Soil and Phytomicrobiome for Plant Disease Suppression and Management under Climate Change: A Review
Abstract
:1. Introduction
2. Agronomic Practices, Phytomicrobiomes, and Plant Diseases
2.1. Tillage
2.2. Mulching
2.3. Monoculture and Polyculture
2.4. Soil Amendments
2.4.1. Fertilization
2.4.2. Treatment with Biofungicides vs. Chemical Pesticides
2.4.3. Biochar
2.4.4. Chitin and Derivatives
2.4.5. Clay Materials: Bentonite
2.4.6. Biocontrol Agents
| Type of Organism | Organism Name | Used as | Targeted Disease or Pathogen | References |
|---|---|---|---|---|
| Bacteria | Bacillus subtilis MBI 600 | PGPR, biocontrol agent | Pythium aphanidermatum, Fusarium oxysporum f. sp. radicis-cucumerinum | [170] |
| Bacillus velezensis HN03 | Biocontrol agent | Fusarium wilt (banana) | [171] | |
| Bacillus thuringiensis JCK-1233 | Biocontrol agent, systemic resistance inducer | Wilt disease of pine caused by the nematode Bursaphelenchus xylophilus | [172] | |
| Bacillus amyloliquefaciens Group, B. amyloliquefaciens, B. velezensis, B. nakamurai, and B. siamensis | Biocontrol agents | Various important plant pathogens such as Alternaria spp., Fusarium spp., Botryosphaeria spp., Botrytris spp., etc. Extensive list is mentioned in the review cited here. | [173] | |
| Paenibacillus polymyxa | Biofertilizer, biocontrol agent | Fusarium graminearum | [164,165,166] | |
| Pseudomonas spp. | PGPR, biofertilizer, biocontrol agent | A broad array of phytopathogens | [167] | |
| Fungi | Ampelomyces spp. | Biofungicide, biocontrol agent | Biocontrol of powdery mildews | [174] |
| Beauveria bassiana | Biocontrol agent, entomopathogenic fungus | Various insect pests | [175,176,177] | |
| Colletotrichum coccodes | Mycoherbicide | Abutilon theophrasti (velvet leaf) | [178] | |
| Coniothyrium minitans | Biocontrol agent | Sclerotinia sclerotiorum | [179] | |
| Metarhizium anisopliae | Growth promoter, biopesticide | Biocontrol of insect pests | [177] | |
| Trichoderma harzianum ZC51 | Biocontrol agent | Fusarium oxysporum | [180] | |
| Trichoderma harzianum SQR-T307 | Biocontrol agent | Fusarium wilt of cucumbers | [181] | |
| Trichoderma asperellum T-34 | Biocontrol agent | F. oxysporum f. sp. lycopersici race 1 causing Fusarium wilt of tomato | [182] |
| Agronomic Practices | Impacts | ||
|---|---|---|---|
| Phytomicrobiome | Pathobiome | ||
| Tillage | Conventional tillage | Reduced diversity and abundance | No disease suppression observed |
| No-Till and minimum tillage | Improved colonization of AMF and increased abundance of PGPR groups like Alphaproteobacteria, Betaproteobacteria, and Bacteroidetes, etc. in NT as compared to CT [15,28,29] | Increase of soil suppressiveness in systems such as Pythium ultimum—Lepidium sativum (cress) and Fusarium graminearum—Triticum aestivum (wheat) [31,32] | |
| Mulching | Plastic film mulches | Long-term mulching increases plant growth and causes surge in AMF (Arbuscular Mycorrhizal Fungi) [37,42] | Physical barrier for pathogens, spores, propagules [38,42,43,44,45] Repellent for insect pests such as whitefly and aphids (carriers of plant viruses) and reduced incidence of insect-transmitted plant diseases [46,47,48] |
| Organic mulches | Favorable to soil environments while providing nutrients to bacterial communities | Reduced severity of Phytophthora root rot with single species mulch [63] | |
| Monoculture and polyculture | Monoculture | Decline in the relative abundance of potentially beneficial microbes (Nitrospira and Trichoderma), Decrease in soil pH and organic matter content Increase in soil electrical conductivity (EC), and salt stress [70] | Accumulation of fungi such as Acrophialophora levis, Aspergillus corrugatus, Asergillus niger, Emericellopsis minima, Fusarium solani, Fusarium oxysporum, Neocosmospora striata, Scedosporium aurantiacum, and Thielavia hyrcaniae in peanut long-term monocropping [71]. Occurrence of severe Fusarium root rot in peas cultivated in continuous rotation in the Canadian prairies associated with reduced soil microbial diversity and lower concentrations of beneficial bacteria and AMF |
| Intercropping | Increase of carbon and nitrogen sequestration [74,75,76] Enrichment in diverse microbial taxa with various ecological functions such as mycorrhiza and/or endophytes, saprophytes, decomposers, bioprotective fungi or PGPR like Streptomyces, Bradyrhizobium, Candidatus Solibacter, Gemmatirosa, and Pseudolabrys [72,74,75,76,77] | Disease suppression: Decrease in maize kernels infections caused by Fusarium verticillioides and Aspergillus flavus along with reduced mycotoxins (fumonisins and aflatoxins) [74,75] Inhibition of Phytophthora capsici (likely attributed to the production of terpene in soil and root exudates) [78] Inhibition of Phytophthora sojae causing Phytophthora blight in soybean through maize root exudates such as cinnamic acid, vanillic acid, ferulic acid, and p-coumaric acid [79] Lower relative abundance of Fusarium oxysporum in the rhizosphere soil of the vanilla plants cultivated with black pepper along with higher relative abundance of potentially beneficial fungal groups such as Trichoderma [80] | |
| Crop rotation | Enrichment with diverse microbial communities | Decreased severity of wheat leaf blotch disease caused by Pyrenophora tritici-repentis with maximum impact in no-till and most diverse crop rotation systems [84] | |
| Soil Amendments | |||
| Fertilization | Chemical fertilizers | Resistance of the phyllosphere microbiome and root endophytes to long-term fertilization although soil microbiome (bacteria, fungi, and more importantly protists) was affected [58,59]. Out-competition of copiotrophic bacteria over oligotrophs | High nitrogen fertilization causing Nitrogen-Induced Susceptibility of biotrophic pathogens (e.g., powdery mildew, downy mildew, leaf rust, stem rot, and rice blast disease), and conversely, reduced infection by nectrotrophic pathogens (e.g., those responsible for take-all, and leaf spot disease) [103,110] |
| Organic fertilizers | Increase in the richness and diversity of the overall bacterial community [113] Higher levels of soil microbial activities, microbial diversity, and richness [16,116,117,118] Shift in microbial community and activity in vineyard pruning waste, with higher relative abundance of Ascomycota and fungal genera such as Fusarium and Zopfiella (known to control Phytophthora root rot in pepper plants) | Decrease soil-borne fungal pathogens such as Fusarium [119] Suppression of Pythium by composted manure or plant residues [120] Alleviation of plant diseases caused by Rhizoctonia, Verticillium, Sclerotinia, Phytophthora, Pythium, Aphanomyces, and Macrophomina in addition to weed control with green manure, especially from Brassica crops [123] Suppression of Fusarium wilt disease with mature compost enriched with biocontrol agents like Bacillus and Trichoderma spp. [113] | |
| Biochar | Decrease in the negative plant–soil feedback by altering the soil bacterial and fungal communities and augmenting the rhizosphere with beneficial bacteria such as Bacillus and Lysobacter [136,137,138,139] | Suppression of the plant pathogens such as Fusarium, Ilyonectria, and Rhizoctonia solani [136,137,138,139] Induction of plant systemic by enriching the root microbiome with PGPR and fungi [137,138,140,141,142,143] | |
| Chitin | Suppression of both soil-borne pathogens, e.g., Verticillium dahliae, and post-harvest pathogenic fungi (e.g., Colletotrichum spp., Botrytis cinerea (grey rot on grapes), Rhizopus stolonifer (black bread mold causing fruit rot)) [148,150] Suppression of potato wart disease, caused by Synchytrium endobioticum, with the use of crab shell (23% chitin) although the population of resting spores was not directly affected [152] | ||
| Bentonite | Increase in soil microbial activity, nutrient cycling and/or shift in fungal communities thanks to the water-holding- and macroaggregate formation capacity of bentonite [153,156] Involved in soil detoxification by absorbing heavy metals like cadmium (Cd) and lead (Pb) from contaminated agricultural soils, thereby increasing soil microbial population [154,155] | Disease suppression of phytopathogens like Alternaria spp., Bipolaris spp., Fusarium spp., Leptosphaeria spp., and Microdochium spp., through increased competition of beneficial microbes [156] Use of bentonite in encapsulation of biocontrol agents like Bacillus subtilis for better survival chances leading to increased activity against Rhizoctonia solani | |
3. Challenges and Pitfalls to the Identification and Use of Phytomicrobiome-Based Approaches
3.1. Conceptual Challenges
3.2. Computational Challenges
3.3. Challenges Associated with the Application of Microorganisms in the Field and the Evaluation of Side Effects
- (1)
- The microorganisms shall be deposited at an internationally recognized culture collection, and the species name of the microorganisms shall be identified unequivocally (no pathogens allowed).
- (2)
- The methods of analysis to identify and quantify them must be validated and shown to be sufficiently specific, correctly calibrated, accurate, and precise.
- (3)
- Their effectiveness in protecting plants from the targeted pests or pathogens must be demonstrated.
- (4)
- They shall not have any unacceptable effects on plants or plant products and on the environment, including fate and distribution in the environment, impact on non-target species, impact on biodiversity and the ecosystem. Risk assessment must fulfill data requirements for active substances, including microorganisms, as described in Regulation (EU) No 544/2011.
4. Agroecosystem Resilience and Adaptation to Climate Change
4.1. Impact of Climate Change on Plant Pathogens
4.1.1. Multiplication
4.1.2. Migration
4.1.3. Evolution
| Causes | Pathogen Names | Disease Name | Crops Affected | Countries Affected | More Comments | References | |
|---|---|---|---|---|---|---|---|
| Multiplication | Due to increased temperature and humidity | Hemileia vastatrix | Coffee rust | Coffee | Colombia and Central America (2008–2013) | Increased temperature increased the pathogen population. | [233] |
| Fusarium graminearum Fusarium culmorum | Fusarium head blight | Wheat | Global | Increased infection due to high abundance of conidia in soil and early anthesis of wheat. | [236,238] | ||
| Migration | Airborne | Phakopsora pachyrhizi | Asian soybean rust | Soybean | US | Hurricane Ivan caused the spread of spores leading to disease outbreak in the largest soybean-producing states. | [247] |
| Insect-borne | Chlorotic mottle virus spread by western corn rootworm, Diabrotica virgifera virgifera | Necrosis | Maize | Europe | Western corn worm is a native American species and is invading Europe. | [256] | |
| New hosts | Botrytis cinerea | Blossom blight | Japanese plums | Chile | First report on Japanese plums in 2013. It infected plums in California in 1960. | [264] | |
| New location | Aspergillus section Flavi | Aflatoxin production | Maize | France, Europe | Originated in America and Africa and reported in France in 2013. | [240,241] | |
| New location Speciation | Phytophthora infestans | Late blight | Potato, tomato | Europe | Led to the Irish famine in 19th century. | [257] | |
| Verticillium longisporum | Verticillium stripe | Canola and other brassica crops | Canada, Europe | Moving polewards. Recently reported in Canada. | [244] | ||
| Botrytis sinoallii | Grey mold | Allium crops | China | New species of Botrytis found in province of China in 2010 due to increasing temperature. | [258] | ||
| Evolution | New strains | Puccinia striiformis f. sp. tritici (Pst) | Stripe (yellow) rust | Wheat | Global | New strains Pst 1 and Pst2 are very aggressive and virulent. Strain adapted to higher temperatures with shorter latency period and increased spore germination percentage. | [251,260] |
| New strains | Puccinia graminis f. sp. tritici (Pgt) Race Ug99 | Stem (black) rust | Wheat | Global | Race Ug99 is the most aggressive that was reported first in Africa and is virulent to the resistant gene Sr31. | [259] |
4.2. Phytomicrobiome Can Modulate Plant’s Response to Climate Change
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMF | Arbuscular Mycorrhizal Fungi |
| AOD | Acute Oat Decline |
| ARG | Antibiotic Resistance Gene |
| CT | Conventional Tillage |
| GH | Greenhouse Gases |
| GTD | Grapevine Truck Disease |
| ISR | Induced Systemic Resistance |
| ITS | Internal Transcribed Spacer |
| MDCs | Meloidogyne-based Disease Complex |
| MT | Minimum Tillage |
| NIS | Nitrogen-Induced Susceptibility |
| NT | No-Till |
| PAW | Plant Available Water |
| PFM | Plastic Film Mulch |
| PGPM | Plant Growth Promoting Microorganisms |
| PGPR | Plant Growth Promoting Rhizobacteria |
| RKN | Root Knot Nematodes |
| RMT | Rhizo-Microbiome Transplants |
| SOC | Soil Organic Carbon |
| SOM | Soil Organic Matter |
| TAD | Take-All Decline |
| TEF | Translated Elongation Factor |
| TREE | Testing, Regulation, Engineering, and Eradication |
| TSWV | Tomato Spotted Virus |
References
- UNO. World Population Prospects Report 2022. Available online: https://population.un.org/wpp/ (accessed on 10 January 2023).
- Plants, S.o.t.W.s.; London (UK): Royal Botanic Gardens, Kew. 2017. Available online: https://www.kew.org/about-us/press-media/state-of-the-worlds-plants-2017 (accessed on 8 March 2023).
- FAO. New Standards to Curb the Global Spread of Plant Pests and Diseases. Available online: https://www.fao.org/news/story/en/item/1187738/icode/ (accessed on 10 January 2023).
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Whipps, J.M.; Lewis, K.; Cooke, R.C. Mycoparasitism and plant disease control. In Fungi in Biological Control Systems; Burge, M.N., Ed.; Manchester University Press: Manchester, UK, 1988. [Google Scholar]
- Stenberg, J.A.; Sundh, I.; Becher, P.G.; Björkman, C.; Dubey, M.; Egan, P.A.; Friberg, H.; Gil, J.F.; Jensen, D.F.; Jonsson, M.; et al. When is it biological control? A framework of definitions, mechanisms, and classifications. J. Pest Sci. 2021, 94, 665–676. [Google Scholar] [CrossRef]
- FAO. Conservation Agriculture. Available online: https://www.fao.org/conservation-agriculture/en/ (accessed on 10 January 2023).
- Morales Moreira, Z.P.; Chen, M.Y.; Yanez Ortuno, D.L.; Haney, C.H. Engineering plant microbiomes by integrating eco-evolutionary principles into current strategies. Curr. Opin. Plant Biol. 2023, 71, 102316. [Google Scholar] [CrossRef]
- Jiang, G.; Zhang, Y.; Gan, G.; Li, W.; Wan, W.; Jiang, Y.; Yang, T.; Zhang, Y.; Xu, Y.; Wang, Y.; et al. Exploring rhizo-microbiome transplants as a tool for protective plant-microbiome manipulation. ISME Commun. 2022, 2, 10. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina, T.; Rio, D.; Jones, C.D.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Sci. New Ser. 2015, 349, 860–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlatter, D.; Kinkel, L.; Thomashow, L.; Weller, D.; Paulitz, T. Disease Suppressive Soils: New Insights from the Soil Microbiome. Phytopathology 2017, 107, 1284–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, D.M.; Raaijmakers, J.M.; Gardener, B.B.M.; Thomashow, L.S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 2002, 40, 309–348. [Google Scholar] [CrossRef] [Green Version]
- Gómez Expósito, R.; De Bruijn, I.; Postma, J.; Raaijmakers, J.M. Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front. Microbiol. 2017, 8, 2529. [Google Scholar] [CrossRef]
- Hartman, K.; van der Heijden, M.G.A.; Wittwer, R.A.; Banerjee, S.; Walser, J.C.; Schlaeppi, K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2018, 6, 14. [Google Scholar] [CrossRef]
- Kraut-Cohen, J.; Zolti, A.; Shaltiel-Harpaz, L.; Argaman, E.; Rabinovich, R.; Green, S.J.; Minz, D. Effects of tillage practices on soil microbiome and agricultural parameters. Sci. Total Environ. 2020, 705, 135791. [Google Scholar] [CrossRef] [PubMed]
- Legrand, F.; Picot, A.; Cobo-Díaz, J.F.; Carof, M.; Chen, W.; Le Floch, G. Effect of tillage and static abiotic soil properties on microbial diversity. Appl. Soil Ecol. 2018, 132, 135–145. [Google Scholar] [CrossRef]
- Nikitin, D.A.; Ivanova, E.A.; Zhelezova, A.D.; Semenov, M.V.; Gadzhiumarov, R.G.; Tkhakakhova, A.K.; Chernov, T.I.; Ksenofontova, N.A.; Kutovaya, O.V. Assessment of the Impact of No-Till and Conventional Tillage Technologies on the Microbiome of Southern Agrochernozems. Eurasian Soil Sci. 2020, 53, 1782–1793. [Google Scholar] [CrossRef]
- Sun, R.; Li, W.; Dong, W.; Tian, Y.; Hu, C.; Liu, B. Tillage changes vertical distribution of soil bacterial and fungal communities. Front. Microbiol. 2018, 9, 699. [Google Scholar] [CrossRef]
- Claassen, R. This document is discoverable and free to researchers across the globe due to the work of AgEcon Search. Help ensure our sustainability. U.S. Dep. Agric. Econ. 2018, 1, 1–22. [Google Scholar]
- Hobbs, P.R.; Sayre, K.; Gupta, R. The role of conservation agriculture in sustainable agriculture. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 543–555. [Google Scholar] [CrossRef]
- Carlos, F.S.; Schaffer, N.; Marcolin, E.; Fernandes, R.S.; Mariot, R.; Mazzurana, M.; Roesch, L.F.W.; Levandoski, B.; de Oliveira Camargo, F.A. A long-term no-tillage system can increase enzymatic activity and maintain bacterial richness in paddy fields. Land Degrad. Dev. 2021, 32, 2257–2268. [Google Scholar] [CrossRef]
- Denardin, L.G.d.O.; Carmona, F.d.C.; Veloso, M.G.; Martins, A.P.; Freitas, T.F.S.d.; Carlos, F.S.; Marcolin, É.; Camargo, F.A.D.O.; Anghinoni, I. No-tillage increases irrigated rice yield through soil quality improvement along time. Soil Tillage Res. 2019, 186, 64–69. [Google Scholar] [CrossRef]
- Kabir, Z. Tillage or no-tillage: Impact on mycorrhizae. Can. J. Plant Sci. 2005, 85, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Palojärvi, A.; Kellock, M.; Parikka, P.; Jauhiainen, L.; Alakukku, L. Tillage System and Crop Sequence Affect Soil Disease Suppressiveness and Carbon Status in Boreal Climate. Front. Microbiol. 2020, 11, 534786. [Google Scholar] [CrossRef]
- Samarendra, H.; Robert, P.; Roland, B.; Liz, D.; Peter, R.; Sarah, D.; Debbie, A. Effect of tillage system and straw management on organic matter dynamics. Agron. Sustain. Dev. 2009, 29, 525–533. [Google Scholar]
- Singh, U.; Choudhary, A.K.; Sharma, S. Agricultural practices modulate the bacterial communities, and nitrogen cycling bacterial guild in rhizosphere: Field experiment with soybean. J. Sci. Food Agric. 2021, 101, 2687–2695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shi, Y.; Dong, Y.; Lapen, D.R.; Liu, J.; Chen, W. Subsoiling and conversion to conservation tillage enriched nitrogen cycling bacterial communities in sandy soils under long-term maize monoculture. Soil Tillage Res. 2022, 215, 105197. [Google Scholar] [CrossRef]
- Aslam, Z.; Yasir, M.; Yoon, H.S.; Jeon, C.O.; Chung, Y.R. Diversity of the bacterial community in the rice rhizosphere managed under conventional and no-tillage practices. J. Microbiol. 2013, 51, 747–756. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, L.; Chen, Q.; Wen, X.; Liu, Y.; Han, J.; Liao, Y. Conservation tillage enhances the stability of the rhizosphere bacterial community responding to plant growth. Agron. Sustain. Dev. 2017, 37, 44. [Google Scholar] [CrossRef]
- Legrand, F.; Chen, W.; Cobo-Díaz, J.F.; Picot, A.; Le Floch, G. Co-occurrence analysis reveal that biotic and abiotic factors influence soil fungistasis against Fusarium graminearum. FEMS Microbiol. Ecol. 2019, 95, fiz056. [Google Scholar] [CrossRef]
- Campos, S.B.; Lisboa, B.B.; Camargo, F.A.; Bayer, C.; Sczyrba, A.; Dirksen, P.; Albersmeier, A.; Kalinowski, J.; Beneduzi, A.; Costa, P.B.; et al. Soil suppressiveness and its relations with the microbial community in a Brazilian subtropical agroecosystem under different management systems. Soil Biol. Biochem. 2016, 96, 191–197. [Google Scholar] [CrossRef]
- Bongiorno, G.; Postma, J.; Bünemann, E.K.; Brussaard, L.; de Goede, R.G.; Mäder, P.; Tamm, L.; Thuerig, B. Soil suppressiveness to Pythium ultimum in ten European long-term field experiments and its relation with soil parameters. Soil Biol. Biochem. 2019, 133, 174–187. [Google Scholar] [CrossRef]
- Kurm, V.; Schilder, M.T.; Haagsma, W.K.; Bloem, J.; Scholten, O.E.; Postma, J. Reduced tillage increases soil biological properties but not suppressiveness against Rhizoctonia solani and Streptomyces scabies. Appl. Soil Ecol. 2022, 181, 104646. [Google Scholar] [CrossRef]
- Bailey, K.L. Diseases under conservation tillage systems. Can. J. Plant Sci. 1996, 76, 635–639. [Google Scholar] [CrossRef] [Green Version]
- Carter, M.R.; Sanderson, J.B. Influence of conservation tillage and rotation length on potato productivity, tuber disease and soil quality parameters on a fine sandy loam in eastern Canada. Soil Tillage Res. 2001, 63, 1–13. [Google Scholar] [CrossRef]
- Paulitz, T.C.; Schroeder, K.L.; Schillinger, W.F. Soilborne pathogens of cereals in an irrigated cropping system: Effects of tillage, residue management, and crop rotation. Plant Dis. 2010, 94, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Liu, Z.; Mou, H.; Li, J.; Zhang, P.; Jia, Z. Impact of farmland mulching practices on the soil bacterial community structure in the semiarid area of the loess plateau in China. Eur. J. Soil Biol. 2019, 92, 8–15. [Google Scholar] [CrossRef]
- Steinmetz, Z.; Wollmann, C.; Schaefer, M.; Buchmann, C.; David, J.; Tröger, J.; Muñoz, K.; Frör, O.; Schaumann, G.E. Plastic mulching in agriculture. Trading short-term agronomic benefits for long-term soil degradation? Sci. Total Environ. 2016, 550, 690–705. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Yu, S.; Zhang, L.; Dong, K.; Feng, B. Mulching practices manipulate the microbial community diversity and network of root-associated compartments in the Loess Plateau. Soil Tillage Res. 2022, 223, 105476. [Google Scholar] [CrossRef]
- Bi, Y.; Qiu, L.; Zhakypbek, Y.; Jiang, B.; Cai, Y.; Sun, H. Combination of plastic film mulching and AMF inoculation promotes maize growth, yield and water use efficiency in the semiarid region of Northwest China. Agric. Water Manag. 2018, 201, 278–286. [Google Scholar] [CrossRef]
- Wang, L.; Li, X.G.; Lv, J.; Fu, T.; Ma, Q.; Song, W.; Wang, Y.P.; Li, F.M. Continuous plastic-film mulching increases soil aggregation but decreases soil pH in semiarid areas of China. Soil Tillage Res. 2017, 167, 46–53. [Google Scholar] [CrossRef]
- Wan, P.; Zhang, N.; Li, Y.; Li, S.; Li, F.M.; Cui, Z.; Zhang, F. Reducing plant pathogens could increase crop yields after plastic film mulching. Sci. Total Environ. 2022, 861, 160615. [Google Scholar] [CrossRef] [PubMed]
- Chalker-Scott, L. Impact of Mulches on Landscape Plants and the Environment—A Review. J. Environ. Hortic. 2007, 25, 239–249. [Google Scholar] [CrossRef]
- Katan, J. Solar Heating by Polyethylene Mulching for the Control of Diseases Caused by Soil-Borne Pathogens. Phytopathology 1976, 66, 683–688. [Google Scholar] [CrossRef]
- Stapleton, J.J.; DeVay, J.E. Soil solarization: A non-chemical approach for management of plant pathogens and pests. Crop Prot. 1986, 5, 190–198. [Google Scholar] [CrossRef]
- Summers, C.G.; Mitchell, J.P.; Stapleton, J.J. Management of aphid-borne viruses and Bemisia argentifolii (Homoptera: Aleyrodidae) in zucchini squash by using UV reflective plastic and wheat straw mulches. Environ. Entomol. 2004, 33, 1447–1457. [Google Scholar] [CrossRef] [Green Version]
- Amare, G.; Desta, B. Coloured plastic mulches: Impact on soil properties and crop productivity. Chem. Biol. Technol. Agric. 2021, 8, 4. [Google Scholar] [CrossRef]
- Brown, J.E.; Dangler, J.M.; Woods, F.M.; Tilt, K.M.; Henshaw, M.D.; Griffey, W.A.; West, M.S.J.H. Delay in mosaic virus onset and aphid vector reduction in summer squash grown on reflective mulches. Hortscience 1993, 28, 895–896. [Google Scholar] [CrossRef] [Green Version]
- Kader, M.A.; Senge, M.; Mojid, M.A.; Ito, K. Recent advances in mulching materials and methods for modifying soil environment. Soil Tillage Res. 2017, 168, 155–166. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Mincer, T.J. Ecology of the plastisphere. Nat. Rev. Microbiol. 2020, 18, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Ossowicki, A.; Yergeau, É.; Vigani, G.; Geissen, V.; Garbeva, P. Plastic mulch film residues in agriculture: Impact on soil suppressiveness, plant growth, and microbial communities. FEMS Microbiol. Ecol. 2022, 98, fiac017. [Google Scholar] [CrossRef]
- Qi, Y.; Yang, X.; Pelaez, A.M.; Huerta Lwanga, E.; Beriot, N.; Gertsen, H.; Garbeva, P.; Geissen, V. Macro- and micro- plastics in soil-plant system: Effects of plastic mulch film residues on wheat (Triticum aestivum) growth. Sci. Total Environ. 2018, 645, 1048–1056. [Google Scholar] [CrossRef]
- Gkoutselis, G.; Rohrbach, S.; Harjes, J.; Obst, M.; Brachmann, A.; Horn, M.A.; Rambold, G. Microplastics accumulate fungal pathogens in terrestrial ecosystems. Sci. Rep. 2021, 11, 13214. [Google Scholar] [CrossRef]
- Sintim, H.Y.; Flury, M. Is Biodegradable Plastic Mulch the Solution to Agriculture’s Plastic Problem? Environ. Sci. Technol. 2017, 51, 1068–1069. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Sun, Z.X.; Feng, L.S.; Zheng, M.Z.; Chi, D.C.; Meng, W.Z.; Hou, Z.Y.; Bai, W.; Li, K.Y. Plastic film mulching for water-efficient agricultural applications and degradable films materials development research. Mater. Manuf. Process. 2015, 30, 143–154. [Google Scholar] [CrossRef]
- García-Orenes, F.; Guerrero, C.; Roldán, A.; Mataix-Solera, J.; Cerdà, A.; Campoy, M.; Zornoza, R.; Bárcenas, G.; Caravaca, F. Soil microbial biomass and activity under different agricultural management systems in a semiarid Mediterranean agroecosystem. Soil Tillage Res. 2010, 109, 110–115. [Google Scholar] [CrossRef]
- Ai, C.; Liang, G.; Sun, J.; Wang, X.; He, P.; Zhou, W. Different roles of rhizosphere effect and long-term fertilization in the activity and community structure of ammonia oxidizers in a calcareous fluvo-aquic soil. Soil Biol. Biochem. 2013, 57, 30–42. [Google Scholar] [CrossRef]
- Sun, A.; Jiao, X.Y.; Chen, Q.; Trivedi, P.; Li, Z.; Li, F.; Zheng, Y.; Lin, Y.; Hu, H.W.; He, J.Z. Fertilization alters protistan consumers and parasites in crop-associated microbiomes. Environ. Microbiol. 2021, 23, 2169–2183. [Google Scholar] [CrossRef]
- Sun, A.; Jiao, X.Y.; Chen, Q.; Wu, A.L.; Zheng, Y.; Lin, Y.X.; He, J.Z.; Hu, H.W. Microbial communities in crop phyllosphere and root endosphere are more resistant than soil microbiota to fertilization. Soil Biol. Biochem. 2021, 153, 108113. [Google Scholar] [CrossRef]
- Ortiz-Cornejo, N.L.; Romero-Salas, E.A.; Navarro-Noya, Y.E.; González-Zúñiga, J.C.; Ramirez-Villanueva, D.A.; Vásquez-Murrieta, M.S.; Verhulst, N.; Govaerts, B.; Dendooven, L.; Luna-Guido, M. Incorporation of bean plant residue in soil with different agricultural practices and its effect on the soil bacteria. Appl. Soil Ecol. 2017, 119, 417–427. [Google Scholar] [CrossRef]
- Sun, X.; Ye, Y.; Guan, Q.; Jones, D.L. Organic mulching masks rhizosphere effects on carbon and nitrogen fractions and enzyme activities in urban greening space. J. Soils Sediments 2021, 21, 1621–1632. [Google Scholar] [CrossRef]
- Sun, X.; Ye, Y.; Liao, J.; Tang, Y.; Wang, D.; Guan, Q. Organic mulching alters the composition, but not the diversity, of rhizosphere bacterial and fungal communities. Appl. Soil Ecol. 2021, 168, 104167. [Google Scholar] [CrossRef]
- Percival, G. Influence of pure mulches on suppressing Phytophthora root rot pathogens. J. Environ. Hortic. 2013, 31, 221–226. [Google Scholar] [CrossRef]
- Andres, C.; Comoé, H.; Beerli, A.; Schneider, M.; Rist, S.; Jacobi, J. Cocoa in Monoculture and Dynamic Agroforestry. In Sustainable Agriculture Reviews; Springer International Publishing: Cham, Switzerland, 2016; pp. 121–153. [Google Scholar] [CrossRef]
- Loh, S.K.; Asubonteng, K.O.; Adanu, S.K. Effects of Monocropping on Land Cover Transitions in the Wet Evergreen Agro-Ecological Zone of Ghana. Land 2022, 11, 1063. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, G.; Cheng, Y.; Shi, P.; Yang, C.; Yang, H.; Xu, Z. Soil acidification in continuously cropped tobacco alters bacterial community structure and diversity via the accumulation of phenolic acids. Sci. Rep. 2019, 9, 12499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.F.; Song, L.X.; Xia, X.J.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Plant-Soil Feedbacks and Soil Sickness: From Mechanisms to Application in Agriculture. J. Chem. Ecol. 2013, 39, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Li, X.G.; Ding, C.F.; Zhang, T.L.; Wang, X.X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Li, H.; Li, C.; Song, X.; Liu, Y.; Gao, Q.; Zheng, R.; Li, J.; Zhang, P.; Liu, X. Impacts of continuous and rotational cropping practices on soil chemical properties and microbial communities during peanut cultivation. Sci. Rep. 2022, 12, 2758. [Google Scholar] [CrossRef]
- Zhao, Q.; Xiong, W.; Xing, Y.; Sun, Y.; Lin, X.; Dong, Y. Long-Term Coffee Monoculture Alters Soil Chemical Properties and Microbial Communities. Sci. Rep. 2018, 8, 6116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Zhang, J.; Liu, H.; Wang, M.; Pan, L.; Chen, N.; Wang, T.; Jing, Y.; Chi, X.; Du, B. Long-term continuously monocropped peanut significantly disturbed the balance of soil fungal communities. J. Microbiol. 2020, 58, 563–573. [Google Scholar] [CrossRef]
- Bowles, T.M.; Jackson, L.E.; Loeher, M.; Cavagnaro, T.R. Ecological intensification and arbuscular mycorrhizas: A meta-analysis of tillage and cover crop effects. J. Appl. Ecol. 2017, 54, 1785–1793. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Chen, D.; Lu, K.; Sun, Z.; Zeng, R. Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant Sci. 2015, 6, 786. [Google Scholar] [CrossRef] [Green Version]
- Mwakilili, A.D.; Mwaikono, K.S.; Herrera, S.L.; Midega, C.A.O.; Magingo, F.; Alsanius, B.; Dekker, T.; Lyantagaye, S.L. Long-term maize-Desmodium intercropping shifts structure and composition of soil microbiome with stronger impact on fungal communities. Plant Soil 2021, 467, 437–450. [Google Scholar] [CrossRef]
- Njeru, N.K.; Midega, C.A.O.; Muthomi, J.W.; Wagacha, J.M.; Khan, Z.R. Impact of push–pull cropping system on pest management and occurrence of ear rots and mycotoxin contamination of maize in western Kenya. Plant Pathol. 2020, 69, 1644–1654. [Google Scholar] [CrossRef]
- Castellano-Hinojosa, A.; Strauss, S.L. Impact of cover crops on the soil microbiome of tree crops. Microorganisms 2020, 8, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Z.; Guo, X.; Xiang, Z.; Liu, D.; Yu, K.; Sun, K.; Yan, B.; Wang, S.; Kang, C.; Xu, Y.; et al. Maize intercropping enriches plant growth-promoting rhizobacteria and promotes both the growth and volatile oil concentration of Atractylodes lancea. Front. Plant Sci. 2022, 13, 1029722. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Y.; Mei, X.; Yang, M.; Huang, H.; Du, F.; Wu, J.; He, Y.; Sun, J.; Wang, H.; et al. Antimicrobial Terpenes Suppressed the Infection Process of Phytophthora in Fennel-Pepper Intercropping System. Front. Plant Sci. 2022, 13, 890534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, Y.; Mei, X.; Li, Y.; Wu, J.; Li, Y.; Wang, H.; Huang, H.; Yang, M.; He, X.; et al. Phenolic Acids Released in Maize Rhizosphere During Maize-Soybean Intercropping Inhibit Phytophthora Blight of Soybean. Front. Plant Sci. 2020, 11, 886. [Google Scholar] [CrossRef]
- Xiong, W.; Zhao, Q.; Xue, C.; Xun, W.; Zhao, J.; Wu, H.; Li, R.; Shen, Q. Comparison of fungal community in black pepper-vanilla and vanilla monoculture systems associated with vanilla Fusarium wilt disease. Front. Microbiol. 2016, 7, 117. [Google Scholar] [CrossRef] [PubMed]
- Modi, D.; Simard, S.; Bérubé, J.; Lavkulich, L.; Hamelin, R.; Grayston, S.J. Long-term effects of stump removal and tree species composition on the diversity and structure of soil fungal communities. FEMS Microbiol. Ecol. 2020, 96, fiaa061. [Google Scholar] [CrossRef] [PubMed]
- Modi, D.; Simard, S.; Lavkulich, L.; Hamelin, R.C.; Grayston, S.J. Stump removal and tree species composition promote a bacterial microbiome that may be beneficial in the suppression of root disease. FEMS Microbiol. Ecol. 2021, 97, fiaa213. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Siddique, K.H.; Liu, K. Cropping systems in agriculture and their impact on soil health—A review. Glob. Ecol. Conserv. 2020, 23, e01118. [Google Scholar] [CrossRef]
- Jalli, M.; Huusela, E.; Jalli, H.; Kauppi, K.; Niemi, M.; Himanen, S.; Jauhiainen, L. Effects of Crop Rotation on Spring Wheat Yield and Pest Occurrence in Different Tillage Systems: A Multi-Year Experiment in Finnish Growing Conditions. Front. Sustain. Food Syst. 2021, 5, 647335. [Google Scholar] [CrossRef]
- Peters, R.D.; Sturz, A.V.; Carter, M.R.; Sanderson, J.B. Developing disease-suppressive soils through crop rotation and tillage management practices. Soil Tillage Res. 2003, 72, 181–192. [Google Scholar] [CrossRef]
- Soonvald, L.; Loit, K.; Runno-Paurson, E.; Astover, A.; Tedersoo, L. Characterising the effect of crop species and fertilisation treatment on root fungal communities. Sci. Rep. 2020, 10, 18741. [Google Scholar] [CrossRef]
- Gan, Y.T.; Miller, P.R.; McConkey, B.G.; Zentner, R.P.; Stevenson, F.C.; McDonald, C.L. Influence of Diverse Cropping Sequences on Durum Wheat Yield and Protein in the Semiarid Northern Great Plains. Agron. J. 2003, 95, 245–252. [Google Scholar] [CrossRef]
- Gahagan, A.C.; Shi, Y.; Radford, D.; Morrison, M.J.; Gregorich, E.; Aris-Brosou, S.; Chen, W. Long-Term Tillage and Crop Rotation Regimes Reshape Soil-Borne Oomycete Communities in Soybean, Corn, and Wheat Production Systems. Plants 2023, 12, 2338. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.J.; Bending, G.D.; Chandler, D.; Hilton, S.; Mills, P. Meeting the demand for crop production: The challenge of yield decline in crops grown in short rotations. Biol. Rev. 2012, 87, 52–71. [Google Scholar] [CrossRef] [PubMed]
- Merz, U.; Falloon, R.E. Review: Powdery Scab of Potato—Increased Knowledge of Pathogen Biology and Disease Epidemiology for Effective Disease Management. Potato Res. 2009, 52, 17–37. [Google Scholar] [CrossRef]
- Nayyar, A.; Hamel, C.; Lafond, G.; Gossen, B.D.; Hanson, K.; Germida, J. Soil microbial quality associated with yield reduction in continuous-pea. Appl. Soil Ecol. 2009, 43, 115–121. [Google Scholar] [CrossRef]
- Da Costa, P.B.; Beneduzi, A.; de Souza, R.; Schoenfeld, R.; Vargas, L.K.; Passaglia, L.M.P. The effects of different fertilization conditions on bacterial plant growth promoting traits: Guidelines for directed bacterial prospection and testing. Plant Soil 2013, 368, 267–280. [Google Scholar] [CrossRef]
- Singh, B.K.; Trivedi, P. Microbiome and the future for food and nutrient security. Microb. Biotechnol. 2017, 10, 50–53. [Google Scholar] [CrossRef]
- EPA. Table 2-8 Inventory of U.S. Greenhouse Gas Emissions and Sinks, 1990 to 2020. EPA Report; 2020. Available online: https://www.epa.gov/ghgemissions/inventory-us-greenhouse-gas-emissions-and-sinks-1990-2019 (accessed on 13 February 2023).
- Diaz, R.J.; Rosenberg, R. Spreading Dead Zones and Consequences for Marine Ecosystems. Science 2008, 321, 926–929. [Google Scholar] [CrossRef] [PubMed]
- Good, A.G.; Beatty, P.H. Fertilizing nature: A tragedy of excess in the commons. PLoS Biol. 2011, 9, e1001124. [Google Scholar] [CrossRef]
- Inselsbacher, E.; Wanek, W.; Ripka, K.; Hackl, E.; Sessitsch, A.; Strauss, J.; Zechmeister-Boltenstern, S. Greenhouse gas fluxes respond to different N fertilizer types due to altered plant-soil-microbe interactions. Plant Soil 2011, 343, 17–35. [Google Scholar] [CrossRef]
- Miransari, M. Soil microbes and plant fertilization. Appl. Microbiol. Biotechnol. 2011, 92, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Oberson, A.; Culvenor, R.A.; Ryan, M.H.; Veneklaas, E.J.; Lambers, H.; Lynch, J.P.; Ryan, P.R.; Delhaize, E.; Smith, F.A.; et al. Strategies and agronomic interventions to improve the phosphorus-use efficiency of farming systems. Plant Soil 2011, 349, 89–120. [Google Scholar] [CrossRef]
- Zhao, Z.B.; He, J.Z.; Geisen, S.; Han, L.L.; Wang, J.T.; Shen, J.P.; Wei, W.X.; Fang, Y.T.; Li, P.P.; Zhang, L.M. Protist communities are more sensitive to nitrogen fertilization than other microorganisms in diverse agricultural soils. Microbiome 2019, 7, 33. [Google Scholar] [CrossRef]
- Zhao, Z.B.; He, J.Z.; Quan, Z.; Wu, C.F.; Sheng, R.; Zhang, L.M.; Geisen, S. Fertilization changes soil microbiome functioning, especially phagotrophic protists. Soil Biol. Biochem. 2020, 148, 107863. [Google Scholar] [CrossRef]
- Van der Bom, F.; Nunes, I.; Raymond, N.S.; Hansen, V.; Bonnichsen, L.; Magid, J.; Nybroe, O.; Jensen, L.S. Long-term fertilisation form, level and duration affect the diversity, structure and functioning of soil microbial communities in the field. Soil Biol. Biochem. 2018, 122, 91–103. [Google Scholar] [CrossRef]
- Huang, H.; Thu, T.N.T.; He, X.; Gravot, A.; Bernillon, S.; Ballini, E.; Morel, J.-B. Increase of fungal pathogenicity and role of plant glutamine in nitrogen-induced susceptibility (NIS) to rice blast. Front. Plant Sci. 2017, 8, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonanomi, G.; Alioto, D.; Minutolo, M.; Marra, R.; Cesarano, G.; Vinale, F. Organic Amendments Modulate Soil Microbiota and Reduce Virus Disease Incidence in the TSWV-Tomato Pathosystem. Pathogens 2020, 9, 379. [Google Scholar] [CrossRef]
- Soulie, M.C.; Koka, S.M.; Floch, K.; Vancostenoble, B.; Barbe, D.; Daviere, A.; Soubigou-Taconnat, L.; Brunaud, V.; Poussereau, N.; Loisel, E.; et al. Plant nitrogen supply affects the Botrytis cinerea infection process and modulates known and novel virulence factors. Mol. Plant Pathol. 2020, 21, 1436–1450. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, F.; Tang, L.; Zheng, Y.; Li, Y.; Christie, P.; Li, L. Wheat powdery mildew and foliar N concentrations as influenced by N fertilization and belowground interactions with intercropped faba bean. Plant Soil 2007, 291, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Devadas, R.; Simpfendorfer, S.; Backhouse, D.; Lamb, D.W. Effect of stripe rust on the yield response of wheat to nitrogen. Crop J. 2014, 2, 201–206. [Google Scholar] [CrossRef] [Green Version]
- Xiong, C.; He, J.Z.; Singh, B.K.; Zhu, Y.G.; Wang, J.T.; Li, P.P.; Zhang, Q.B.; Han, L.L.; Shen, J.P.; Ge, A.H.; et al. Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environ. Microbiol. 2021, 23, 1907–1924. [Google Scholar] [CrossRef]
- Dordas, C. Role of nutrients in controlling plant diseases in sustainable agriculture. A review. Agron. Sustain. Dev. 2008, 28, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Wang, M.; Mur, L.A.J.; Shen, Q.; Guo, S. Unravelling the Roles of Nitrogen Nutrition in Plant Disease Defences. Int. J. Mol. Sci. 2020, 21, 572. [Google Scholar] [CrossRef] [Green Version]
- Tao, C.; Li, R.; Xiong, W.; Shen, Z.; Liu, S.; Wang, B.; Ruan, Y.; Geisen, S.; Shen, Q.; Kowalchuk, G.A. Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 2020, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Medina-Sauza, R.M.; Álvarez-Jiménez, M.; Delhal, A.; Reverchon, F.; Blouin, M.; Guerrero-Analco, J.A.; Cerdán, C.R.; Guevara, R.; Villain, L.; Barois, I. Earthworms Building Up Soil Microbiota, a Review. Front. Environ. Sci. 2019, 7, 81. [Google Scholar] [CrossRef] [Green Version]
- Xiong, W.; Guo, S.; Jousset, A.; Zhao, Q.; Wu, H.; Li, R.; Kowalchuk, G.A.; Shen, Q. Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol. Biochem. 2017, 114, 238–247. [Google Scholar] [CrossRef]
- Garcia-Franco, N.; Albaladejo, J.; Almagro, M.; Martínez-Mena, M. Beneficial effects of reduced tillage and green manure on soil aggregation and stabilization of organic carbon in a Mediterranean agroecosystem. Soil Tillage Res. 2015, 153, 66–75. [Google Scholar] [CrossRef]
- Hati, K.M.; Swarup, A.; Mishra, B.; Manna, M.C.; Wanjari, R.H.; Mandal, K.G.; Misra, A.K. Impact of long-term application of fertilizer, manure and lime under intensive cropping on physical properties and organic carbon content of an Alfisol. Geoderma 2008, 148, 173–179. [Google Scholar] [CrossRef]
- Bhunia, S.; Bhowmik, A.; Mallick, R.; Mukherjee, J. Agronomic Efficiency of Animal-Derived Organic Fertilizers and Their Effects on Biology and Fertility of Soil: A Review. Agronomy 2021, 11, 823. [Google Scholar] [CrossRef]
- Liang, Y.; Yang, Y.; Yang, C.; Shen, Q.; Zhou, J.; Yang, L. Soil enzymatic activity and growth of rice and barley as influenced by organic manure in an anthropogenic soil. Geoderma 2003, 115, 149–160. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, D.; Cheng, H.; Ren, L.; Jin, X.; Fang, W.; Yan, D.; Li, Y.; Wang, Q.; Cao, A. Organic fertilizers activate soil enzyme activities and promote the recovery of soil beneficial microorganisms after dazomet fumigation. J. Environ. Manag. 2022, 309, 114666. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.-C.; Li, H.-Y.; Lin, Z.-A.; Zhao, B.-Q.; Sun, Z.-B.; Yuan, L.; Xu, J.-K.; Li, Y.-Q. Long-term fertilization alters soil properties and fungal community composition in fluvo-aquic soil of the North China Plain. Sci. Rep. 2020, 10, 7198. [Google Scholar] [CrossRef]
- Noble, R. Risks and benefits of soil amendment with composts in relation to plant pathogens. Australas. Plant Pathol. 2011, 40, 157–167. [Google Scholar] [CrossRef]
- Blaya, J.; Marhuenda, F.C.; Pascual, J.A.; Ros, M. Microbiota characterization of compost using omics approaches opens new perspectives for Phytophthora root rot control. PLoS ONE 2016, 11, e0158048. [Google Scholar] [CrossRef] [Green Version]
- Termorshuizen, A.J.; van Rijn, E.; van der Gaag, D.J.; Alabouvette, C.; Chen, Y.; Lagerlöf, J.; Malandrakis, A.A.; Paplomatas, E.J.; Rämert, B.; Ryckeboer, J.; et al. Suppressiveness of 18 composts against 7 pathosystems: Variability in pathogen response. Soil Biol. Biochem. 2006, 38, 2461–2477. [Google Scholar] [CrossRef]
- Larkin, R.P. Green manures and plant disease management. CABI Rev. 2013, 8, 1–10. [Google Scholar] [CrossRef]
- Chen, Q.L.; Cui, H.L.; Su, J.Q.; Penuelas, J.; Zhu, Y.G. Antibiotic Resistomes in Plant Microbiomes. Trends Plant Sci. 2019, 24, 530–541. [Google Scholar] [CrossRef]
- Udikovic-Kolic, N.; Wichmann, F.; Broderick, N.A.; Handelsman, J. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. USA 2014, 111, 15202–15207. [Google Scholar] [CrossRef]
- Marutescu, L.G.; Jaga, M.; Postolache, C.; Barbuceanu, F.; Milita, N.M.; Romascu, L.M.; Schmitt, H.; de Roda Husman, A.M.; Sefeedpari, P.; Glaeser, S.; et al. Insights into the impact of manure on the environmental antibiotic residues and resistance pool. Front. Microbiol. 2022, 13, 3489. [Google Scholar] [CrossRef]
- Hahn, M. The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J. Chem. Biol. 2014, 7, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, N.J.; Bass, C.; Dixon, A.; Neve, P. The evolutionary origins of pesticide resistance. Biol. Rev. 2019, 94, 135–155. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Zhao, J.; Jiang, J.; Chen, S.; Guan, Z.; Chen, F.; Fang, W.; Zhao, S. Assessing the influence of fumigation and bacillus subtilis-based biofungicide on the microbiome of chrysanthemum rhizosphere. Agriculture 2019, 9, 255. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Zhao, S.; Zhang, K.; Zhao, J.; Jiang, J.; Chen, F.; Fang, W. Evaluation of soil-applied chemical fungicide and biofungicide for control of the fusarium wilt of chrysanthemum and their effects on rhizosphere soil microbiota. Agriculture 2018, 8, 184. [Google Scholar] [CrossRef] [Green Version]
- Markarova, A.E.; Markarova, M.Y.; Razin, O.A.; Nadezhkin, S.M. The microorganisms natural consortia effectiveness in the white cabbage crop cultivation. IOP Conf. Ser. Earth Environ. Sci. 2022, 953, 012035. [Google Scholar] [CrossRef]
- Noel, Z.A.; Longley, R.; Benucci, G.M.N.; Trail, F.; Chilvers, M.I.; Bonito, G. Non-target impacts of fungicide disturbance on phyllosphere yeasts in conventional and no-till management. ISME Commun. 2022, 2, 19. [Google Scholar] [CrossRef]
- Pang, G.; Cai, F.; Li, R.; Zhao, Z.; Li, R.; Gu, X.; Shen, Q.; Chen, W. Trichoderma-enriched organic fertilizer can mitigate microbiome degeneration of monocropped soil to maintain better plant growth. Plant Soil 2017, 416, 181–192. [Google Scholar] [CrossRef]
- Beesley, L.; Moreno-Jiménez, E.; Gomez-Eyles, J.L. Effects of biochar and greenwaste compost amendments on mobility, bioavailability and toxicity of inorganic and organic contaminants in a multi-element polluted soil. Environ. Pollut. 2010, 158, 2282–2287. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, B.; Zhu, L.; Xing, B. Effects and mechanisms of biochar-microbe interactions in soil improvement and pollution remediation: A review. Environ. Pollut. 2017, 227, 98–115. [Google Scholar] [CrossRef]
- Jaiswal, A.K.; Elad, Y.; Graber, E.R.; Frenkel, O. Rhizoctonia solani suppression and plant growth promotion in cucumber as affected by biochar pyrolysis temperature, feedstock and concentration. Soil Biol. Biochem. 2014, 69, 110–118. [Google Scholar] [CrossRef]
- Jaiswal, A.K.; Elad, Y.; Paudel, I.; Graber, E.R.; Cytryn, E.; Frenkel, O. Linking the Belowground Microbial Composition, Diversity and Activity to Soilborne Disease Suppression and Growth Promotion of Tomato Amended with Biochar. Sci. Rep. 2017, 7, 44382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, A.K.; Frenkel, O.; Elad, Y.; Lew, B.; Graber, E.R. Non-monotonic influence of biochar dose on bean seedling growth and susceptibility to Rhizoctonia solani: The “Shifted Rmax-Effect”. Plant Soil 2015, 395, 125–140. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Z.; Yang, K.; Wang, P.; Wang, H.; Guo, L.; Zhu, S.; Zhu, Y.; He, X. Biochar Application Alleviated Negative Plant-Soil Feedback by Modifying Soil Microbiome. Front. Microbiol. 2020, 11, 799. [Google Scholar] [CrossRef]
- Bakker, P.A.H.M.; Doornbos, R.F.; Zamioudis, C.; Berendsen, R.L.; Pieterse, C.M.J. Induced systemic resistance and the rhizosphere microbiome. Plant Pathol. J. 2013, 29, 136–143. [Google Scholar] [CrossRef] [Green Version]
- Mehari, Z.H.; Elad, Y.; Rav-David, D.; Graber, E.R.; Meller Harel, Y. Induced systemic resistance in tomato (Solanum lycopersicum) against Botrytis cinerea by biochar amendment involves jasmonic acid signaling. Plant Soil 2015, 395, 31–44. [Google Scholar] [CrossRef]
- Samain, E.; Aussenac, T.; Selim, S. The Effect of Plant Genotype, Growth Stage, and Mycosphaerella graminicola Strains on the Efficiency and Durability of Wheat-Induced Resistance by Paenibacillus sp. Strain B2. Front. Plant Sci. 2019, 10, 587. [Google Scholar] [CrossRef] [Green Version]
- Samain, E.; van Tuinen, D.; Jeandet, P.; Aussenac, T.; Selim, S. Biological control of septoria leaf blotch and growth promotion in wheat by Paenibacillus sp. strain B2 and Curtobacterium plantarum strain EDS. Biol. Control 2017, 114, 87–96. [Google Scholar] [CrossRef]
- Brtnicky, M.; Datta, R.; Holatko, J.; Bielska, L.; Gusiatin, Z.M.; Kucerik, J.; Hammerschmiedt, T.; Danish, S.; Radziemska, M.; Mravcova, L.; et al. A critical review of the possible adverse effects of biochar in the soil environment. Sci. Total Environ. 2021, 796, 148756. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.; Hussain, Q.; Khan, K.S.; Al-Wabel, M.I.; Afeng, Z.; Akmal, M.; Ijaz, S.S.; Aziz, R.; Shah, G.A.; Mehdi, S.M.; et al. Prospects of biochar in alkaline soils to mitigate climate change. In Environment, Climate, Plant and Vegetation Growth; Springer: Cham, Switzerland, 2020; pp. 133–149. [Google Scholar]
- Brassard, P.; Godbout, S.; Raghavan, V. Soil biochar amendment as a climate change mitigation tool: Key parameters and mechanisms involved. J. Environ. Manag. 2016, 181, 484–497. [Google Scholar] [CrossRef]
- Chu-hsi, H.; Jui-lien, H.; Rong-huei, C. Wastewater Treatment with Chitosan. 2008. Available online: https://www.semanticscholar.org/paper/Wastewater-treatment-with-chitosan-Chu-hsi-Jui-lien/d1624a03c425324c247a5d772b117612d7255a27 (accessed on 15 February 2023).
- Hernandez-Lauzardo, A.N.; Bautista-Baños, S.; Velazquez-Del Valle, M.G.; Méndez-Montealvo, M.; Sánchez-Rivera, M.; Bello-Perez, L.A. Antifungal effects of chitosan with different molecular weights on in vitro development of Rhizopus stolonifer (Ehrenb.: Fr.) Vuill. Carbohydr. Polym. 2008, 73, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Stiling, P.; Cornelissen, T. What makes a successful biocontrol agent? A meta-analysis of biological control agent performance. Biol. Control 2005, 34, 236–246. [Google Scholar] [CrossRef]
- El Ghaouth, A.; Arul, J.; Grenier, J.; Asselin, A. Effect of chitosan and other polyions on chitin deacetylase in Rhizopus stolonifer. Exp. Mycol. 1992, 16, 173–177. [Google Scholar] [CrossRef]
- Cretoiu, M.S.; Korthals, G.W.; Visser, J.H.M.; van Elsas, J.D. Chitin Amendment Increases Soil Suppressiveness toward Plant Pathogens and Modulates the Actinobacterial and Oxalobacteraceal Communities in an Experimental Agricultural Field. Appl. Environ. Microbiol. 2013, 79, 5291–5301. [Google Scholar] [CrossRef] [Green Version]
- Chisnall Hampson, M.; Coombes, J.W. Use of crabsheli meal to control potato wart in Newfoundland. Can. J. Plant Pathol. 1991, 13, 97–105. [Google Scholar] [CrossRef]
- Mi, J.; Gregorich, E.G.; Xu, S.; McLaughlin, N.B.; Liu, J. Effect of bentonite as a soil amendment on field water-holding capacity, and millet photosynthesis and grain quality. Sci. Rep. 2020, 10, 18282. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Xu, Y.; Liang, X.; Wang, L. In situ stabilization remediation of cadmium (Cd) and lead (Pb) co-contaminated paddy soil using bentonite. Appl. Clay Sci. 2015, 105–106, 200–206. [Google Scholar] [CrossRef]
- Xu, Y.; Liang, X.; Xu, Y.; Qin, X.; Huang, Q.; Wang, L.; Sun, Y. Remediation of Heavy Metal-Polluted Agricultural Soils Using Clay Minerals: A Review. Pedosphere 2017, 27, 193–204. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, W.; Zhao, B.; Phillips, L.A.; Zhou, Y.; Lapen, D.R.; Liu, J. Sandy soils amended with bentonite induced changes in soil microbiota and fungistasis in maize fields. Appl. Soil Ecol. 2020, 146, 103378. [Google Scholar] [CrossRef]
- CFIA. Import and Release of Biological Control Agents into Canada. Available online: https://inspection.canada.ca/plant-health/invasive-species/biological-control-agents/eng/1514956211166/1514956212112 (accessed on 19 February 2023).
- He, D.C.; He, M.H.; Amalin, D.M.; Liu, W.; Alvindia, D.G.; Zhan, J. Biological Control of Plant Diseases: An Evolutionary and Eco-Economic Consideration. Pathogens 2021, 10, 1311. [Google Scholar] [CrossRef]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological Control of Plant Pathogens: A Global Perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.; Ianiri, G.; Del Grosso, C.; Barone, G.; De Curtis, F.; Castoria, R.; Lima, G. Advances and Perspectives in the Use of Biocontrol Agents against Fungal Plant Diseases. Horticulturae 2022, 8, 577. [Google Scholar] [CrossRef]
- Wagemans, J.; Holtappels, D.; Vainio, E.; Rabiey, M.; Marzachì, C.; Herrero, S.; Ravanbakhsh, M.; Tebbe, C.C.; Ogliastro, M.; Ayllón, M.A.; et al. Going Viral: Virus-Based Biological Control Agents for Plant Protection. Annu. Rev. Phytopathol. 2022, 60, 21–42. [Google Scholar] [CrossRef]
- Agriculture, F.M.O. Qu’est-ce que le Biocontrôle? Available online: https://agriculture.gouv.fr/quest-ce-que-le-biocontrole (accessed on 22 May 2023).
- Tymon, L.S.; Morgan, P.; Gundersen, B.; Inglis, D.A. Potential of endophytic fungi collected from Cucurbita pepo roots grown under three different agricultural mulches as antagonistic endophytes to Verticillium dahliae in western Washington. Microbiol. Res. 2020, 240, 126535. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Copolovici, D.; Copolovici, L.; Teder, T.; Nevo, E.; Behers, L. Paenibacillus polymyxa biofilm polysaccharides antagonise Fusarium graminearum. Sci. Rep. 2019, 9, 662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abd El Daim, I.A.; Häggblom, P.; Karlsson, M.; Stenström, E.; Timmusk, S. Paenibacillus polymyxa A26 Sfp-type PPTase inactivation limits bacterial antagonism against Fusarium graminearum but not of F. culmorum in kernel assay. Front. Plant Sci. 2015, 6, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moussa, T.A.A.; Almaghrabi, O.A.; Abdel-Moneim, T.S. Biological control of the wheat root rot caused by Fusarium graminearum using some PGPR strains in Saudi Arabia. Ann. Appl. Biol. 2013, 163, 72–81. [Google Scholar] [CrossRef]
- Walsh, U.F.; Morrissey, J.P.; O’Gara, F. Pseudomonas for biocontrol of phytopathogens: From functional genomics to commercial exploitation. Curr. Opin. Biotechnol. 2001, 12, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Afridi, M.S.; Javed, M.A.; Ali, S.; De Medeiros, F.H.V.; Ali, B.; Salam, A.; Sumaira; Marc, R.A.; Alkhalifah, D.H.M.; Selim, S.; et al. New opportunities in plant microbiome engineering for increasing agricultural sustainability under stressful conditions. Front. Plant Sci. 2022, 13, 899464. [Google Scholar] [CrossRef] [PubMed]
- Perrone, G.; Ferrara, M.; Medina, A.; Pascale, M.; Magan, N. Toxigenic fungi and mycotoxins in a climate change scenario: Ecology, genomics, distribution, prediction and prevention of the risk. Microorganisms 2020, 8, 1496. [Google Scholar] [CrossRef]
- Samaras, A.; Nikolaidis, M.; Antequera-Gómez, M.L.; Cámara-Almirón, J.; Romero, D.; Moschakis, T.; Amoutzias, G.D.; Karaoglanidis, G.S. Whole genome sequencing and root colonization studies reveal novel insights in the biocontrol potential and growth promotion by Bacillus subtilis MBI 600 on cucumber. Front. Microbiol. 2021, 11, 600393. [Google Scholar] [CrossRef]
- Wu, X.; Shan, Y.; Li, Y.; Li, Q.; Wu, C. The soil nutrient environment determines the strategy by which Bacillus velezensis HN03 suppresses Fusarium wilt in banana plants. Front. Plant Sci. 2020, 11, 599904. [Google Scholar] [CrossRef]
- Park, A.R.; Jeong, S.-I.; Jeon, H.W.; Kim, J.; Kim, N.; Ha, M.T.; Mannaa, M.; Kim, J.; Lee, C.W.; Min, B.S.; et al. A diketopiperazine, cyclo-(L-Pro-L-Ile), derived from Bacillus thuringiensis JCK-1233 controls pine wilt disease by elicitation of moderate hypersensitive reaction. Front. Plant Sci. 2020, 11, 1023. [Google Scholar] [CrossRef]
- Wang, S.-Y.; Herrera-Balandrano, D.D.; Wang, Y.-X.; Shi, X.-C.; Chen, X.; Jin, Y.; Liu, F.-Q.; Laborda, P. Biocontrol Ability of the Bacillus amyloliquefaciens Group, B. amyloliquefaciens, B. velezensis, B. nakamurai, and B. siamensis, for the Management of Fungal Postharvest Diseases: A Review. J. Agric. Food Chem. 2022, 70, 6591–6616. [Google Scholar] [CrossRef] [PubMed]
- Prahl, R.E.; Khan, S.; Deo, R.C. Ampelomyces mycoparasites of powdery mildews—A review. Can. J. Plant Pathol. 2023, 1–14. [Google Scholar] [CrossRef]
- Ownley, B.H.; Griffin, M.R.; Klingeman, W.E.; Gwinn, K.D.; Moulton, J.K.; Pereira, R.M. Beauveria bassiana: Endophytic colonization and plant disease control. J. Invertebr. Pathol. 2008, 98, 267–270. [Google Scholar] [CrossRef]
- Rodríguez-González, Á.; Carro-Huerga, G.; Mayo-Prieto, S.; Lorenzana, A.; Gutiérrez, S.; Peláez, H.J.; Casquero, P.A. Investigations of Trichoderma spp. and Beauveria bassiana as biological control agent for Xylotrechus arvicola, a major insect pest in Spanish vineyards. J. Econ. Entomol. 2018, 111, 2585–2591. [Google Scholar]
- Liu, Y.; Yang, Y.; Wang, B. Entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae play roles of maize (Zea mays) growth promoter. Sci. Rep. 2022, 12, 15706. [Google Scholar] [CrossRef] [PubMed]
- Babalola, O.O. Pectinase and cellulase enhance the control of Abutilon theophrasti by Colletotrichum coccodes. Biocontrol Sci. Technol. 2007, 17, 53–61. [Google Scholar] [CrossRef]
- De Vrije, T.; Antoine, N.; Buitelaar, R.M.; Bruckner, S.; Dissevelt, M.; Durand, A.; Gerlagh, M.; Jones, E.E.; Lüth, P.; Oostra, J.; et al. The fungal biocontrol agent Coniothyrium minitans: Production by solid-state fermentation, application and marketing. Appl. Microbiol. Biotechnol. 2001, 56, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhou, L.; Din, I.U.; Arafat, Y.; Li, Q.; Wang, J.; Wu, T.; Wu, L.; Wu, H.; Qin, X.; et al. Antagonistic activity of Trichoderma spp. against Fusarium oxysporum in rhizosphere of Radix pseudostellariae triggers the expression of host defense genes and improves its growth under long-term monoculture system. Front. Microbiol. 2021, 12, 579920. [Google Scholar] [CrossRef]
- Yang, X.; Chen, L.; Yong, X.; Shen, Q. Formulations can affect rhizosphere colonization and biocontrol efficiency of Trichoderma harzianum SQR-T037 against Fusarium wilt of cucumbers. Biol. Fertil. Soils 2011, 47, 239–248. [Google Scholar] [CrossRef]
- Cotxarrera, L.; Trillas-Gay, M.; Steinberg, C.; Alabouvette, C. Use of sewage sludge compost and Trichoderma asperellum isolates to suppress Fusarium wilt of tomato. Soil Biol. Biochem. 2002, 34, 467–476. [Google Scholar] [CrossRef]
- Vayssier-Taussat, M.; Albina, E.; Citti, C.; Cosson, J.-F.; Jacques, M.-A.; Lebrun, M.; Le Loir, Y.; Ogliastro, M.; Petit, M.-A.; Roumagnac, P.; et al. Shifting the paradigm from pathogens to pathobiome: New concepts in the light of meta-omics. Front. Cell. Infect. Microbiol. 2014, 4, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bass, D.; Stentiford, G.; Wang, H.-C.; Koskella, B.; Tyler, C. The Pathobiome in Animal and Plant Diseases. Trends Ecol. Evol. 2019, 34, 996–1008. [Google Scholar] [CrossRef] [Green Version]
- Mannaa, M.; Seo, Y.-S. Plants under the Attack of Allies: Moving towards the Plant Pathobiome Paradigm. Plants 2021, 10, 125. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.E.; Kim, M.-S.; Lalande, B.; Klopfenstein, N.B. Chapter 15—Pathobiome and microbial communities associated with forest tree root diseases. In Forest Microbiology; Asiegbu, F.O., Kovalchuk, A., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 277–292. [Google Scholar] [CrossRef]
- Droby, S.; Zhimo, V.Y.; Wisniewski, M.; Freilich, S. The pathobiome concept applied to postharvest pathology and its implication on biocontrol strategies. Postharvest Biol. Technol. 2022, 189, 111911. [Google Scholar] [CrossRef]
- Lamelas, A.; Desgarennes, D.; López-Lima, D.; Villain, L.; Alonso-Sánchez, A.; Artacho, A.; Latorre, A.; Moya, A.; Carrión, G. The Bacterial Microbiome of Meloidogyne-Based Disease Complex in Coffee and Tomato. Front. Plant Sci. 2020, 11, 136. [Google Scholar] [CrossRef]
- Doonan, J.; Broberg, M.; Denman, S.; McDonald, J. Host–microbiota–insect interactions drive emergent virulence in a complex tree disease. Proc. R. Soc. B Biol. Sci. 2020, 287, 20200956. [Google Scholar] [CrossRef] [PubMed]
- Bruez, E.; Vallance, J.; Gautier, A.; Laval, V.; Compant, S.; Maurer, W.; Sessitsch, A.; Lebrun, M.-H.; Rey, P. Major changes in grapevine wood microbiota are associated with the onset of esca, a devastating trunk disease. Environ. Microbiol. 2020, 22, 5189–5206. [Google Scholar] [CrossRef]
- Haidar, R.; Yacoub, A.; Pinard, A.; Roudet, J.; Fermaud, M.; Rey, P. Synergistic effects of water deficit and wood- inhabiting bacteria on pathogenicity of the grapevine trunk pathogen Neofusicoccum parvum. Phytopathol. Mediterr. 2021, 59, 473–484. [Google Scholar] [CrossRef]
- Haidar, R.; Yacoub, A.; Vallance, J.; Compant, S.; Antonielli, L.; Saad, A.; Habenstein, B.; Kauffmann, B.; Grélard, A.; Loquet, A.; et al. Bacteria associated with wood tissues of Esca-diseased grapevines: Functional diversity and synergy with Fomitiporia mediterranea to degrade wood components. Environ. Microbiol. 2021, 23, 6104–6121. [Google Scholar] [CrossRef] [PubMed]
- Cobo-Díaz, J.F.; Baroncelli, R.; Le Floch, G.; Picot, A. Combined Metabarcoding and Co-occurrence Network Analysis to Profile the Bacterial, Fungal and Fusarium Communities and Their Interactions in Maize Stalks. Front. Microbiol. 2019, 10, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauvert, C.; Fort, T.; Calonnec, A.; Faivre d’Arcier, J.; Chancerel, E.; Massot, M.; Chiquet, J.; Robin, S.; Bohan, D.A.; Vallance, J. Microbial association networks give relevant insights into plant pathobiomes. BioRxiv 2020. [Google Scholar] [CrossRef]
- Qiu, Z.; Verma, J.P.; Liu, H.; Wang, J.; Batista, B.D.; Kaur, S.; de Araujo Pereira, A.P.; Macdonald, C.A.; Trivedi, P.; Weaver, T.; et al. Response of the plant core microbiome to Fusarium oxysporum infection and identification of the pathobiome. Environ. Microbiol. 2022, 24, 4652–4669. [Google Scholar] [CrossRef]
- Goberna, M.; Verdú, M. Cautionary notes on the use of co-occurrence networks in soil ecology. Soil Biol. Biochem. 2022, 166, 108534. [Google Scholar] [CrossRef]
- Picot, A.; Doster, M.; Islam, M.-S.; Callicott, K.; Ortega-Beltran, A.; Cotty, P.; Michailides, T. Distribution and incidence of atoxigenic Aspergillus flavus VCG in tree crop orchards in California: A strategy for identifying potential antagonists, the example of almonds. Int. J. Food Microbiol. 2018, 265, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Deroo, W.; De Troyer, L.; Dumoulin, F.; De Saeger, S.; De Boevre, M.; Vandenabeele, S.; De Gelder, L.; Audenaert, K. A Novel In Planta Enrichment Method Employing Fusarium graminearum-Infected Wheat Spikes to Select for Competitive Biocontrol Bacteria. Toxins 2022, 14, 222. [Google Scholar] [CrossRef]
- Gnonlonfoun, E.; Fotin, G.; Risler, A.; Elfassy, A.; Schwebel, S.; Schmitt, M.; Borges, F.; Mangavel, C.; Revol-Junelles, A.-M.; Fick, M.; et al. Inhibition of the Growth of Fusarium tricinctum and Reduction of Its Enniatin Production by Erwinia gerundensis Isolated from Barley Kernels. J. Am. Soc. Brew. Chem. 2023, 81, 340–350. [Google Scholar] [CrossRef]
- Gerbore, J.; Benhamou, N.; Vallance, J.; Le Floch, G.; Grizard, D.; Regnault-Roger, C.; Rey, P. Biological control of plant pathogens: Advantages and limitations seen through the case study of Pythium oligandrum. Environ. Sci. Pollut. Res. 2014, 21, 4847–4860. [Google Scholar] [CrossRef]
- Cobo-Díaz, J.F.; Baroncelli, R.; Le Floch, G.; Picot, A. A novel metabarcoding approach to investigate Fusarium species composition in soil and plant samples. FEMS Microbiol. Ecol. 2019, 95, fiz084. [Google Scholar] [CrossRef]
- Boutigny, A.-L.; Gautier, A.; Basler, R.; Dauthieux, F.; Leite, S.; Valade, R.; Aguayo, J.; Ioos, R.; Laval, V. Metabarcoding targeting the EF1 alpha region to assess Fusarium diversity on cereals. PLoS ONE 2019, 14, e0207988. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Radford, D.; Hambleton, S. Towards improved detection and identification of rust fungal pathogens in environmental samples using a metabarcoding approach. Phytopathology 2022, 112, 535–548. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Bolchacova, E.; Voigt, K.; Crous, P.W.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Tiwari, K.K.; Chaudhary, K.; Pratap, D. Genic molecular markers in fungi: Availability and utility for bioprospection. In Molecular Markers in Mycology: Diagnostics and Marker Developments; Springer: Cham, Switzerland, 2017; pp. 151–176. [Google Scholar]
- Li, S.; Deng, Y.; Wang, Z.; Zhang, Z.; Kong, X.; Zhou, W.; Yi, Y.; Qu, Y. Exploring the accuracy of amplicon-based internal transcribed spacer markers for a fungal community. Mol. Ecol. Resour. 2020, 20, 170–184. [Google Scholar] [CrossRef]
- Aime, M.C.; McTaggart, A.R.; Mondo, S.J.; Duplessis, S. Phylogenetics and phylogenomics of rust fungi. Adv. Genet. 2017, 100, 267–307. [Google Scholar]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef] [Green Version]
- Belair, M.; Pensec, F.; Jany, J.-L.; Le Floch, G.; Picot, A. Profiling Walnut Fungal Pathobiome Associated with Walnut Dieback Using Community-Targeted DNA Metabarcoding. Plants 2023, 12, 2383. [Google Scholar] [CrossRef]
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, rapidly, OTUs with galaxy solution. Bioinformatics 2018, 34, 1287–1294. [Google Scholar] [CrossRef] [Green Version]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, V.G. Pitfalls in relative abundance estimation using eDNA metabarcoding. Mol. Ecol. Resour. 2018, 18, 923–926. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Wang, S.; Du, X.; He, Q.; Liu, Y.; Wang, Z.; Feng, K.; Li, Y.; Deng, Y. ddPCR surpasses classical qPCR technology in quantitating bacteria and fungi in the environment. Mol. Ecol. Resour. 2022, 22, 2587–2598. [Google Scholar] [CrossRef] [PubMed]
- Tkacz, A.; Hortala, M.; Poole, P.S. Absolute quantitation of microbiota abundance in environmental samples. Microbiome 2018, 6, 110. [Google Scholar] [CrossRef] [Green Version]
- Zemb, O.; Achard, C.S.; Hamelin, J.; De Almeida, M.-L.; Gabinaud, B.; Cauquil, L.; Verschuren, L.M.G.; Godon, J.-J. Absolute quantitation of microbes using 16S rRNA gene metabarcoding: A rapid normalization of relative abundances by quantitative PCR targeting a 16S rRNA gene spike-in standard. MicrobiologyOpen 2020, 9, e977. [Google Scholar] [CrossRef] [PubMed]
- Sevim, V.; Lee, J.; Egan, R.; Clum, A.; Hundley, H.; Lee, J.; Everroad, R.C.; Detweiler, A.M.; Bebout, B.M.; Pett-Ridge, J.; et al. Shotgun metagenome data of a defined mock community using Oxford Nanopore, PacBio and Illumina technologies. Sci. Data 2019, 6, 285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, J.; Alm, E.J. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [Green Version]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, M.; Xiong, C.; Gao, C.; Tsui, C.K.; Wang, M.-M.; Zhou, X.; Zhang, A.-M.; Cai, L.J.M. Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome 2021, 9, 187. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing Causality: Opportunities of Synthetic Communities for Plant Microbiome Research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef]
- Qiu, Z.; Egidi, E.; Liu, H.; Kaur, S.; Singh, B.K. New frontiers in agriculture productivity: Optimised microbial inoculants and in situ microbiome engineering. Biotechnol. Adv. 2019, 37, 107371. [Google Scholar] [CrossRef] [PubMed]
- Mawarda, P.C.; Le Roux, X.; Dirk van Elsas, J.; Salles, J.F. Deliberate introduction of invisible invaders: A critical appraisal of the impact of microbial inoculants on soil microbial communities. Soil Biol. Biochem. 2020, 148, 107874. [Google Scholar] [CrossRef]
- Jack, C.N.; Petipas, R.H.; Cheeke, T.E.; Rowland, J.L.; Friesen, M.L. Microbial inoculants: Silver bullet or microbial Jurassic Park? Trends Microbiol. 2021, 29, 299–308. [Google Scholar] [CrossRef]
- Liu, X.; Roux, X.; Salles, J. The Legacy of Microbial Inoculants in Agroecosystems and Potential for Tackling Climate Change Challenges. iScience 2022, 25, 103821. [Google Scholar] [CrossRef]
- Moore, J.A.M.; Abraham, P.E.; Michener, J.K.; Muchero, W.; Cregger, M.A. Ecosystem consequences of introducing plant growth promoting rhizobacteria to managed systems and potential legacy effects. New Phytol. 2022, 234, 1914–1918. [Google Scholar] [CrossRef]
- Heimpel, G.E.; Cock, M.J.W. Shifting paradigms in the history of classical biological control. BioControl 2018, 63, 27–37. [Google Scholar] [CrossRef]
- Ehrlich, K. Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: Advantages and limitations. Front. Microbiol. 2014, 5, 50. [Google Scholar] [CrossRef]
- Olarte, R.A.; Horn, B.W.; Dorner, J.W.; Monacell, J.T.; Singh, R.; Stone, E.A.; Carbone, I. Effect of sexual recombination on population diversity in aflatoxin production by Aspergillus flavus and evidence for cryptic heterokaryosis. Mol. Ecol. 2012, 21, 1453–1476. [Google Scholar] [CrossRef] [PubMed]
- Molo, M.S.; White, J.B.; Cornish, V.; Gell, R.M.; Baars, O.; Singh, R.; Carbone, M.A.; Isakeit, T.; Wise, K.A.; Woloshuk, C.P.; et al. Asymmetrical lineage introgression and recombination in populations of Aspergillus flavus: Implications for biological control. PLoS ONE 2022, 17, e0276556. [Google Scholar] [CrossRef] [PubMed]
- OECD Guidance to the Environmental Safety Evaluation of Microbial Biocontrol Agents. 2012. Available online: https://www.oecd.org/env/oecd-guidance-to-the-environmental-safety-evaluation-of-microbial-biocontrol-agents-9789264221659-en.htm (accessed on 22 May 2023).
- Raza, M.M.; Bebber, D.P. Climate change and plant pathogens. Curr. Opin. Microbiol. 2022, 70, 102233. [Google Scholar] [CrossRef] [PubMed]
- Avelino, J.; Cristancho, M.; Georgiou, S.; Imbach, P.; Aguilar, L.; Bornemann, G.; Läderach, P.; Anzueto, F.; Hruska, A.J.; Morales, C. The coffee rust crises in Colombia and Central America (2008–2013): Impacts, plausible causes and proposed solutions. Food Secur. 2015, 7, 303–321. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Baquerizo, M.; Guerra, C.A.; Cano-Díaz, C.; Egidi, E.; Wang, J.T.; Eisenhauer, N.; Singh, B.K.; Maestre, F.T. The proportion of soil-borne pathogens increases with warming at the global scale. Nat. Clim. Chang. 2020, 10, 550–554. [Google Scholar] [CrossRef]
- West, J.S.; Holdgate, S.; Townsend, J.A.; Edwards, S.G.; Jennings, P.; Fitt, B.D.L. Impacts of changing climate and agronomic factors on fusarium ear blight of wheat in the UK. Fungal Ecol. 2012, 5, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Johns, L.E.; Bebber, D.P.; Gurr, S.J.; Brown, N.A. Emerging health threat and cost of Fusarium mycotoxins in European wheat. Nat. Food 2022, 3, 1014–1019. [Google Scholar] [CrossRef]
- Grosdidier, M.; Ioos, R.; Marçais, B. Do higher summer temperatures restrict the dissemination of Hymenoscyphus fraxineus in France? For. Pathol. 2018, 48, e12426. [Google Scholar] [CrossRef]
- Madgwick, J.W.; West, J.S.; White, R.P.; Semenov, M.A.; Townsend, J.A.; Turner, J.A.; Fitt, B.D.L. Impacts of climate change on wheat anthesis and fusarium ear blight in the UK. Eur. J. Plant Pathol. 2011, 130, 117–131. [Google Scholar] [CrossRef] [Green Version]
- Bezner Kerr, R.; Hasegawa, T.; Lasco, R.; Bhatt, I.; Deryng, D.; Farrell, A.; Gurney-Smith, H.; Ju, H.; Lluch-Cota, S.; Meza, F.; et al. Food, Fibre, and Other Ecosystem Products. In Climate Change 2022: Impacts, Adaptation and Vulnerability. Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Pörtner, H.-O., Roberts, D.C., Tignor, M., Poloczanska, E.S., Mintenbeck, K., Alegría, A., Craig, M., Langsdorf, S., Löschke, S., Möller, V., et al., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2022; pp. 713–906. [Google Scholar] [CrossRef]
- Battilani, P.; Toscano, P.; Van Der Fels-Klerx, H.J.; Moretti, A.; Camardo Leggieri, M.; Brera, C.; Rortais, A.; Goumperis, T.; Robinson, T. Aflatoxin B 1 contamination in maize in Europe increases due to climate change. Sci. Rep. 2016, 6, 24328. [Google Scholar] [CrossRef] [Green Version]
- Bailly, S.; El Mahgubi, A.; Carvajal-Campos, A.; Lorber, S.; Puel, O.; Oswald, I.P.; Bailly, J.D.; Orlando, B. Occurrence and identification of Aspergillus section flavi in the context of the emergence of aflatoxins in french maize. Toxins 2018, 10, 525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrone, G.; Gallo, A.; Logrieco, A.F. Biodiversity of Aspergillus section Flavi in Europe in relation to the management of aflatoxin risk. Front. Microbiol. 2014, 5, 377. [Google Scholar] [CrossRef] [Green Version]
- Bebber, D.P.; Ramotowski, M.A.T.; Gurr, S.J. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Change 2013, 3, 985–988. [Google Scholar] [CrossRef]
- Zou, Z.; Bisht, V.; Fernando, W.G.D. Identification and Characterization of Verticillium longisporum Lineage A1/D1 from Brassica Crops in Manitoba, Canada. Int. J. Mol. Sci. 2020, 21, 3499. [Google Scholar] [CrossRef]
- Dixon, G.R. Climate change-impact on crop growth and food production, and plant pathogens. Can. J. Plant Pathol. 2012, 34, 362–379. [Google Scholar] [CrossRef]
- Olesen, J.E.; Bindi, M. Consequences of climate change for European agricultural productivity. Land Use Policy 2002, 16, 239–262. [Google Scholar]
- Rosa CR, E.; Spehar, C.R.; Liu, J.Q. Asian Soybean Rust Resistance: An Overview. J. Plant Pathol. Microbiol. 2015, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Ozkan, H.E.; Zhu, H.; Derksen, R.C.; Guler, H.; Krause, C. Evaluation of various spraying equipment for effective application of fungicides to control Asian soybean rust. Asp Appl. Biol. 2006, 77. Available online: https://pubag.nal.usda.gov/catalog/16392 (accessed on 16 February 2023).
- Brown, J.K.M.; Hovmoller, M.S. Aerial Dispersal of Pathogens on the Global and Continental Scales and Its Impact on Plant Disease. Science 2002, 297, 537–541. [Google Scholar] [CrossRef] [Green Version]
- McIntosh, R.A.; Pretorius, Z.A. Borlaug Global Rust Initiative provides momentum for wheat rust research. Euphytica 2011, 179, 1–2. [Google Scholar] [CrossRef]
- Hovmøller, M.S.; Thach, T.; Justesen, A.F. Global dispersal and diversity of rust fungi in the context of plant health. Curr. Opin. Microbiol. 2023, 71, 102243. [Google Scholar] [CrossRef]
- Chen, W.; Hambleton, S.; Seifert, K.A.; Carisse, O.; Diarra, M.S.; Peters, R.D.; Lowe, C.; Chapados, J.T.; Lévesque, C.A. Assessing performance of spore samplers in monitoring aeromycobiota and fungal plant pathogen diversity in Canada. Appl. Environ. Microbiol. 2018, 84, e02601-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Martin, C.; Teigell-Perez, N.; Valladares, B.; Griffin, D.W. The Global Dispersion of Pathogenic Microorganisms by Dust Storms and Its Relevance to Agriculture. Adv. Agron. 2014, 127, 1–41. [Google Scholar] [CrossRef]
- Dietzel, K.; Valle, D.; Fierer, N.; U’Ren, J.M.; Barberán, A. Geographical Distribution of Fungal Plant Pathogens in Dust Across the United States. Front. Ecol. Evol. 2019, 7, 304. [Google Scholar] [CrossRef] [Green Version]
- Dhar, A.; Parrott, L.; Hawkins, C.D.B. Aftermath of Mountain Pine Beetle Outbreak in British Columbia: Stand Dynamics, Management Response and Ecosystem Resilience. Forests 2016, 7, 171. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, P.; Ammunét, T.; Barton, M.; Battisti, A.; Eigenbrode, S.D.; Jepsen, J.U.; Kalinkat, G.; Neuvonen, S.; Niemelä, P.; Terblanche, J.S.; et al. Complex responses of global insect pests to climate warming. Front. Ecol. Environ. 2020, 18, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, S.B.; Cohen, B.A.; Fry, W.E. Panglobal distribution of a single clonal lineage of the Irish potato famine fungus. Proc. Natl. Acad. Sci. USA 1994, 91, 11591–11595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, L.; Li, G.-Q.; Yang, L.; Jiang, D.-H.; Zhuang, W.-Y.; Huang, H.-C. Botrytis sinoallii: A new species of the grey mould pathogen on Allium crops in China. Mycoscience 2010, 51, 421–431. [Google Scholar] [CrossRef]
- Singh, R.P.; Hodson, D.P.; Huerta-Espino, J.; Jin, Y.; Bhavani, S.; Njau, P.; Herrera-Foessel, S.; Singh, P.K.; Singh, S.; Govindan, V. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 2011, 49, 465–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hovmøller, M.S.; Walter, S.; Justesen, A.F. Escalating Threat of Wheat Rusts. Science 2010, 329, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubbard, A.; Lewis, C.M.; Yoshida, K.; Ramirez-Gonzalez, R.H.; De Vallavieille-Pope, C.; Thomas, J.; Kamoun, S.; Bayles, R.; Uauy, C.; Saunders, D.G. Field pathogenomics reveals the emergence of a diverse wheat yellow rust population. Genome Biol. 2015, 16, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gold, K.M. Plant disease sensing: Studying plant-pathogen interactions at scale. Msystems 2021, 6, e01228-21. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Camino, C.; Beck, P.S.A.; Calderon, R.; Hornero, A.; Hernández-Clemente, R.; Kattenborn, T.; Montes-Borrego, M.; Susca, L.; Morelli, M.; et al. Previsual symptoms of Xylella fastidiosa infection revealed in spectral plant-trait alterations. Nat. Plants 2018, 4, 432–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrada, E.E.; Zoffoli, J.P.; Diaz, G.A.; Latorre, B.A. First report of Botrytis cinerea causing blossom blight on Japanese plums in Chile. Plant Dis. 2015, 99, 887. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Jajoo, A.; Mathur, S. Role of arbuscular mycorrhizal fungi as an underground saviuor for protecting plants from abiotic stresses. Physiol. Mol. Biol. Plants 2021, 27, 2589–2603. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [Green Version]
- Itakura, M.; Uchida, Y.; Akiyama, H.; Hoshino, Y.T.; Shimomura, Y.; Morimoto, S.; Tago, K.; Wang, Y.; Hayakawa, C.; Uetake, Y.J.N.C.C. Mitigation of nitrous oxide emissions from soils by Bradyrhizobium japonicum inoculation. Nat. Clim. Change 2013, 3, 208–212. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Nazari, M.; Antar, M.; Msimbira, L.A.; Naamala, J.; Lyu, D.; Rabileh, M.; Zajonc, J.; Smith, D.L. PGPR in Agriculture: A Sustainable Approach to Increasing Climate Change Resilience. Front. Sustain. Food Syst. 2021, 5, 667546. [Google Scholar] [CrossRef]
- Ma, Z.; Guo, D.; Xu, X.; Lu, M.; Bardgett, R.D.; Eissenstat, D.M.; McCormack, M.L.; Hedin, L.O. Evolutionary history resolves global organization of root functional traits. Nature 2018, 555, 94–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergmann, J.; Weigelt, A.; van Der Plas, F.; Laughlin, D.C.; Kuyper, T.W.; Guerrero-Ramirez, N.; Valverde-Barrantes, O.J.; Bruelheide, H.; Freschet, G.T.; Iversen, C.M.; et al. The fungal collaboration gradient dominates the root economics space in plants. Sci. Adv. 2020, 6, eaba3756. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Guo, L.; Xu, X.; Zhang, L.; Zhang, K.; Chen, M.; Zhao, Y.; Burkey, K.O.; Shew, H.D.; Zobel, R.W.; et al. Warming and elevated ozone induce tradeoffs between fine roots and mycorrhizal fungi and stimulate organic carbon decomposition. Sci. Adv. 2021, 7, eabe9256. [Google Scholar] [CrossRef]

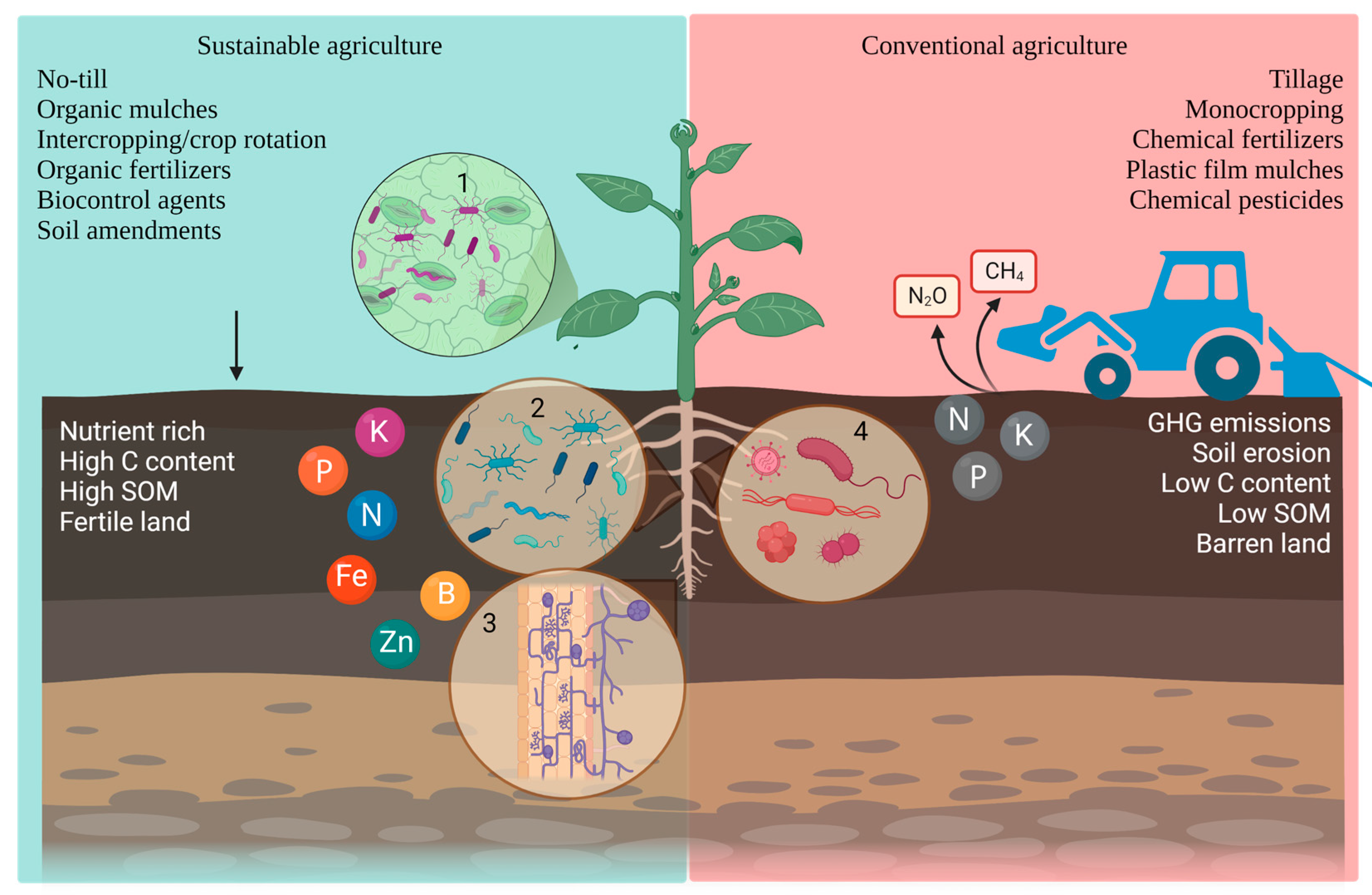
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 Picot and His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada. Submitted for possible open access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Modi, D.; Picot, A. Soil and Phytomicrobiome for Plant Disease Suppression and Management under Climate Change: A Review. Plants 2023, 12, 2736. https://doi.org/10.3390/plants12142736
Chen W, Modi D, Picot A. Soil and Phytomicrobiome for Plant Disease Suppression and Management under Climate Change: A Review. Plants. 2023; 12(14):2736. https://doi.org/10.3390/plants12142736
Chicago/Turabian StyleChen, Wen, Dixi Modi, and Adeline Picot. 2023. "Soil and Phytomicrobiome for Plant Disease Suppression and Management under Climate Change: A Review" Plants 12, no. 14: 2736. https://doi.org/10.3390/plants12142736
APA StyleChen, W., Modi, D., & Picot, A. (2023). Soil and Phytomicrobiome for Plant Disease Suppression and Management under Climate Change: A Review. Plants, 12(14), 2736. https://doi.org/10.3390/plants12142736






