Ning, X.; Sun, Y.; Wang, C.; Zhang, W.; Sun, M.; Hu, H.; Liu, J.; Yang, L. A Rice CPYC-Type Glutaredoxin OsGRX20 in Protection against Bacterial Blight, Methyl Viologen and Salt Stresses. Front. Plant Sci. 2018, 9, 111. [Google Scholar] [CrossRef]
- Ding, S.; He, F.; Tang, W.; Du, H.; Wang, H. Identification of Maize CC-Type Glutaredoxins That Are Associated with Response to Drought Stress. Genes 2019, 10, 610. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J.M. Sodium in plants: Perception, signalling, and regulation of sodium fluxes. J. Exp. Bot. 2013, 65, 849–858. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Shams, M.; Khadivi, A. Mechanisms of salinity tolerance and their possible application in the breeding of vegetables. BMC Plant Biol. 2023, 23, 139. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.P.; Zhu, J.K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef]
- Bassil, E.; Blumwald, E. The ins and outs of intracellular ion homeostasis: NHX-type cation/ H+ transporters. Curr. Opin. Plant Biol. 2014, 22, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef] [PubMed]
- Davletova, S.; Schlauch, K.; Coutu, J.; Mittler, R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005, 139, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Le, C.T.; Brumbarova, T.; Ivanov, R.; Stoof, C.; Weber, E.; Mohrbacher, J.; Fink-Straube, C.; Bauer, P. ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) Interacts with FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) Linking Iron Deficiency and Oxidative Stress Responses. Plant Physiol. 2016, 170, 540–557. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Niu, M.X.; Feng, C.H.; Li, H.G.; Su, Y.; Su, W.L.; Pang, H.; Yang, Y.; Yu, X.; Wang, H.L.; et al. PeSTZ1 confers salt stress tolerance by scavenging the accumulation of ROS through regulating the expression of PeZAT12 and PeAPX2 in Populus. Tree Physiol. 2020, 40, 1292–1311. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhang, S.; Xie, L.; Zhu, Q.; Tan, Z.; Bian, J.; Sun, L.; Chen, J. The secretory pathway and the actomyosin motility system are required for plasmodesmatal localization of the P7-1 of rice black-streaked dwarf virus. Arch. Virol. 2013, 158, 1055–1064. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Li, X.; Su, M.; Shi, J.; Zhang, Q.; Liu, X. LeGRXS14 Reduces Salt Stress Tolerance in Arabidopsis thaliana. Plants 2023, 12, 2320. https://doi.org/10.3390/plants12122320
Liu L, Li X, Su M, Shi J, Zhang Q, Liu X. LeGRXS14 Reduces Salt Stress Tolerance in Arabidopsis thaliana. Plants. 2023; 12(12):2320. https://doi.org/10.3390/plants12122320
Chicago/Turabian StyleLiu, Lulu, Xiaofei Li, Mengke Su, Jiaping Shi, Qing Zhang, and Xunyan Liu. 2023. "LeGRXS14 Reduces Salt Stress Tolerance in Arabidopsis thaliana" Plants 12, no. 12: 2320. https://doi.org/10.3390/plants12122320
APA StyleLiu, L., Li, X., Su, M., Shi, J., Zhang, Q., & Liu, X. (2023). LeGRXS14 Reduces Salt Stress Tolerance in Arabidopsis thaliana. Plants, 12(12), 2320. https://doi.org/10.3390/plants12122320



