45S rDNA Diversity In Natura as One Step towards Ribosomal Heterogeneity in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
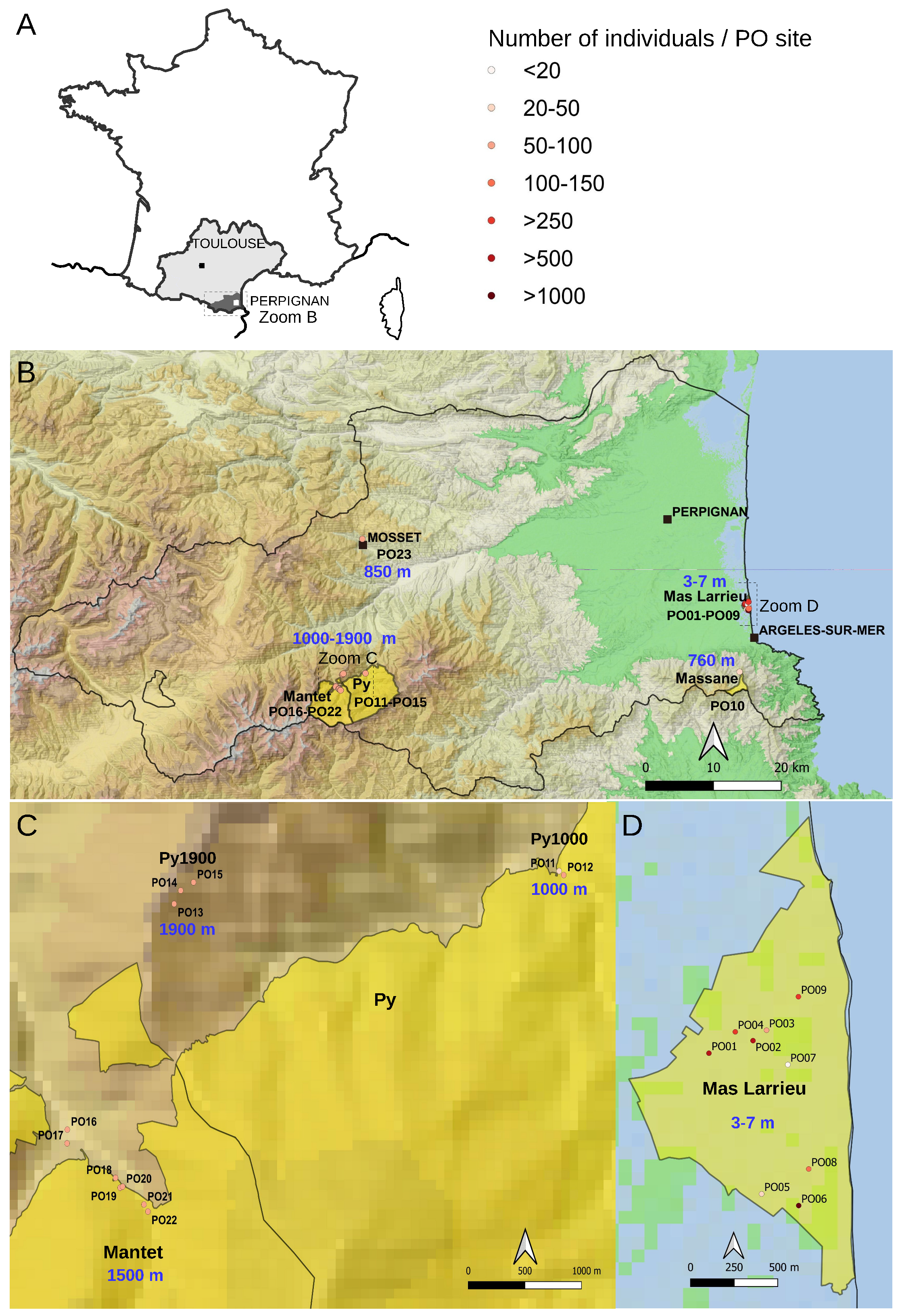
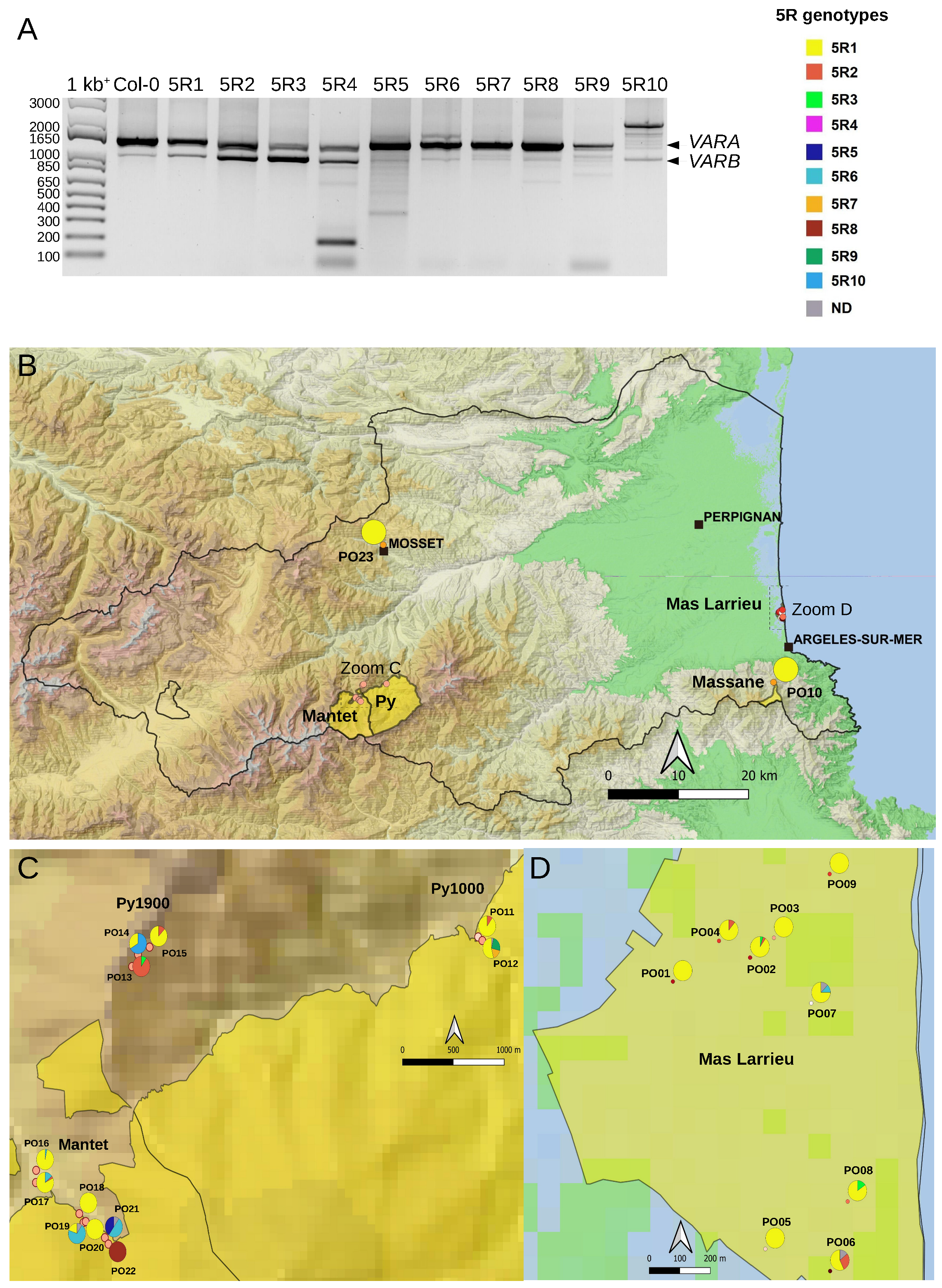
2.1. The At66 Metapopulation
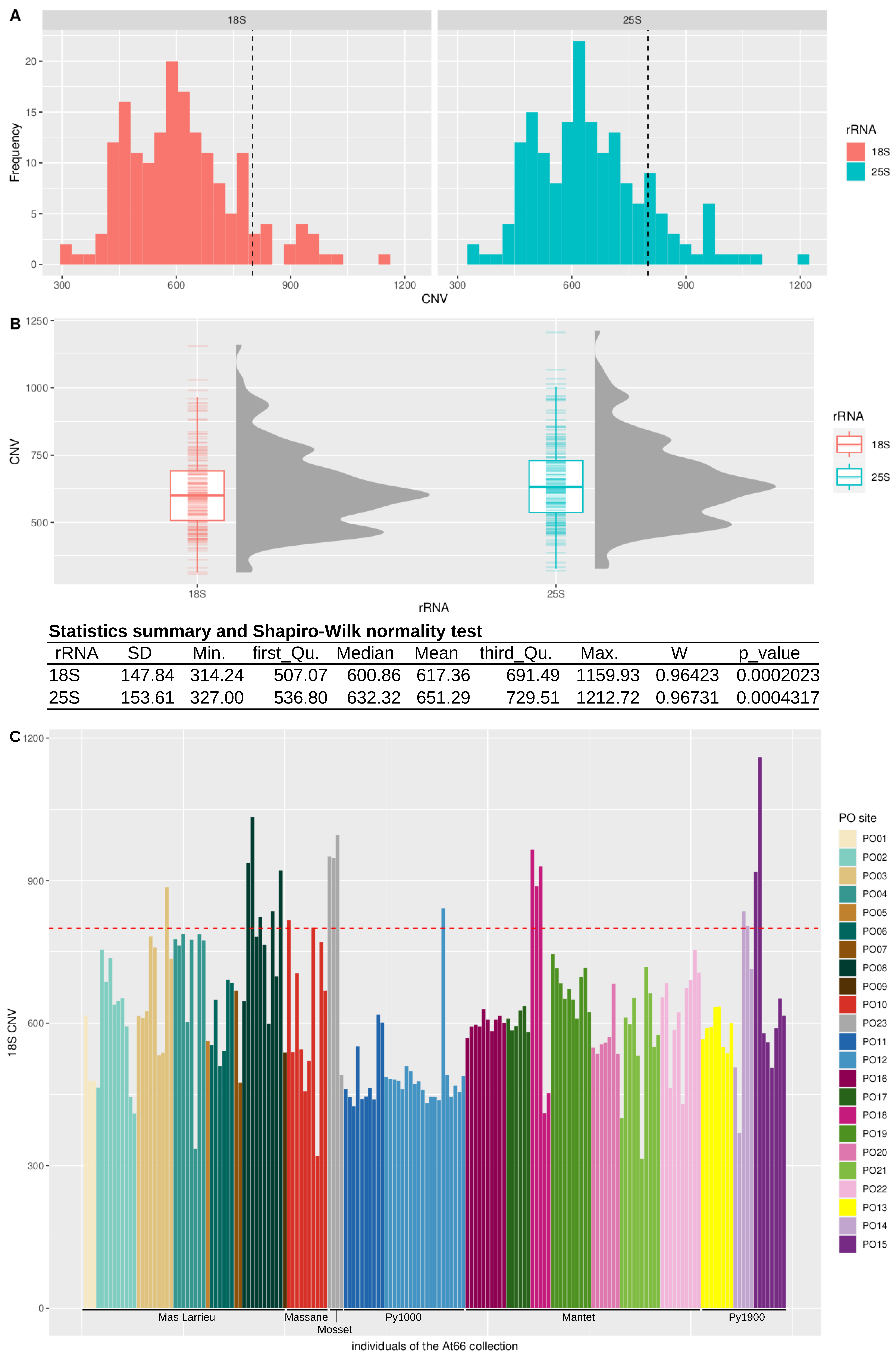
2.2. Copy Number Variation in At66
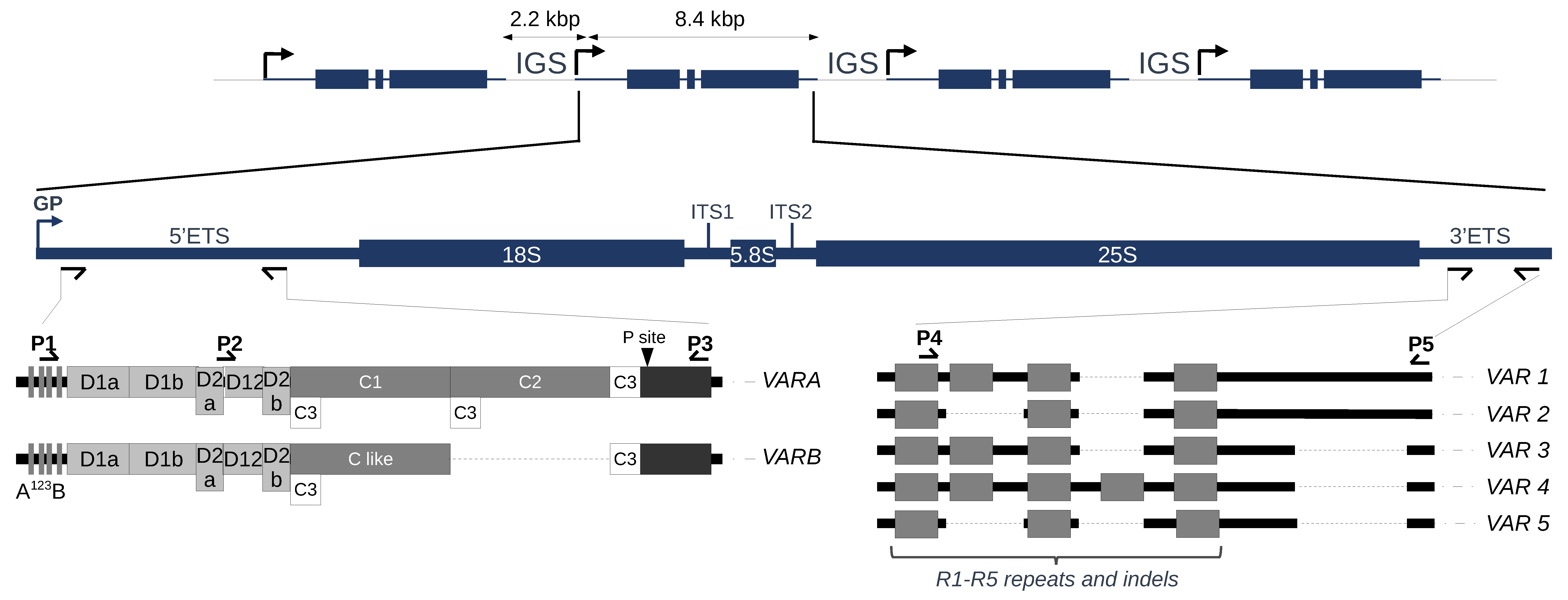
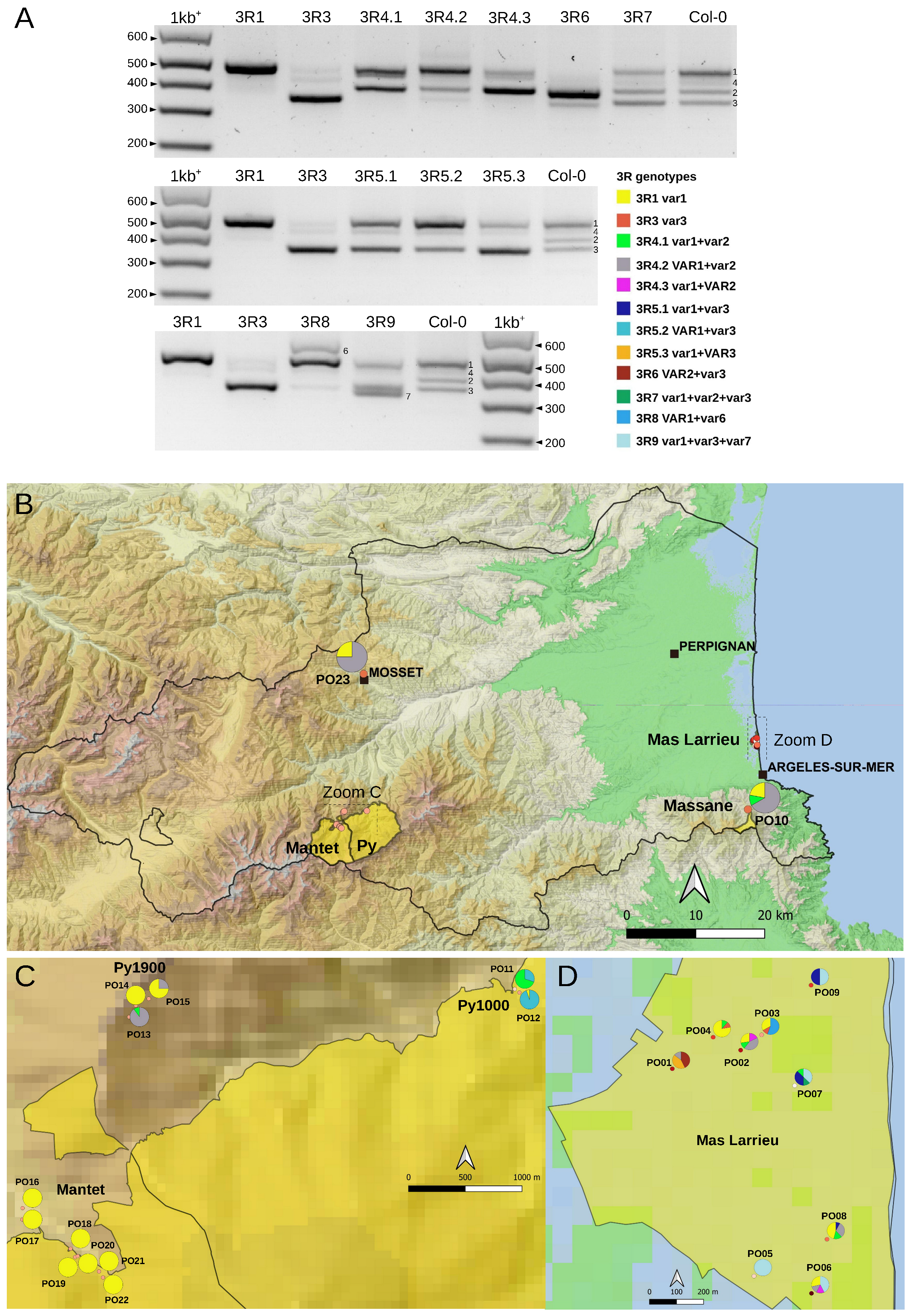
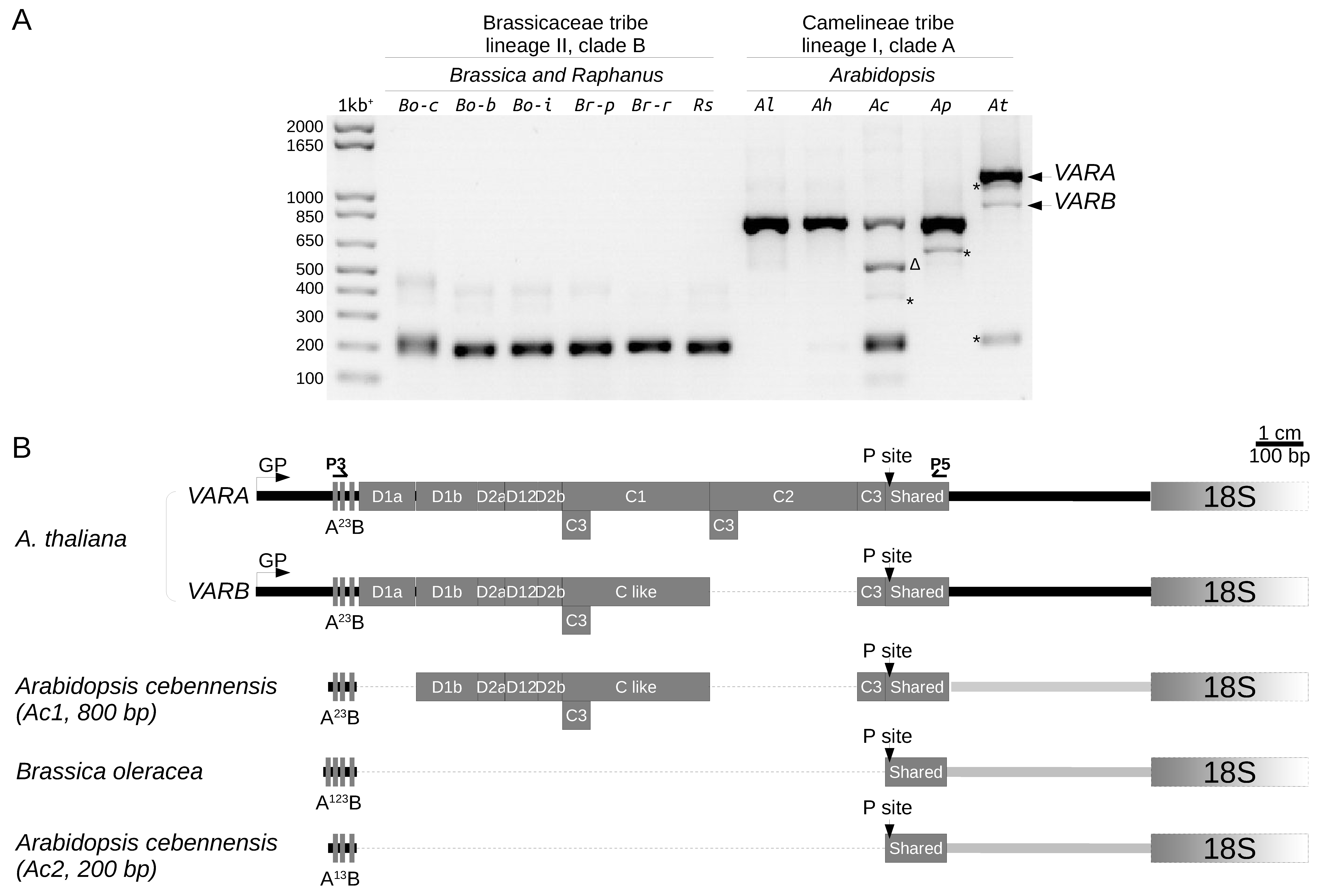
2.3. Variation at the 3′ETS in At66
2.4. Variation at the 5′ETS in At66
2.5. 5′ETS Length Variation in Brassicaceae Species
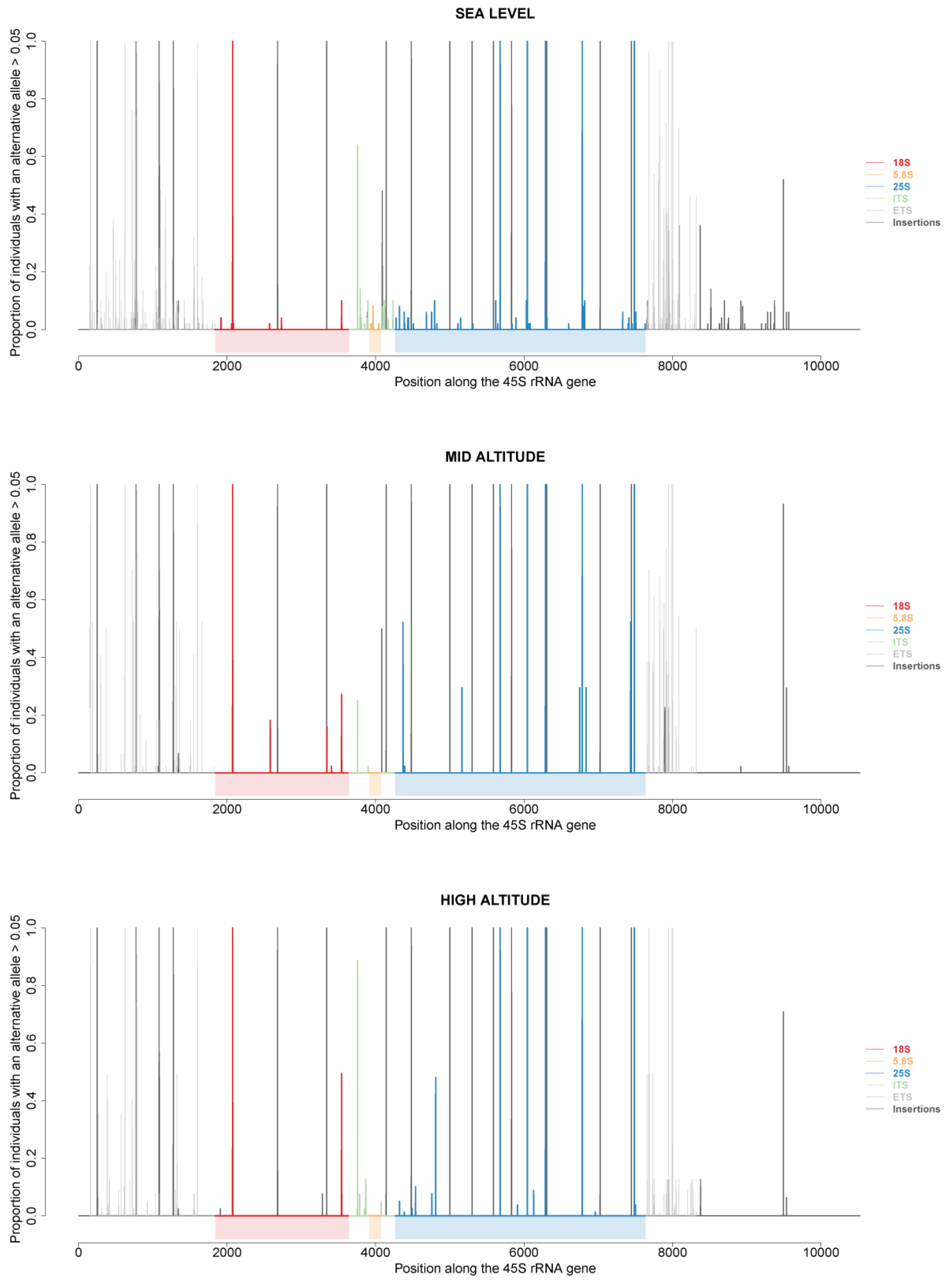
2.6. 45S rRNA Gene Polymorphisms in At66
3. Discussion
4. Materials and Methods
4.1. Sampling Individuals of At66 and Growth Conditions
4.2. Additionnal Plant Material
4.3. Estimating rDNA Copy Number Variation and Detection of rDNA Variants through NGS
4.4. 5′ and 3′ETS Genotyping
4.5. Expression of New VAR6 and VAR7 3′ETS Variants
4.6. Primer List
- P1, 5′-CTTTTCCGGGCACTTTTCCGG-3′
- P2, 5′-CTTGCGCACGAAATACCGAG-3′
- P3, 5′-GTTCGGCGTATGAGTGGTGATCGG-3′
- P4, 5′-GACAGACTTGTCCAAAACGCCCACC-3′
- P5, 5′-CTGGTCGAGGAATCCTGGACGATT-3′
- P6, 5′-CCTCGGACCCGGTAAAC-3′
- P7, 5′-TGTTCACCCACCAATAGGGAA-3′
- P8, 5′-GGCGGTCCGAACGACCGTTCCGCC-3′
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carroll, A.J. The Arabidopsis cytosolic ribosomal proteome: From form to function. Front. Plant Sci. 2013, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Seidel, F.; Beine-Golovchuk, O.; Hsieh, Y.-C.; Kopka, J. Systematic review of plant ribosome heterogeneity and specialization. Front. Plant Sci. 2020, 11, 948. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Vásquez, J.; Delseny, M. Ribosome biogenesis in plants: From functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell 2019, 31, 1945–1967. [Google Scholar] [CrossRef] [PubMed]
- Gruendler, P.; Unfried, I.; Pascher, K.; Schweizer, D. rDNA intergenic region from Arabidopsis thaliana: Structural analysis, intraspecific variation and functional implications. J. Mol. Biol. 1991, 221, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Gruendler, P.; Unfried, I.; Pointner, R.; Schweizer, D. Nucleotide sequence of the 25S-18S ribosomal gene spacer from Arabidopsis thaliana. Nucleic Acids Res. 1989, 17, 6395–6396. [Google Scholar] [CrossRef]
- Unfried, I.; Gruendler, P. Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 1990, 18, 4011. [Google Scholar] [CrossRef]
- Unfried, I.; Stocker, U.; Gruendler, P. Nucleotide sequence of the 18S rRNA gene from Arabidopsis thaliana Col0. Nucleic Acids Res. 1989, 17, 7513. [Google Scholar] [CrossRef]
- Copenhaver, G.P.; Doelling, J.H.; Gens, J.S.; Pikaard, C.S. Use of RFLPs larger than 100 kbp to map the position and internal organization of the nucleolus organizer region on chromosome 2 in Arabidopsis thaliana. Plant J. 1995, 7, 273–286. [Google Scholar] [CrossRef]
- Havlová, K.; Dvořáčková, M.; Peiro, R.; Abia, D.; Mozgová, I.; Vansáčová, L.; Gutierrez, C.; Fajkus, J. Variation of 45S rDNA intergenic spacers in Arabidopsis thaliana. Plant Mol. Biol. 2016, 92, 457–471. [Google Scholar] [CrossRef]
- Rabanal, F.A.; Mandáková, T.; Soto-Jiménez, L.M.; Greenhalgh, R.; Parrott, D.L.; Lutzmayer, S.; Steffen, J.G.; Nizhynska, V.; Mott, R.; Lysak, M.A.; et al. Epistatic and allelic interactions control expression of ribosomal RNA gene clusters in Arabidopsis thaliana. Genome Biol. 2017, 18, 75. [Google Scholar] [CrossRef]
- Rabanal, F.A. Characterization of the different levels of variation in 45S rRNA genes. Methods Mol. Biol. 2022, 2484, 363–379. [Google Scholar] [PubMed]
- Sims, J.; Sestini, G.; Elgert, C.; von Haeseler, A.; Schlögelhofer, P. Sequencing of the Arabidopsis NOR2 reveals its distinct organization and tissue-specific rRNA ribosomal variants. Nat. Commun. 2021, 12, 387. [Google Scholar] [CrossRef]
- Copenhaver, G.P.; Pikaard, C.S. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J. 1996, 9, 259–272. [Google Scholar] [CrossRef]
- Copenhaver, G.P.; Pikaard, C.S. Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant J. 1996, 9, 273–282. [Google Scholar] [CrossRef]
- McClintock, B. The relation of a particular chromosomal element to the development of the nucleoli in Zea mays. Z. Zellforsch. 1934, 21, 294–326. [Google Scholar] [CrossRef]
- McStay, B. Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination. Genes Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhara, C.; Mohannath, G.; Blevins, T.; Pontvianne, F.; Pikaard, C.S. Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev. 2016, 30, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Rabanal, F.A.; Nizhynska, V.; Mandáková, T.; Novikova, P.Y.; Lysak, M.A.; Mott, R.; Nordborg, M. Unstable Inheritance of 45S rRNA genes in Arabidopsis thaliana. G3 Genes Genomes Genet. 2017, 7, 1201–1209. [Google Scholar] [CrossRef]
- Long, Q.; Rabanal, F.A.; Meng, D.; Huber, C.D.; Farlow, A.; Platzer, A.; Zhang, Q.; Vilhjálmsson, B.J.; Korte, A.; Nizhynska, V.; et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 2013, 45, 884–890. [Google Scholar] [CrossRef]
- Pontvianne, F.; Abou-Ellail, M.; Douet, J.; Comella, P.; Matia, I.; Chandrasekhara, C.; Debures, A.; Blevins, T.; Cooke, R.; Medina, F.J.; et al. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001225. [Google Scholar] [CrossRef]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.B.; Schneeberger, K. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat. Commun. 2020, 11, 989. [Google Scholar] [CrossRef] [PubMed]
- Michael, T.P.; Jupe, F.; Bemm, F.; Motley, S.T.; Sandoval, J.P.; Lanz, C.; Loudet, O.; Weigel, D.; Ecker, J.R. High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat. Commun. 2018, 9, 541. [Google Scholar] [CrossRef] [PubMed]
- Caparros-Ruiz, D.; Lahmy, S.; Piersanti, S.; Echeverría, M. Two ribosomal DNA-binding factors interact with a cluster of motifs on the 5′ external transcribed spacer, upstream from the primary pre-rRNA processing site in a higher plant. Eur. J. Biochem. 1997, 247, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Vasquez, J.; Caparros-Ruiz, D.; Barneche, F.; Echeverría, M. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 2004, 24, 7284–7297. [Google Scholar] [CrossRef]
- Abou-Ellail, M.; Cooke, R.; Sáez-Vásquez, J. Variations in a team: Major and minor variants of Arabidopsis thaliana rDNA genes. Nucleus 2011, 2, 294–299. [Google Scholar] [CrossRef]
- Bartoli, C.; Frachon, L.; Barret, M.; Rigal, M.; Huard-Chauveau, C.; Mayjonade, B.; Zanchetta, C.; Bouchez, O.; Roby, D.; Carrère, S.; et al. In situ relationships between microbiota and potential pathobiota in Arabidopsis thaliana. ISME J. 2018, 12, 2024–2038. [Google Scholar] [CrossRef]
- Duruflé, H.; Ranocha, P.; Mbadinga Mbadinga, D.L.; Déjean, S.; Bonhomme, M.; San Clemente, H.; Viudes, S.; Eljebbawi, A.; Delorme-Hinoux, V.; Sáez-Vásquez, J.; et al. Phenotypic trait variation as a response to altitude-related constraints in Arabidopsis populations. Front. Plant Sci. 2019, 10, 430. [Google Scholar] [CrossRef]
- Frachon, L.; Bartoli, C.; Carrère, S.; Bouchez, O.; Chaubet, A.; Gautier, M.; Roby, D.; Roux, F. A genomic map of climate adaptation in Arabidopsis thaliana at a micro-geographic scale. Front. Plant Sci. 2018, 9, 967. [Google Scholar] [CrossRef]
- Busoms, S.; Teres, J.; Huang, X.Y.; Bomblies, K.; Danku, J.; Douglas, A.; Weigel, D.; Poschenrieder, C.; Salt, D.E. Salinity is an agent of divergent selection driving local adaptation of Arabidopsis to coastal habitats. Plant Physiol. 2015, 168, 915–929. [Google Scholar] [CrossRef]
- Montesinos-Navarro, A.; Wig, J.; Picó, F.X.; Tonsor, S.J. Arabidopsis thaliana populations show clinal variation in a climatic gradient associated with altitude. New Phytol. 2011, 189, 282–294. [Google Scholar] [CrossRef]
- Montesinos, A.; Tonsor, S.J.; Alonso-Blanco, C.; Picó, F.X. Demographic and genetic patterns of variation among populations of Arabidopsis thaliana from contrasting native environments. PLoS ONE 2009, 4, e7213. [Google Scholar] [CrossRef] [PubMed]
- Schmuths, H.; Meister, A.; Horres, R.; Bachmann, K. Genome size variation among accessions of Arabidopsis thaliana. Ann. Bot. 2004, 93, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Davison, J.; Tyagi, A.; Comai, L. Large-scale polymorphism of heterochromatic repeats in the DNA of Arabidopsis thaliana. BMC Plant Biol. 2007, 7, 44. [Google Scholar] [CrossRef]
- Prokopowich, C.D.; Gregory, T.R.; Crease, T.J. The correlation between rDNA copy number and genome size in eukaryotes. Genome 2003, 46, 48–50. [Google Scholar] [CrossRef] [PubMed]
- Earley, K.W.; Pontvianne, F.; Wierzbicki, A.T.; Blevins, T.; Tucker, S.; Costa-Nunes, P.; Pontes, O.; Pikaard, C.S. Mechanisms of HDA6-mediated rRNA gene silencing: Suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 2010, 24, 1119–1132. [Google Scholar] [CrossRef]
- Darriere, T.; Jobet, E.; Zavala, D.; Escande, M.L.; Durut, N.; De Bures, A.; Blanco-Herrera, F.; Vidal, E.A.; Rompais, M.; Carapito, C.; et al. Upon heat stress processing of ribosomal RNA precursors into mature rRNAs is compromised after cleavage at primary P site in Arabidopsis thaliana. RNA Biol. 2022, 19, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genomics. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of Fixed and Random Effect Models for powerful and efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Rosato, M.; Kovařík, A.; Garilleti, R.; Rosselló, J.A. Conserved Organisation of 45S rDNA Sites and rDNA Gene Copy Number among Major Clades of Early Land Plants. PLoS ONE 2016, 11, e0162544. [Google Scholar] [CrossRef] [PubMed]
- Sims, J.; Copenhaver, G.P.; Schlögelhofer, P. Meiotic DNA repair in the nucleolus employs a Nonhomologous End-Joining Mechanism. Plant Cell 2019, 31, 2259–2275. [Google Scholar] [CrossRef] [PubMed]
- Schindele, A.; Gehrke, F.; Schmidt, C.; Rohrig, S.; Dorn, A.; Puchta, H. Using CRISPR-Kill for organ specific cell elimination by cleavage of tandem repeats. Nat. Commun. 2022, 13, 1502. [Google Scholar] [CrossRef] [PubMed]
- Datson, P.M.; Murray, B.G. Ribosomal DNA locus evolution in Nemesia: Transposition rather than structural rearrangement as the key mechanism? Chromosome Res. 2006, 14, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Pavlištová, V.; Dvořáčková, M.; Jež, M.; Mozgová, I.; Mokroš, P.; Fajkus, J. Phenotypic reversion in fas mutants of Arabidopsis thaliana by reintroduction of FAS genes: Variable recovery of telomeres with major spatial rearrangements and transcriptional reprogramming of 45S rDNA genes. Plant J. Cell Mol. Biol. 2016, 88, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Picart-Picolo, A.; Grob, S.; Picault, N.; Franek, M.; Llauro, C.; Halter, T.; Maier, T.R.; Jobet, E.; Descombin, J.; Zhang, P.; et al. Large tandem duplications affect gene expression, 3D organization, and plant-pathogen response. Genome Res. 2020, 30, 1583–1592. [Google Scholar] [CrossRef]
- Lopez, F.B.; Fort, A.; Tadini, L.; Probst, A.V.; McHale, M.; Friel, J.; Ryder, P.; Pontvianne, F.; Pesaresi, P.; Sulpice, R.; et al. Gene dosage compensation of rRNA transcript levels in Arabidopsis thaliana lines with reduced ribosomal gene copy number. Plant Cell 2021, 33, 1135–1150. [Google Scholar] [CrossRef]
- Lopez, F.B.; McKeown, P.C.; Fort, A.; Brychkova, G.; Spillane, C. The boys are back in town: Rethinking the function of ribosomal DNA repeats in the genomic era. Mol. Plant 2023, 16, 514–516. [Google Scholar] [CrossRef]
- Durut, N.; Abou-Ellail, M.; Pontvianne, F.; Das, S.; Kojima, H.; Ukai, S.; De Bures, A.; Comella, P.; Nidelet, S.; Rialle, S.; et al. A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis. Plant Cell 2014, 26, 1330–1344. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high-resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Heming, N.M.; Dambros, C.; Gutiérrez, E. ENMwizard: Advanced Techniques for Ecological Niche Modeling Made Easy. 2018. Available online: https://github.com/HemingNM/ENMwizard/ (accessed on 13 July 2023).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 12 July 2023).
- Posit Team. RStudio: Integrated Development Environment for R; Posit Software, PBC: Boston, MA, USA, 2022. Available online: http://www.posit.co/ (accessed on 12 July 2023).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Barton, K. MuMIn: Multi-Model Inference. R Package. 2022. Available online: https://cran.r-project.org/web/packages/MuMIn/MuMIn.pdf (accessed on 12 July 2023).
- Azevedo-Favory, J.; Gaspin, C.; Ayadi, L.; Montacié, C.; Marchand, V.; Jobet, E.; Rompais, M.; Carapito, C.; Motorin, Y.; Sáez-Vásquez, J. Mapping rRNA 2′-O-methylations and identification of C/D snoRNAs in Arabidopsis thaliana plants. RNA Biol. 2021, 18, 1760–1777. [Google Scholar] [CrossRef]
- Wender, N.J.; Polisetty, C.R.; Donohue, K. Density-dependent processes influencing the evolutionary dynamics of dispersal: A functional analysis of seed dispersal in Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 2005, 92, 960–971. [Google Scholar] [CrossRef]
- Vaillancourt, B.; Buell, C.R. High molecular weight DNA isolation method from diverse plant species for use with Oxford Nanopore sequencing. bioRxiv 2019, 783159. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup, The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Gouy, M.; Gautier, C. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996, 12, 543–548. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delorme-Hinoux, V.; Mbodj, A.; Brando, S.; De Bures, A.; Llauro, C.; Covato, F.; Garrigue, J.; Guisset, C.; Borrut, J.; Mirouze, M.; et al. 45S rDNA Diversity In Natura as One Step towards Ribosomal Heterogeneity in Arabidopsis thaliana. Plants 2023, 12, 2722. https://doi.org/10.3390/plants12142722
Delorme-Hinoux V, Mbodj A, Brando S, De Bures A, Llauro C, Covato F, Garrigue J, Guisset C, Borrut J, Mirouze M, et al. 45S rDNA Diversity In Natura as One Step towards Ribosomal Heterogeneity in Arabidopsis thaliana. Plants. 2023; 12(14):2722. https://doi.org/10.3390/plants12142722
Chicago/Turabian StyleDelorme-Hinoux, Valérie, Assane Mbodj, Sophie Brando, Anne De Bures, Christel Llauro, Fabrice Covato, Joseph Garrigue, Claude Guisset, Jacques Borrut, Marie Mirouze, and et al. 2023. "45S rDNA Diversity In Natura as One Step towards Ribosomal Heterogeneity in Arabidopsis thaliana" Plants 12, no. 14: 2722. https://doi.org/10.3390/plants12142722
APA StyleDelorme-Hinoux, V., Mbodj, A., Brando, S., De Bures, A., Llauro, C., Covato, F., Garrigue, J., Guisset, C., Borrut, J., Mirouze, M., Reichheld, J.-P., & Sáez-Vásquez, J. (2023). 45S rDNA Diversity In Natura as One Step towards Ribosomal Heterogeneity in Arabidopsis thaliana. Plants, 12(14), 2722. https://doi.org/10.3390/plants12142722






