Analysis of the Tomato mTERF Gene Family and Study of the Stress Resistance Function of SLmTERF-13
Abstract
1. Introduction
2. Results
2.1. Identification of mTERF Gene Family Members and Construction of an Evolutionary Tree of Tomato
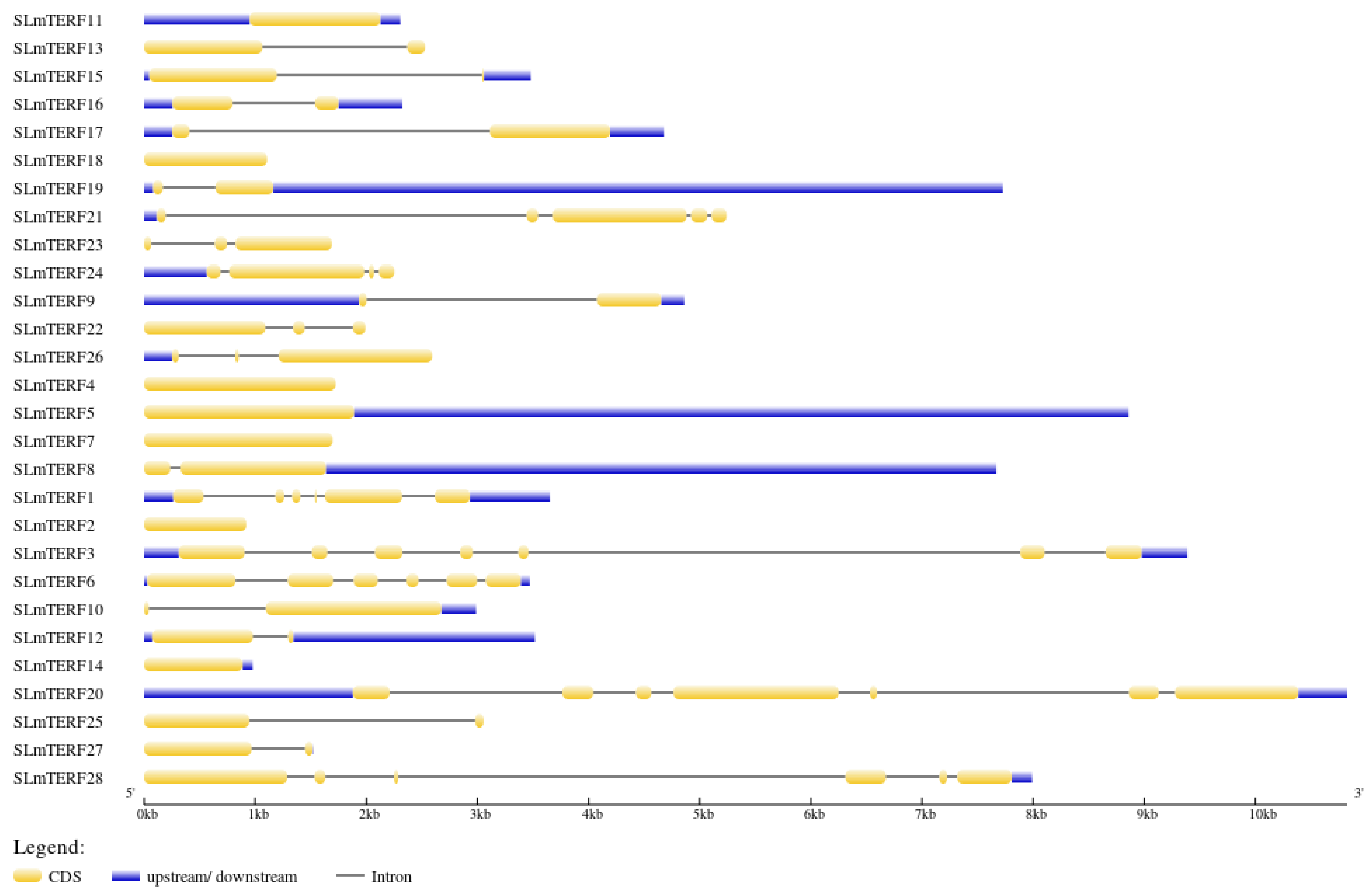
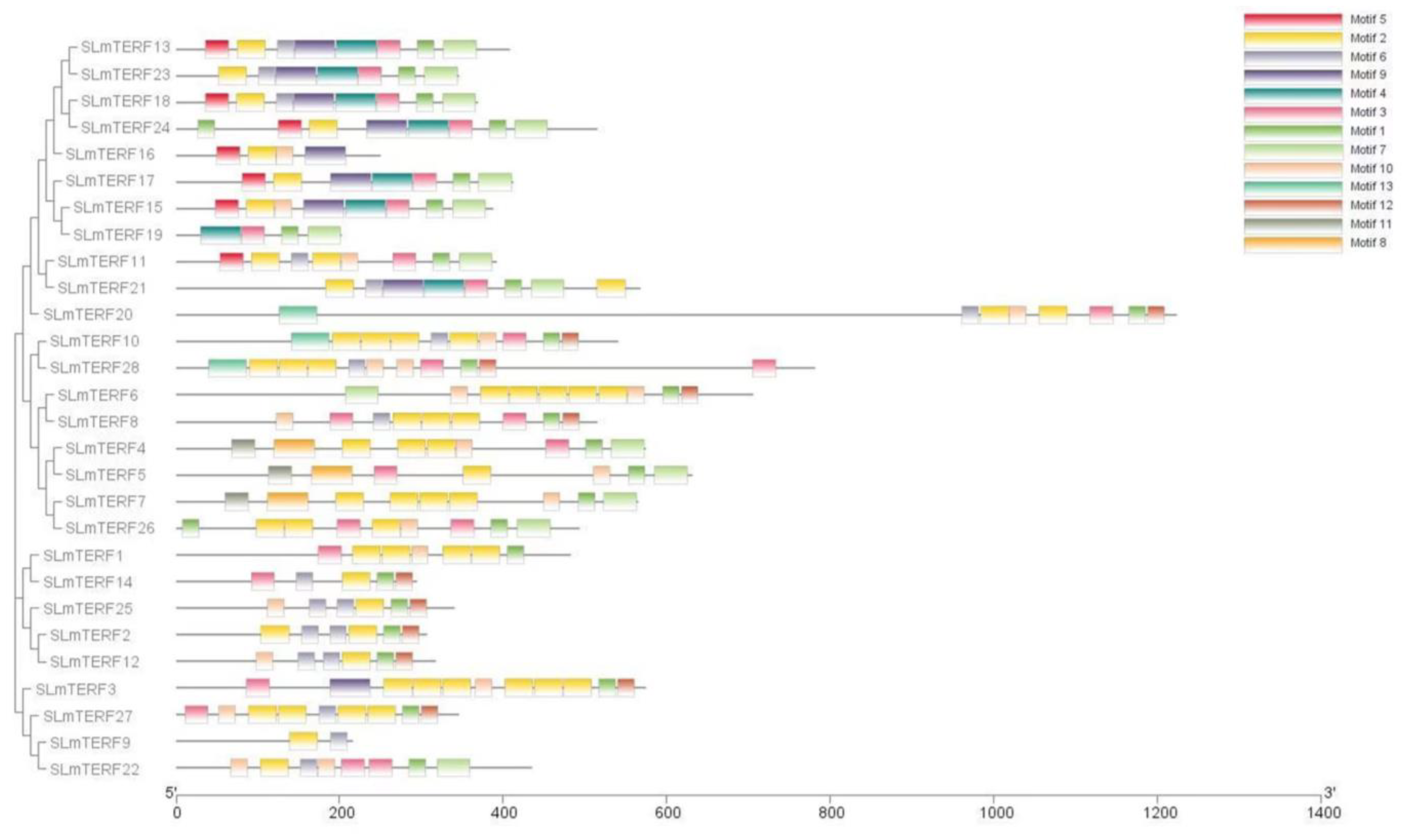
2.2. mTERF Gene Structure, Protein Secondary Structure, and Motif Analysis in Tomato
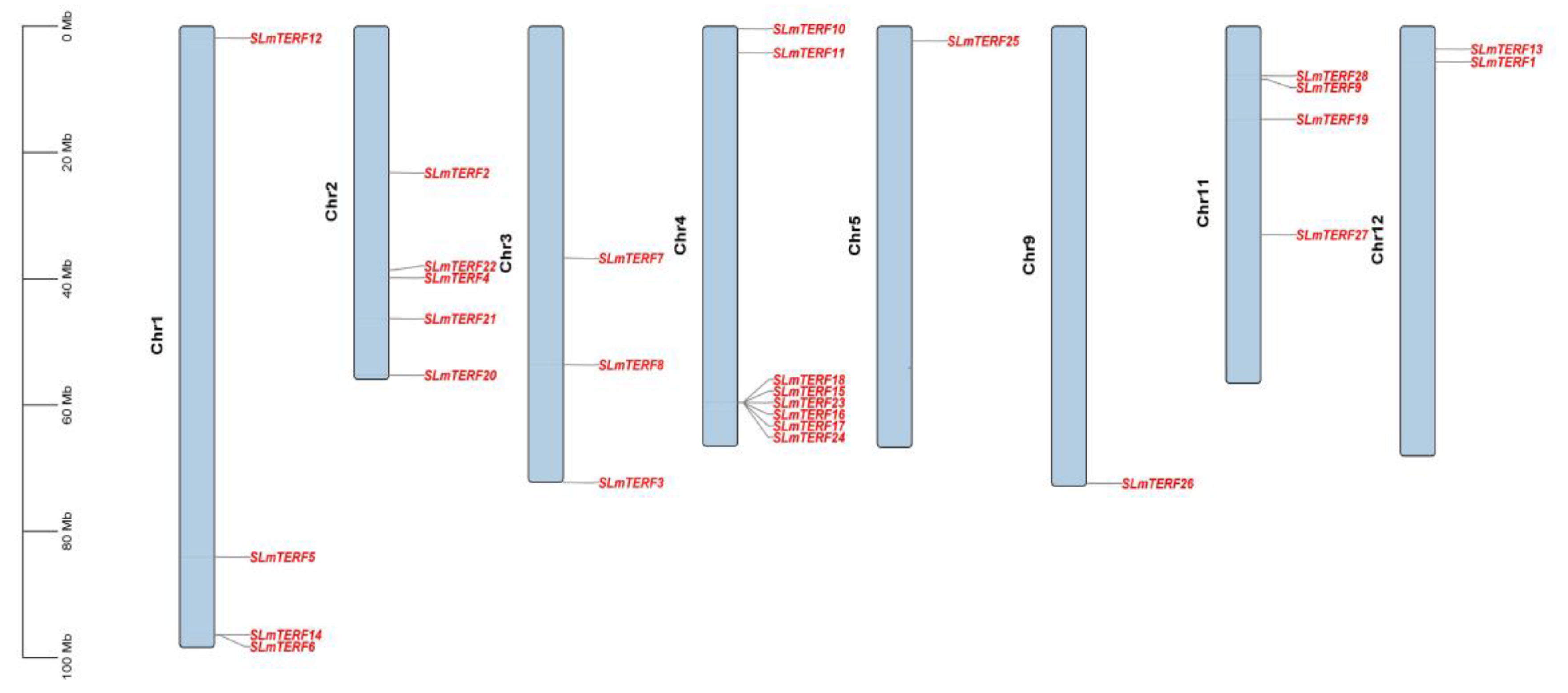
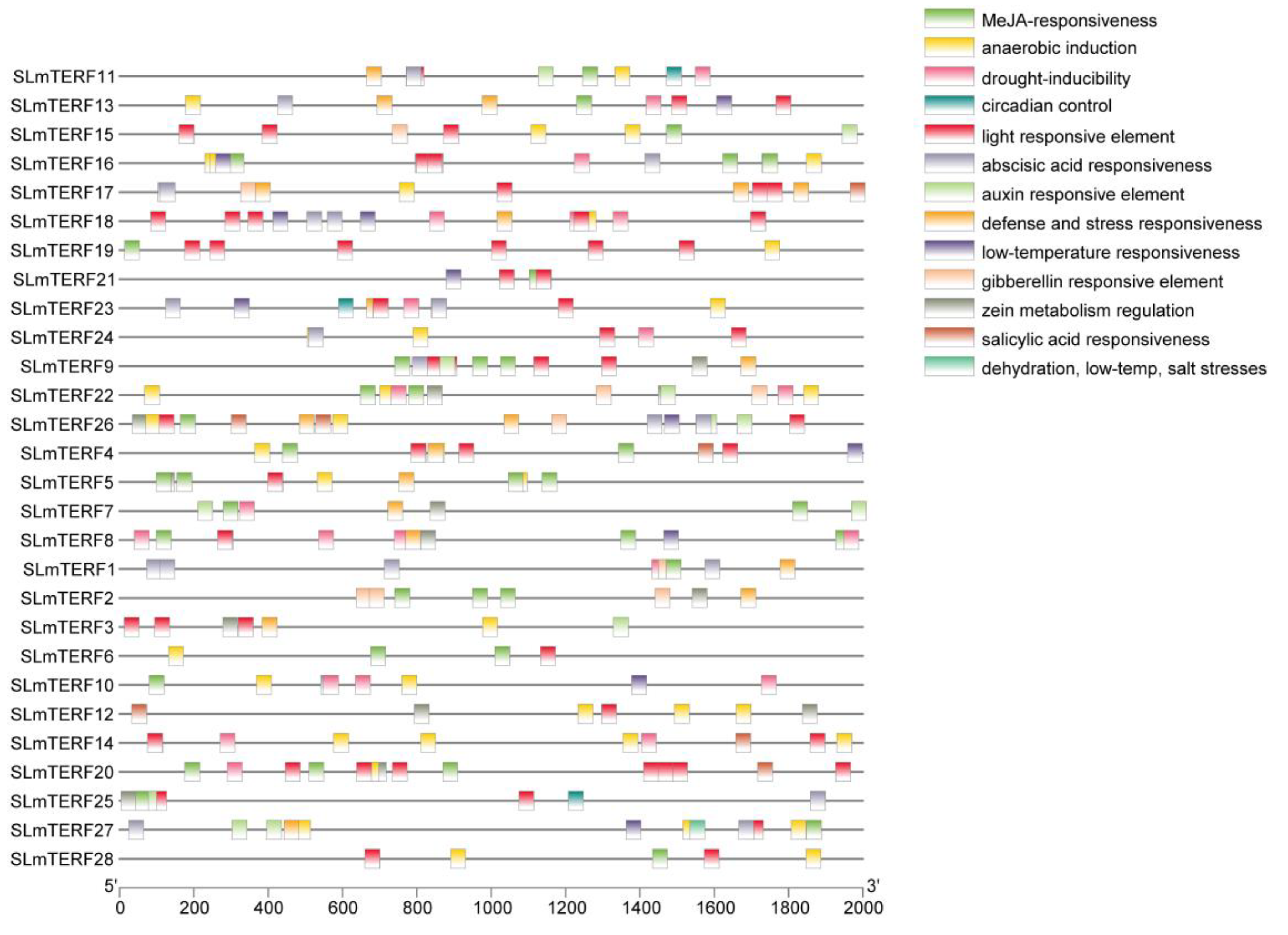
2.3. Chromosome Localization Prediction, Subcellular Localization, and Cis-Acting Element Analysis
2.4. Tissue Specificity Analysis of the mTERF Gene in Tomato
2.5. Analysis of mTERF Gene Expression in Tomato under Abiotic Stress
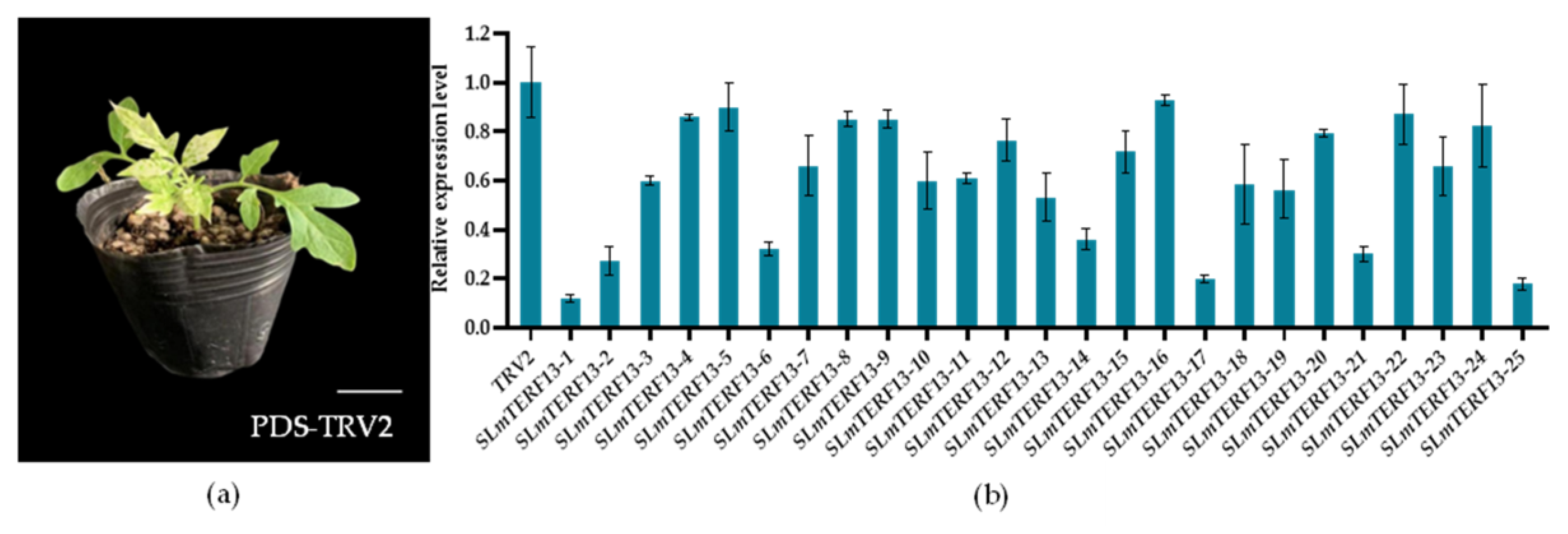
2.6. Cloning and Subcellular Localization of the SLmTERF13 Gene
2.7. Functional Analysis of the SLmTERF13 Gene
2.7.1. Phenotypic Analysis of Stress Resistance of Gene-Silenced Plants
2.7.2. Physiological Index Measurements
2.7.3. Active Oxygen Staining
2.7.4. Chlorophyll Content and Electrical Conductivity Determination
3. Discussion
4. Materials and Methods
4.1. Identification of mTERF Gene Family Members and Construction of an mTERF Evolutionary Tree in Tomato
4.2. Analysis of the Physical and Chemical Properties of Tomato mTERF Proteins
4.3. Gene Structure, Protein Secondary Structure, and Motif Analysis of Tomato mTERFs
4.4. Chromosome Localization Prediction, Subcellular Localization, and Cis-Acting Element Analysis of Tomato mTERFs
4.5. Plant Materials and Treatment Methods
4.6. cDNA Synthesis and qRT-PCR
4.7. Gene Silencing and Stress Resistance Treatments in Silenced Plants
4.8. Physiological Index Measurements
4.8.1. Determination of Superoxide Dismutase (SOD) and Peroxidase (POD) Activities and Proline (Pro) and Malondialdehyde (MDA) Contents
4.8.2. Reactive Oxygen Staining Observation
4.8.3. Determination of Chlorophyll Content and Electrical Conductivity
4.9. Subcellular Localization of SLmTERF13 Protein
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gray, M.W.; Burger, G.; Lang, B.F. Mitochondrial Evolution. Science 1999, 283, 1476–1481. [Google Scholar] [CrossRef]
- Lang, B.F.; Gray, M.W.; Burger, G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 1999, 33, 351–397. [Google Scholar] [CrossRef]
- Roberti, M.; Polosa, P.L.; Bruni, F.; Manzari, C.; Deceglie, S.; Gadaleta, M.N.; Cantatore, P. The MTERF family proteins: Mitochondrial transcription regulators and beyond. Biochim. Biophys. Acta 2009, 1787, 303–311. [Google Scholar] [CrossRef]
- Quesada, V. The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiol. Plant 2016, 157, 389–399. [Google Scholar] [CrossRef]
- Kruse, B.; Narasimhan, N.; Attardi, G. Termination of transcription in human mitochondria: Identification and purification of a DNA binding protein factor that promotes termination. Cell 1989, 58, 391–397. [Google Scholar] [CrossRef]
- Linder, T.; Park, C.B.; Asin-Cayuela, J.; Pellegrini, M.; Larsson, N.-G.; Falkenberg, M.; Samuelsson, T.; Gustafsson, C.M. A family of putative transcription termination factors shared amongst metazoans and plants. Curr. Genet. 2005, 48, 265–269. [Google Scholar] [CrossRef]
- Leister, D.; Kleine, T. Extending the Repertoire of mTERF Proteins with Functions in Organellar Gene Expression. Mol. Plant 2020, 13, 817–819. [Google Scholar] [CrossRef]
- Kleine, T. Arabidopsis thaliana mTERF proteins: Evolution and functional classification. Front. Plant Sci. 2012, 3, 233. [Google Scholar] [CrossRef]
- Tang, B.; Xie, L.; Yi, T.; Lv, J.; Yang, H.; Cheng, X.; Liu, F.; Zou, X. Genome-Wide Identification and Characterization of the Mitochondrial Transcription Termination Factors (mTERFs) in Capsicum annuum L. Int. J. Mol. Sci. 2019, 21, 269. [Google Scholar] [CrossRef]
- Yin, X.; Gao, Y.; Song, S.; Hassani, D.; Lu, J. Identification, characterization and functional analysis of grape (Vitis vinifera L.) mitochondrial transcription termination factor (mTERF) genes in responding to biotic stress and exogenous phytohormone. BMC Genom. 2021, 22, 136. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, M.; Zhang, X.; Li, Y.; Zhang, J.; Zhao, H.; Kong, F.; Zheng, Y.; Qiu, F. Genome-wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS ONE 2014, 9, e94126. [Google Scholar] [CrossRef]
- Babiychuk, E.; Vandepoele, K.; Wissing, J.; Garcia-Diaz, M.; De Rycke, R.; Akbari, H.; Joubes, J.; Beeckman, T.; Jansch, L.; Frentzen, M.; et al. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl. Acad. Sci. USA 2011, 108, 6674–6679. [Google Scholar] [CrossRef]
- Robles, P.; Quesada, V. Research Progress in the Molecular Functions of Plant mTERF Proteins. Cells 2021, 10, 205. [Google Scholar] [CrossRef]
- Wobbe, L.; Nixon, P.J. The mTERF protein MOC1 terminates mitochondrial DNA transcription in the unicellular green alga Chlamydomonas reinhardtii. Nucleic Acids Res. 2013, 41, 6553–6567. [Google Scholar] [CrossRef]
- Robles, P.; Micol, J.L.; Quesada, V. Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses. PLoS ONE 2012, 7, e42924. [Google Scholar] [CrossRef]
- Zhang, X.L. Preliminary Study on the Function of mTERF Protein PDE191 in Arabidopsis Thaliana. Master’s Thesis, Shanghai Normal University, Shanghai, China, 2011. Available online: https://kns.cnki.net/kcms2/article/abstract?v=3uoqIhG8C475KOm_zrgu4lQARvep2SAkWGEmc0QetxDh64Dt3veMp9WlHuIlcOUegHxiiC9JpkuwF0vFy65JH81Abp2071Q4&uniplatform=NZKPT (accessed on 5 July 2020). (In Chinese).
- Romani, I.; Manavski, N.; Morosetti, A.; Tadini, L.; Maier, S.; Kuhn, K.; Ruwe, H.; Schmitz-Linneweber, C.; Wanner, G.; Leister, D.; et al. A Member of the Arabidopsis Mitochondrial Transcription Termination Factor Family Is Required for Maturation of Chloroplast Transfer RNAIle(GAU). Plant Physiol. 2015, 169, 627–646. [Google Scholar] [CrossRef]
- Kim, M.; Lee, U.; Small, I.; des Francs-Small, C.C.; Vierling, E. Mutations in an Arabidopsis mitochondrial transcription termination factor-related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell 2012, 24, 3349–3365. [Google Scholar] [CrossRef]
- Hammani, K.; Barkan, A. An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res. 2014, 42, 5033–5042. [Google Scholar] [CrossRef]
- Lu, Y.; Zhu, H. The regulation of nutrient and flavor metabolism in tomato fruit. Veg. Res. 2022, 2, 1–14. [Google Scholar] [CrossRef]
- Bruno, A.; Durante, M.; Marrese, P.P.; Migoni, D.; Laus, M.N.; Pace, E.; Pastore, D.; Mita, G.; Piro, G.; Lenucci, M.S. Shades of red: Comparative study on supercritical CO2 extraction of lycopene-rich oleoresins from gac, tomato and watermelon fruits and effect of the α-cyclodextrin clathrated extracts on cultured lung adenocarcinoma cells’ viability. J. Food Compos. Anal. 2018, 65, 23–32. [Google Scholar] [CrossRef]
- Muscatello, M.R.A.; Zoccali, R.A.; Bruno, A. Citrus Fruit Polyphenols and Flavonoids: Applications to Psychiatric Disorders. In Polyphenols: Mechanisms of Action in Human Health and Disease; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 119–131. [Google Scholar]
- Baker, S.S.; Wilhelm, K.S.; Thomashow, M.F. The 5’-region of Arabidopsis thaliana corl5a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 1994, 24, 701–713. [Google Scholar] [CrossRef]
- Khong, G.; Richaud, F.; Coudert, Y.; Pati, P.K.; Santi, C.; Perin, C.; Breitler, J.C.; Meynard, D.; Vinh Do, N.; Guiderdoni, E.; et al. Modulating rice stress tolerance by transcription factors. Biotechnol. Genet. Eng. Rev. 2008, 25, 381–403. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Mundy, J. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc. Natl. Acad. Sci. USA 1990, 87, 1406–1410. [Google Scholar] [CrossRef]
- Mundy, J.; Chua, N.H. Abscisic acid and water: Tress induce the expression of a novel rice gene. EMBO J. 1988, 7, 2279–2286. [Google Scholar] [CrossRef]
- Jiang, C.; Iu, B.; Singh, J. Requirement of a CCGAC cis -acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 1996, 30, 679–684. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Turkoglu, M.; Yanikoğlu, B.; Hanbay, D. PlantDiseaseNet: Convolutional neural network ensemble for plant disease and pest detection. Signal Image Video Process. 2021, 16, 301–309. [Google Scholar] [CrossRef]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008, 36, D1009–D1014. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Nordahl, P.T.; Søren, B.; Gunnar, V.H.; Henrik, N. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar]
- Bo, H.; Jinpu, J.; An-Yuan, G.; He, Z.; Jingchu, L.; Ge, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, A.; Khan, A.A.; Cheema, H.; Aslam, U.; Malik, W. Biology. Transcriptome analysis of pigment related genes in colored cotton. Int. J. Agric. 2015, 17, 205–210. [Google Scholar]
- Niu, Y.L.; Jiang, X.M.; Xu, X.Y. Bioinformatics analysis of GRAS transcription factor family in tomato. Chin. Agric. Sci. Bull. 2016, 32, 106–116. (In Chinese) [Google Scholar]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Rosli, H.G.; Martin, G.B.; Mueller, L.A. The SGN VIGS Tool: User-Friendly Software to Design Virus-Induced Gene Silencing (VIGS) Constructs for Functional Genomics. Mol. Plant 2015, 8, 486–488. [Google Scholar] [CrossRef]
- Lin, Y.; Guo, W.Z.; Xu, Z.H.; Jia, Z.K. Study on cold resistance of Privet macrophylla and changes of leaf relative conductivity in winter. Tianjin Agric. Sci. 2012, 18, 145–149. (In Chinese) [Google Scholar]









| GENE ID | GENE NAME | Number of Amino Acids (aa) | Molecular Weight (Da) | Theoretical pI | Aliphatic Index | GRAVY | Instability Index |
|---|---|---|---|---|---|---|---|
| Solyc12g015640.2.1 | SLmTERF1 | 482 | 56,021.91 | 9.23 | 87.34 | −0.494 | 44.09 |
| Solyc02g021430.1.1 | SLmTERF2 | 306 | 34,876.53 | 8.78 | 93.33 | −0.148 | 46.43 |
| Solyc03g124040.3.1 | SLmTERF3 | 574 | 65,951.33 | 9.18 | 97.65 | −0.090 | 52.19 |
| Solyc02g069320.1.1 | SLmTERF4 | 574 | 65,300.50 | 8.76 | 96.11 | −0.147 | 35.39 |
| Solyc01g090500.3.1 | SLmTERF5 | 630 | 72,679.44 | 8.18 | 88.83 | −0.133 | 46.50 |
| Solyc01g109630.3.1 | SLmTERF6 | 705 | 81,980.16 | 8.96 | 89.67 | −0.283 | 45.02 |
| Solyc03g063390.1.1 | SLmTERF7 | 565 | 65,319.42 | 9.12 | 101.42 | −0.107 | 40.83 |
| Solyc03g081300.3.1 | SLmTERF8 | 514 | 57,846.31 | 7.65 | 103.17 | 0.015 | 42.70 |
| Solyc11g017430.2.1 | SLmTERF9 | 215 | 24,380.79 | 6.82 | 79.30 | −0.385 | 48.39 |
| Solyc04g005630.3.1 | SLmTERF10 | 540 | 61,573.69 | 7.61 | 97.22 | −0.150 | 45.02 |
| Solyc04g011700.2.1 | SLmTERF11 | 392 | 44,616.77 | 9.78 | 107.09 | 0.017 | 38.51 |
| Solyc01g007750.3.1 | SLmTERF12 | 317 | 36,039.24 | 9.18 | 95.52 | −0.008 | 59.20 |
| Solyc12g010650.1.1 | SLmTERF13 | 407 | 47,343.82 | 9.59 | 102.65 | −0.046 | 26.94 |
| Solyc01g109550.2.1 | SLmTERF14 | 294 | 34,528.11 | 9.32 | 87.55 | −0.233 | 45.58 |
| Solyc04g072510.3.1 | SLmTERF15 | 387 | 43,999.61 | 9.17 | 105.99 | 0.015 | 35.54 |
| Solyc04g072530.2.1 | SLmTERF16 | 250 | 28,271.08 | 9.28 | 102.88 | 0.130 | 30.46 |
| Solyc04g072540.3.1 | SLmTERF17 | 412 | 46,777.19 | 9.77 | 103.11 | 0.025 | 37.75 |
| Solyc04g072500.1.1 | SLmTERF18 | 368 | 42,229.61 | 9.73 | 101.11 | −0.042 | 28.78 |
| Solyc11g022600.2.1 | SLmTERF19 | 202 | 24,224.60 | 9.69 | 86.39 | −0.253 | 58.63 |
| Solyc02g093950.3.1 | SLmTERF20 | 1222 | 134,350.96 | 7.99 | 38.02 | 0.103 | 101.82 |
| Solyc02g082010.2.1 | SLmTERF21 | 567 | 64,440.59 | 9.55 | 97.09 | 0.053 | 46.81 |
| Solyc02g067960.2.1 | SLmTERF22 | 435 | 50,317.11 | 9.58 | 97.52 | −0.116 | 41.46 |
| Solyc04g072520.2.1 | SLmTERF23 | 346 | 39,620.67 | 9.60 | 96.88 | −0.178 | 30.62 |
| Solyc04g072550.2.1 | SLmTERF24 | 514 | 59,546.28 | 9.55 | 104.59 | 0.076 | 32.91 |
| Solyc05g007840.3.1 | SLmTERF25 | 340 | 38,787.68 | 8.86 | 110.59 | 0.106 | 43.82 |
| Solyc09g097920.2.1 | SLmTERF26 | 493 | 56,722.41 | 8.87 | 100.75 | −0.100 | 41.31 |
| Solyc11g044360.2.1 | SLmTERF27 | 345 | 39,458.31 | 9.25 | 98.23 | −0.170 | 38.39 |
| Solyc11g017050.2.1 | SLmTERF28 | 781 | 87,309.93 | 5.67 | 98.00 | −0.100 | 39.10 |
| ID | Alpha Helix | Extended Strand | Random Coil |
|---|---|---|---|
| SLmTERF11 | 49.23% | 15.82% | 34.95% |
| SLmTERF13 | 52.09% | 14.74% | 33.17% |
| SLmTERF15 | 50.39% | 11.63% | 37.98% |
| SLmTERF16 | 40.00% | 19.60% | 40.40% |
| SLmTERF17 | 45.87% | 14.81% | 39.32% |
| SLmTERF18 | 44.29% | 20.65% | 35.05% |
| SLmTERF19 | 45.05% | 12.38% | 42.57% |
| SLmTERF21 | 51.32% | 14.99% | 33.69% |
| SLmTERF23 | 41.91% | 17.34% | 40.75% |
| SLmTERF24 | 51.36% | 15.95% | 32.68% |
| SLmTERF9 | 35.35% | 14.88% | 49.77% |
| SLmTERF22 | 59.54% | 6.90% | 33.56% |
| SLmTERF26 | 46.04% | 14.60% | 39.35% |
| SLmTERF4 | 41.64% | 15.85% | 42.51% |
| SLmTERF5 | 44.60% | 12.54% | 42.86% |
| SLmTERF7 | 47.79% | 13.10% | 39.12% |
| SLmTERF8 | 43.00% | 14.40% | 42.61% |
| SLmTERF1 | 30.71% | 19.92% | 49.38% |
| SLmTERF2 | 38.89% | 15.36% | 45.75% |
| SLmTERF3 | 48.43% | 12.37% | 49.38% |
| SLmTERF6 | 46.95% | 14.47% | 38.58% |
| SLmTERF10 | 48.15% | 7.96% | 43.89% |
| SLmTERF12 | 39.43% | 15.14% | 45.43% |
| SLmTERF14 | 42.86% | 13.61% | 43.54% |
| SLmTERF20 | 49.18% | 13.83% | 36.99% |
| SLmTERF25 | 37.94% | 18.82% | 43.24% |
| SLmTERF27 | 49.86% | 15.65% | 34.49% |
| SLmTERF28 | 38.80% | 13.83% | 47.38% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, A.; Ge, S.; Zhou, B.; Wang, Z.; Zhou, L.; Zhang, Z.; Yan, X.; Wang, Y.; Li, D.; Zhang, H.; et al. Analysis of the Tomato mTERF Gene Family and Study of the Stress Resistance Function of SLmTERF-13. Plants 2023, 12, 2862. https://doi.org/10.3390/plants12152862
Su A, Ge S, Zhou B, Wang Z, Zhou L, Zhang Z, Yan X, Wang Y, Li D, Zhang H, et al. Analysis of the Tomato mTERF Gene Family and Study of the Stress Resistance Function of SLmTERF-13. Plants. 2023; 12(15):2862. https://doi.org/10.3390/plants12152862
Chicago/Turabian StyleSu, Ao, Siyu Ge, Boyan Zhou, Ziyu Wang, Liping Zhou, Ziwei Zhang, Xiaoyu Yan, Yu Wang, Dalong Li, He Zhang, and et al. 2023. "Analysis of the Tomato mTERF Gene Family and Study of the Stress Resistance Function of SLmTERF-13" Plants 12, no. 15: 2862. https://doi.org/10.3390/plants12152862
APA StyleSu, A., Ge, S., Zhou, B., Wang, Z., Zhou, L., Zhang, Z., Yan, X., Wang, Y., Li, D., Zhang, H., Xu, X., & Zhao, T. (2023). Analysis of the Tomato mTERF Gene Family and Study of the Stress Resistance Function of SLmTERF-13. Plants, 12(15), 2862. https://doi.org/10.3390/plants12152862






