Subcellular Proteomics to Elucidate Soybean Response to Abiotic Stress
Abstract
1. Introduction
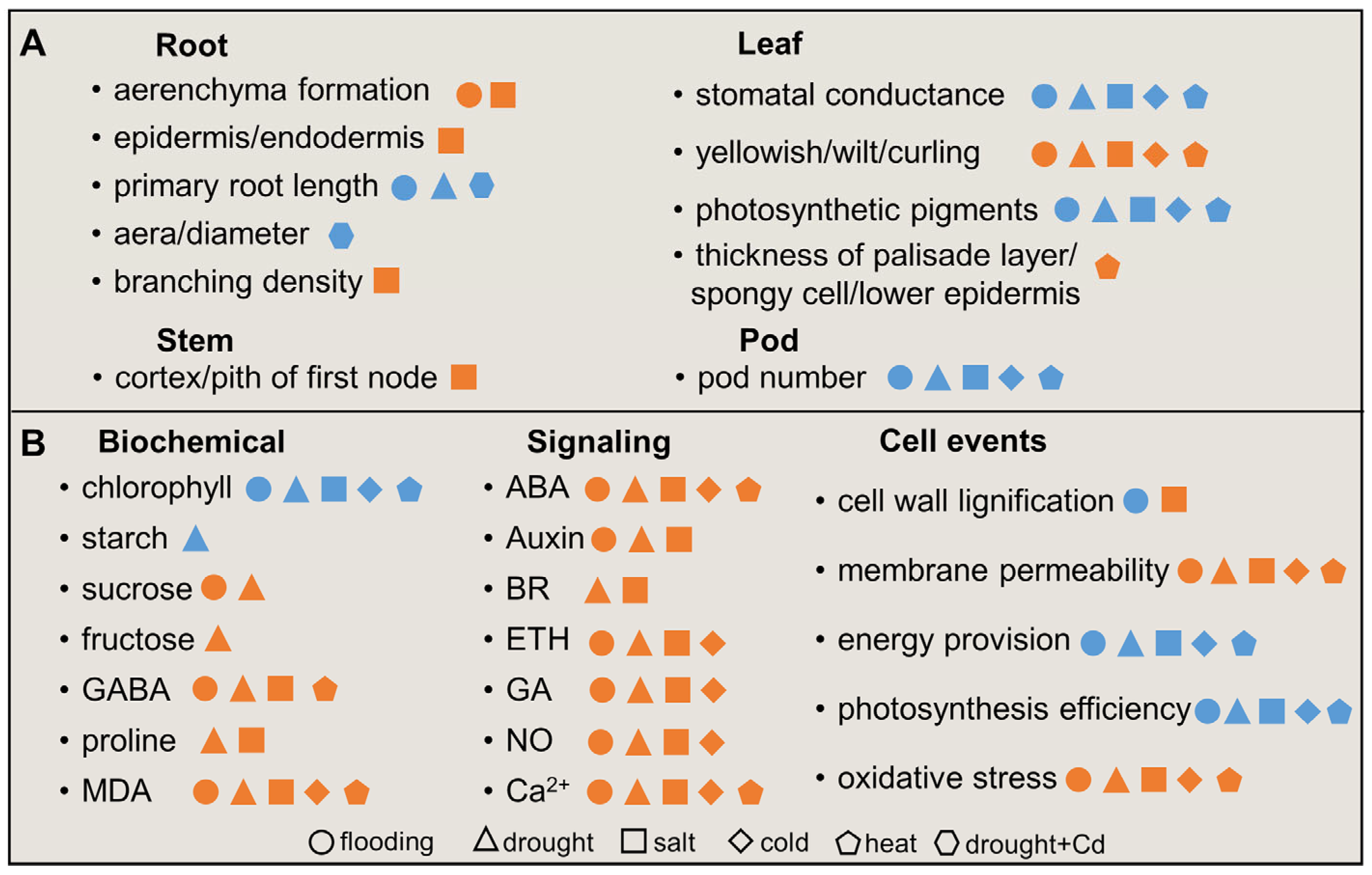
2. Morphophysiological Changes of Soybean under Abiotic Stress
2.1. Morphophysiological Changes of Soybean under Flooding Stress
2.2. Morphophysiological Changes of Soybean under Drought Stress
2.3. Morphophysiological Changes of Soybean under Salt Stress
2.4. Morphophysiological Changes of Soybean under Low-Temperature Stress
2.5. Morphophysiological Changes of Soybean under High-Temperature Stress
2.6. Morphophysiological Changes of Soybean under Other Stresses
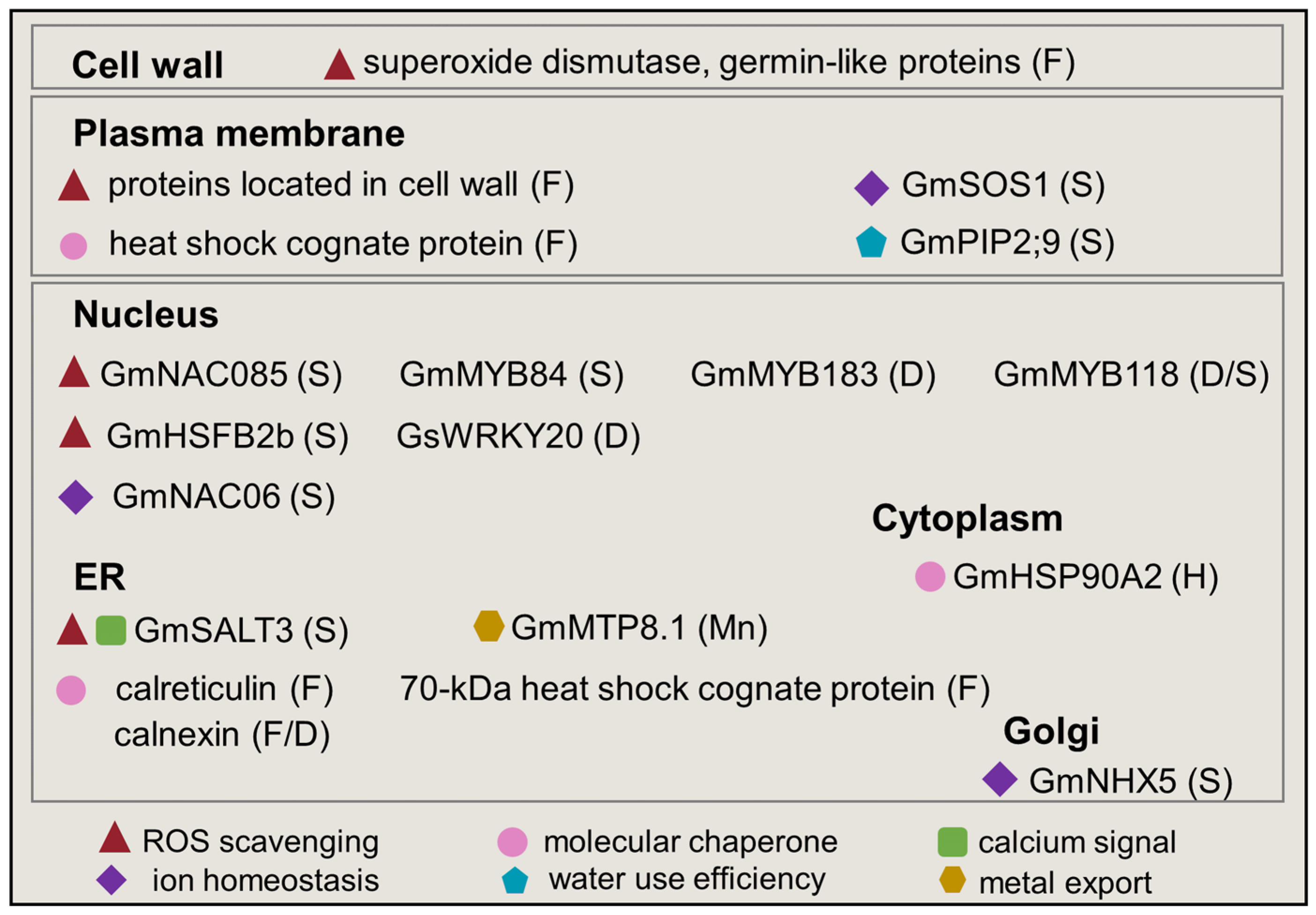
3. Subcellular Response in Soybean under Abiotic Stress
3.1. The Response of Endoplasmic Reticulum in Soybean under Abiotic Stress
3.2. The Response of Chloroplast in Soybean under Abiotic Stress
3.3. The Response of Mitochondria in Soybean under Abiotic Stress
3.4. The Response of Nucleus in Soybean under Abiotic Stress
3.5. The Response of Plasma Membrane in Soybean under Abiotic Stress
3.6. The Response of Cell Wall in Soybean under Abiotic Stress
3.7. The Response of Other Organelles in Soybean under Abiotic Stress
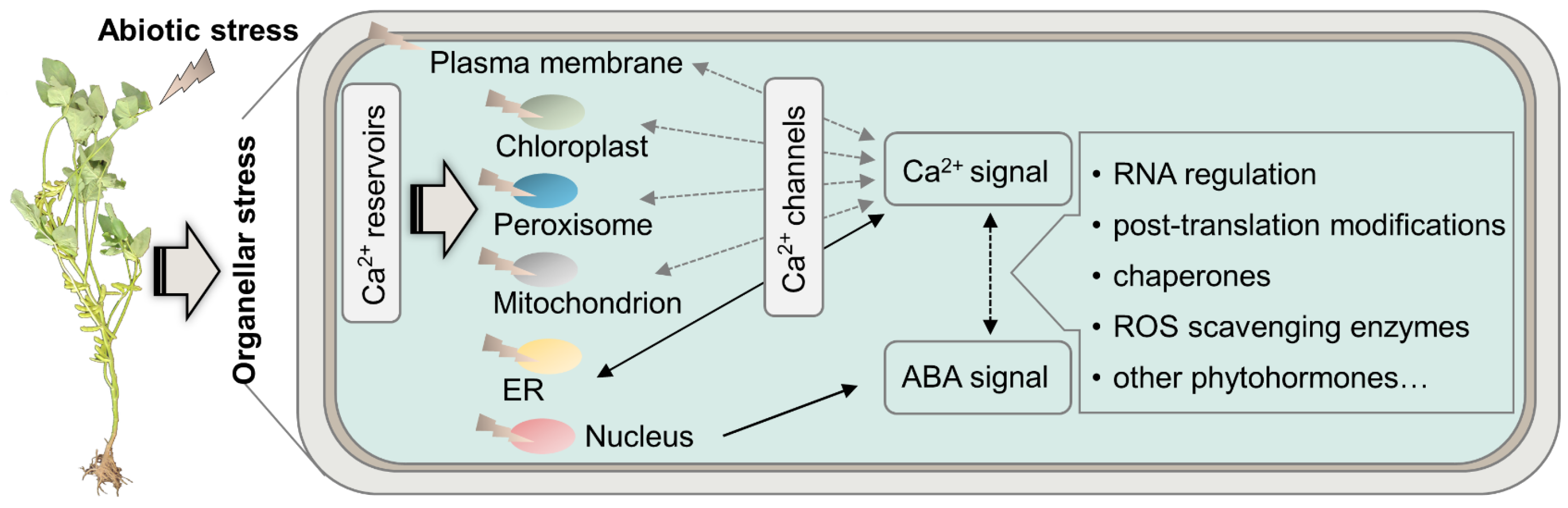
4. Proteins Regulated Organellar Stress among Subcellular Compartments in Soybean under Abiotic Stress
4.1. Reactive Oxygen Species
4.2. Molecular Chaperons
4.3. Other Proteins
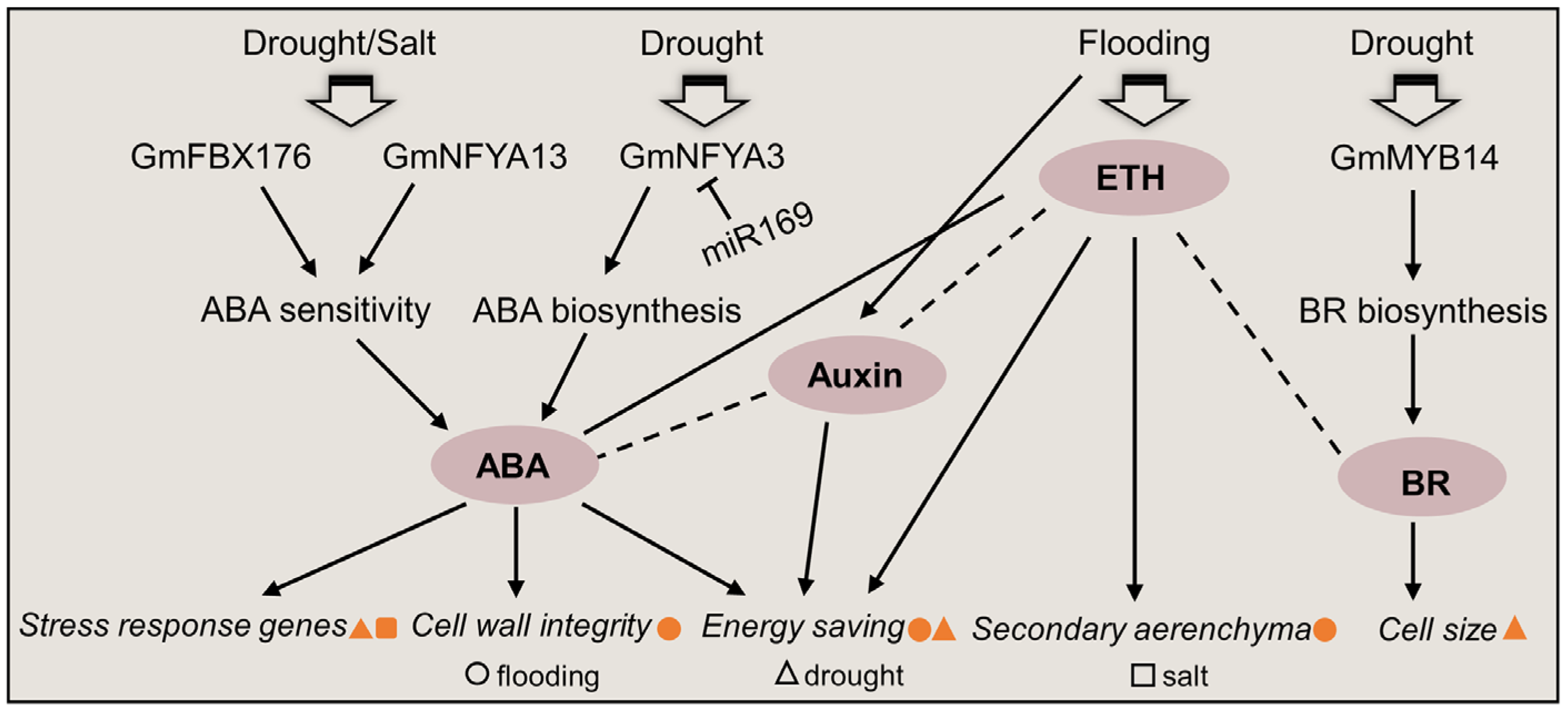
5. Phytohormone Signals in Soybean under Abiotic Stress
6. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chae, J.H.; Ha, B.K.; Chung, G.; Park, J.E.; Park, E.; Ko, J.M.; Shannon, J.G.; Song, J.T.; Lee, J.D. Identification of environmentally stable wild soybean genotypes with high alpha-linolenic acid concentration. Crop Sci. 2015, 55, 1629–1636. [Google Scholar] [CrossRef]
- Nakai, S.; Fujita, M.; Kamei, Y. Health promotion effects of soy isoflavones. J. Nutr. Sci. Vitaminol. 2020, 66, 502–507. [Google Scholar] [CrossRef]
- Schlenker, W.; Roberts, M.J. Nonlinear temperature effects indicate severe damages to US crop yields under climate change. Proc. Natl. Acad. Sci. USA 2009, 106, 15594–15598. [Google Scholar] [CrossRef] [PubMed]
- Shannon, J.G.; Stevens, W.E.; Wiebold, W.J.; McGraw, R.L.; Sleper, D.A. Breeding soybeans for improved tolerance to flooding. In Proceedings of the 35th Soybean Seed Research Conference American Seed Trade Association, Chicago, IL, USA, 7–9 December 2005. [Google Scholar]
- Specht, J.E.; Hume, D.J.; Kumudini, S.V. Soybean yield potential—A genetic and physiological perspective. Crop Sci. 1999, 39, 1560–1570. [Google Scholar] [CrossRef]
- Katerji, N.; Van Hoorn, J.W.; Hamdy, A.; Mastrorilli, M. Salt tolerance classification of crops according to soil salinity and to water stress day index. Agr. Water Manag. 2000, 43, 99–109. [Google Scholar] [CrossRef]
- Deryng, D.; Conway, D.; Ramankutty, N.; Price, J.; Warren, R. Global crop yield response to extreme heat stress under multiple climate change futures. Environ. Res. Lett. 2014, 9, 034011. [Google Scholar] [CrossRef]
- Vezza, M.E.; Llanes, A.; Travaglia, C.; Agostini, E.; Talano, M.A. Arsenic stress effects on root water absorption in soybean plants: Physiological and morphological aspects. Plant Physiol. Biochem. 2018, 123, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Li, P.S.; Yu, T.F.; He, G.H.; Chen, M.; Zhou, Y.B.; Chai, S.C.; Xu, Z.S.; Ma, Y.Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar] [CrossRef]
- Rao, S.S.; El-Habbak, M.H.; Havens, W.M.; Singh, A.; Zheng, D.; Vaughn, L.; Haudenshield, J.S.; Hartman, G.L.; Korban, S.S.; Ghabrial, S.A. Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol. Plant Pathol. 2014, 15, 145–160. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, W.; Qin, M.; Li, S.; Qiao, M.; Liu, Z.; Xiang, F. Drought tolerance conferred in soybean (Glycine max. L.) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 2017, 58, 1764–1776. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liu, X.; He, X.; Xu, L.; Huang, Y.; Shao, H.; Zhang, D.; Tang, B.; Ma, H. The soybean basic helix-loop-helix transcription factor ORG3-like enhances cadmium tolerance via increased iron and reduced cadmium uptake and transport from roots to shoots. Front. Plant Sci. 2017, 8, 1098. [Google Scholar] [CrossRef]
- Zhao, M.J.; Yin, L.J.; Ma, J.; Zheng, J.C.; Wang, Y.X.; Lan, J.H.; Fu, J.D.; Chen, M.; Xu, Z.S.; Ma, Y.Z. The roles of GmERF135 in improving salt tolerance and decreasing ABA sensitivity in soybean. Front. Plant Sci. 2019, 10, 940. [Google Scholar] [CrossRef]
- Chen, L.; Yang, H.; Fang, Y.; Guo, W.; Chen, H.; Zhang, X.; Dai, W.; Chen, S.; Hao, Q.; Yuan, S.; et al. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 2021, 19, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Ketehouli, T.; Zhou, Y.G.; Dai, S.Y.; Carther, K.F.I.; Sun, D.Q.; Li, Y.; Nguyen, Q.V.H.; Xu, H.; Wang, F.W.; Liu, W.C.; et al. A soybean calcineurin B-like protein-interacting protein kinase, GmPKS4, regulates plant responses to salt and alkali stresses. J. Plant Physiol. 2021, 256, 153331. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Deng, X.; Huang, Y.; Fang, X.; Zhang, J.; Wan, H.; Yang, C. Comparison of subcellular distribution and chemical forms of cadmium among four soybean cultivars at young seedlings. Environ. Sci. Pollut. Res. Int. 2015, 22, 19584–19595. [Google Scholar] [CrossRef]
- Chen, W.; Yao, Q.; Patil, G.B.; Agarwal, G.; Deshmukh, R.K.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.J.; Song, L.; et al. Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef]
- Chung, W.S.; Lee, S.H.; Kim, J.C.; Heo, W.D.; Kim, M.C.; Park, C.Y.; Park, H.C.; Lim, C.O.; Kim, W.B.; Harper, J.F.; et al. Identification of a calmodulin-regulated soybean Ca2+-ATPase (SCA1) that is located in the plasma membrane. Plant Cell 2000, 12, 1393–1407. [Google Scholar] [CrossRef]
- Yang, L.; Wu, K.; Gao, P.; Liu, X.; Li, G.; Wu, Z. GsLRPK, a novel cold-activated leucine-rich repeat receptor-like protein kinase from Glycine soja, is a positive regulator to cold stress tolerance. Plant Sci. 2014, 215–216, 19–28. [Google Scholar] [CrossRef]
- Sun, X.; Cai, X.; Yin, K.; Gu, L.; Shen, Y.; Hu, B.; Wang, Y.; Chen, Y.; Zhu, Y.; Jia, B.; et al. Wild soybean SNARE proteins BET1s mediate the subcellular localization of the cytoplasmic receptor-like kinases CRCK1s to modulate salt stress responses. Plant J. 2021, 105, 771–785. [Google Scholar] [CrossRef]
- Ahsan, N.; Nanjo, Y.; Sawada, H.; Kohno, Y.; Komatsu, S. Ozone stress-induced proteomic changes in leaf total soluble and chloroplast proteins of soybean reveal that carbon allocation is involved in adaptation in the early developmental stage. Proteomics 2010, 10, 2605–2619. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Hu, C.; Tan, Q.; Qin, S.; Sun, X. Subcellular distribution of molybdenum, ultrastructural and antioxidative responses in soybean seedlings under excess molybdenum stress. Plant Physiol. Biochem. 2018, 123, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, G.; Komatsu, S. Insights into the response of soybean mitochondrial proteins to various sizes of aluminum oxide nanoparticles under flooding stress. J. Proteome Res. 2016, 15, 4464–4475. [Google Scholar] [CrossRef]
- Finger-Teixeira, A.; Ishii-Iwamoto, E.L.; Marchiosi, R.; Coelho, É.M.P.; Constantin, R.P.; Dos Santos, W.D.; Soares, A.R.; Ferrarese-Filho, O. Cadmium uncouples mitochondrial oxidative phosphorylation and induces oxidative cellular stress in soybean roots. Environ. Sci. Pollut. Res. Int. 2021, 28, 67711–67723. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Komatsu, S. Gel-free/label-free proteomic analysis of endoplasmic reticulum proteins in soybean root tips under flooding and drought stresses. J. Proteome Res. 2016, 15, 2211–2227. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Liu, Y.; Yu, K.; Zhou, Y. GmHMA3 sequesters Cd to the root endoplasmic reticulum to limit translocation to the stems in soybean. Plant Sci. 2018, 270, 23–29. [Google Scholar] [CrossRef]
- Komatsu, S.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Kono, Y.; Nishimura, M. Proteomic and biochemical analyses of the mechanism of tolerance in mutant soybean responding to flooding stress. Int. J. Mol. Sci. 2021, 22, 9046. [Google Scholar] [CrossRef]
- Li, J.; Dong, R.; Jia, Y.; Huang, J.; Zou, X.; An, N.; Song, J.; Chen, Z. Characterization of metal tolerance proteins and functional analysis of GmMTP8.1 involved in manganese tolerance in soybean. Front. Plant Sci. 2021, 12, 683813. [Google Scholar] [CrossRef]
- Wang, X.; Komatsu, S. Plant subcellular proteomics: Application for exploring optimal cell function in soybean. J. Proteom. 2016, 143, 45–56. [Google Scholar] [CrossRef]
- Komatsu, S.; Hashiguchi, A. Subcellular proteomics: Application to elucidation of flooding-response mechanisms in soybean. Proteomes 2018, 6, 13. [Google Scholar] [CrossRef]
- Vogel, S.; Gebbers, R.; Oertel, M.; Kramer, E. Evaluating soil-borne causes of biomass variability in grassland by remote and proximal sensing. Sensors 2019, 19, 4593. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Lazin, R.; Shen, X.; Anagnostou, E. Estimation of flood-damaged cropland area using a convolutional neural network. Environ. Res. Lett. 2021, 16, 054011. [Google Scholar] [CrossRef]
- Sayama, T.; Nakazaki, T.; Ishikawa, G.; Yagasaki, K.; Yamada, N.; Hirota, N.; Hirata, K.; Yoshikawa, T.; Saito, H.; Teraishi, M.; et al. QTL analysis of seed-flooding tolerance in soybean (Glycine max [L.] Merr.). Plant Sci. 2009, 176, 514–521. [Google Scholar] [CrossRef]
- Suematsu, K.; Abiko, T.; Nguyen, V.L.; Mochizuki, T. Phenotypic variation in root development of 162 soybean accessions under hypoxia condition at the seedling stage. Plant Prod. Sci. 2017, 20, 323–335. [Google Scholar] [CrossRef]
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef]
- Wang, X.; Komatsu, S. Review: Proteomic techniques for the development of flood-tolerant soybean. Int. J. Mol. Sci. 2020, 21, 7497. [Google Scholar] [CrossRef]
- Cao, W.; Zhou, J.; Yuan, Y.; Ye, H.; Nguyen, H.T.; Chen, J.; Zhou, J. Quantifying variation in soybean due to flood using a low-cost 3D imaging system. Sensors 2019, 19, 2682. [Google Scholar] [CrossRef]
- Wu, C.; Zeng, A.; Chen, P.; Hummer, W.; Mokua, J.; Shannon, J.G.; Nguyen, H.T. Evaluation and development of flood-tolerant soybean cultivars. Plant Breed. 2017, 136, 913–923. [Google Scholar] [CrossRef]
- Maranna, S.; Nataraj, V.; Kumawat, G.; Chandra, S.; Rajesh, V.; Ramteke, R.; Patel, R.M.; Ratnaparkhe, M.B.; Husain, S.M.; Gupta, S.; et al. Breeding for higher yield, early maturity, wider adaptability and waterlogging tolerance in soybean (Glycine max L.): A case study. Sci. Rep. 2021, 11, 22853. [Google Scholar] [CrossRef]
- Kausar, R.; Wang, X.; Komatsu, S. Crop proteomics under abiotic stress: From data to insights. Plants 2022, 11, 2877. [Google Scholar] [CrossRef]
- Yeung, E.; Bailey-Serres, J.; Sasidharan, R. After the deluge: Plant revival post-flooding. Trends Plant Sci. 2019, 24, 443–454. [Google Scholar] [CrossRef]
- Fenta, B.A.; Beebe, S.E.; Kunert, K.J.; Burridge, J.D.; Barlow, K.M.; Lynch, J.P.; Foyer, C.H. Field phenotyping of soybean roots for drought stress tolerance. Agronomy 2014, 4, 418–435. [Google Scholar] [CrossRef]
- Ergo, V.V.; Veas, R.E.; Vega, C.R.C.; Lascano, R.; Carrera, C.S. Leaf photosynthesis and senescence in heated and droughted field-grown soybean with contrasting seed protein concentration. Plant Physiol. Biochem. 2021, 166, 437–447. [Google Scholar] [CrossRef]
- Kulundžić, A.M.; Josipović, A.; Kočar, M.M.; Vuletić, M.V.; Dunić, J.A.; Varga, I.; Cesar, V.; Sudarić, A.; Lepeduš, H. Physiological insights on soybean response to drought. Agr. Water Manag. 2022, 268, 107620. [Google Scholar] [CrossRef]
- Molinari, M.D.C.; Fuganti-Pagliarini, R.; Marcolino-Gomes, J.; Barbosa, D.D.; Marin, S.R.R.; Mertz-Henning, L.M.; Nepomuceno, A.L.; Rech, E.L. Flower and pod genes involved in soybean sensitivity to drought. J. Plant Interact. 2021, 16, 187–200. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, P.; Bai, Y.; Wang, Y.; Wan, H.; Liu, C.; Ni, Z. The soybean F-box protein GmFBX176 regulates ABA-mediated responses to drought and salt stress. Environ. Exp. Bot. 2020, 176, 104056. [Google Scholar] [CrossRef]
- Wang, X.; Song, S.; Wang, X.; Liu, J.; Dong, S. Transcriptomic and metabolomic analysis of seedling-stage soybean responses to PEG-simulated drought stress. Int. J. Mol. Sci. 2022, 23, 6869. [Google Scholar] [CrossRef]
- Ma, X.J.; Yu, T.F.; Li, X.H.; Cao, X.Y.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Zhang, J.H.; et al. Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants. BMC Plant Biol. 2020, 20, 123. [Google Scholar] [CrossRef]
- Ma, X.J.; Fu, J.D.; Tang, Y.M.; Yu, T.F.; Yin, Z.G.; Chen, J.; Zhou, Y.B.; Chen, M.; Xu, Z.S.; Ma, Y.Z. GmNFYA13 improves salt and drought tolerance in transgenic soybean plants. Front. Plant Sci. 2020, 11, 587244. [Google Scholar] [CrossRef]
- Sun, M.; Li, Y.; Zheng, J.; Wu, D.; Li, C.; Li, Z.; Zang, Z.; Zhang, Y.; Fang, Q.; Li, W.; et al. A nuclear factor Y-B transcription factor, GmNFYB17, regulates resistance to drought stress in soybean. Int. J. Mol. Sci. 2022, 23, 7242. [Google Scholar] [CrossRef]
- Li, B.; Zheng, J.C.; Wang, T.T.; Min, D.H.; Wei, W.L.; Chen, J.; Zhou, Y.B.; Chen, M.; Xu, Z.S.; Ma, Y.Z. Expression analyses of soybean VOZ transcription factors and the role of GmVOZ1G in drought and salt stress tolerance. Int. J. Mol. Sci. 2020, 21, 2177. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Chuong, N.N.; Hoa, T.T.K.; Doan, H.; Van, P.H.P.; Trang, L.D.M.; Huyen, P.N.T.; Le, D.T.; Tran, L.P.; Thao, N.P. The drought-mediated soybean GmNAC085 functions as a positive regulator of plant response to salinity. Int. J. Mol. Sci. 2021, 22, 8986. [Google Scholar] [CrossRef]
- Wei, W.; Liang, D.W.; Bian, X.H.; Shen, M.; Xiao, J.H.; Zhang, W.K.; Ma, B.; Lin, Q.; Lv, J.; Chen, X.; et al. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 2019, 1002, 384–398. [Google Scholar] [CrossRef]
- Ning, W.; Zhai, H.; Yu, J.; Liang, S.; Yang, X.; Xing, X.; Huo, J.; Pang, T.; Yang, Y.; Bai, X. Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol. Breed. 2017, 37, 1–10. [Google Scholar] [CrossRef]
- Kan, G.; Ning, L.; Li, Y.; Hu, Z.; Zhang, W.; He, X.; Yu, D. Identification of novel loci for salt stress at the seed germination stage in soybean. Breed. Sci. 2016, 66, 530–541. [Google Scholar] [CrossRef]
- Shi, M.; Liao, X.; Qian, Y.; Zhang, W.; Li, Y.; Bhat, J.; Kan, G.; Yu, D. Linkage and association mapping of wild soybean (Glycine soja) seeds germinating under salt stress. J. Integr. Agr. 2022, 21, 2833–2847. [Google Scholar] [CrossRef]
- Ji, W.; Cong, R.; Li, S.; Li, R.; Qin, Z.; Li, Y.; Zhou, X.; Chen, S.; Li, J. Comparative proteomic analysis of soybean leaves and roots by iTRAQ provides insights into response mechanisms to short-term salt stress. Front. Plant Sci. 2016, 7, 573. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, N.; Yang, J.; Guo, H.; Liu, Z.; Zheng, X.; Li, S.; Xiang, F. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 2020, 291, 110326. [Google Scholar] [CrossRef]
- Ledesma, F.; Lopez, C.; Ortiz, D.; Chen, P.; Korth, K.; Ishibashi, T.; Zeng, A.; Orazaly, M.; Florez-Palacios, L. A simple greenhouse method for screening salt tolerance in soybean. Crop Sci. 2016, 56, 585–594. [Google Scholar] [CrossRef]
- Do, T.; Vuong, T.; Dunn, D.; Smothers, S.; Patil, G.; Yungbluth, D.; Chen, P.; Scaboo, A.; Xu, D.; Carter, T.; et al. Mapping and confirmation of loci for salt tolerance in a novel soybean germplasm, Fiskeby III. Theor. Appl. Genet. 2018, 131, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, L.; Qu, Y.; Chen, J.; Liu, X.; Hong, H.; Liu, Z.; Chang, R.; Gilliham, M.; Qiu, L.; et al. GmSALT3, which confers improved soybean salt tolerance in the field, increases leaf Cl− exclusion prior to Na+ exclusion but does not improve early vigor under salinity. Front. Plant Sci. 2016, 7, 1485. [Google Scholar] [CrossRef]
- Do, T.; Chen, H.; Hien, V.; Hamwieh, A.; Yamada, T.; Sato, T.; Yan, Y.; Cong, H.; Shono, M.; Suenaga, K.; et al. Ncl synchronously regulates Na+, K+, and Cl− in soybean and greatly increases the grain yield in saline field conditions. Sci. Rep. 2016, 6, 19147. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, W.; Cao, Q.; Zheng, X.; Yang, J.; Xue, T.; Sun, W.; Du, X.; Wang, L.; Wang, J.; et al. WinRoots: A high-throughput cultivation and phenotyping system for plant phenomics studies under soil stress. Front. Plant Sci. 2022, 12, 794020. [Google Scholar] [CrossRef]
- Phang, T.H.; Shao, G.; Lam, H.M. Salt tolerance in soybean. J. Integr. Plant Biol. 2008, 50, 1196–1212. [Google Scholar] [CrossRef]
- He, Y.; Fu, J.; Yu, C.; Wang, X.; Jiang, Q.; Hong, J.; Lu, K.; Xue, G.; Yan, C.; James, A.; et al. Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. J. Exp. Bot. 2015, 66, 6877–6889. [Google Scholar] [CrossRef]
- Wei, P.; Wang, L.; Liu, A.; Yu, B.; Lam, H.M. GmCLC1 confers enhanced salt tolerance through regulating chloride accumulation in soybean. Front. Plant Sci. 2016, 7, 1082. [Google Scholar] [CrossRef]
- Jia, Q.; Li, M.W.; Zheng, C.; Xu, Y.; Sun, S.; Li, Z.; Wong, F.L.; Song, J.; Lin, W.W.; Li, Q.; et al. The soybean plasma membrane-localized cation/H+ exchanger GmCHX20a plays a negative role under salt stress. Physiol. Plant. 2021, 171, 714–727. [Google Scholar] [CrossRef]
- Liu, X.; Liu, F.; Zhang, L.; Cheng, C.; Wei, P.; Yu, B. GsCLC-c2 from wild soybean confers chloride/salt tolerance to transgenic Arabidopsis and soybean composite plants by regulating anion homeostasis. Physiol. Plant. 2021, 172, 1867–1879. [Google Scholar] [CrossRef]
- Sun, T.; Ma, N.; Wang, C.; Fan, H.; Wang, M.; Zhang, J.; Cao, J.; Wang, D. A Golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front. Plant Sci. 2021, 12, 638340. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, J.; Zhang, T.; Xu, T.; Yang, L.; Li, X.; Ji, F.; Gao, Y.; Ali, S.; Zhang, Q.; et al. A putative plasma membrane Na+/H+ antiporter GmSOS1 is critical for salt stress tolerance in Glycine max. Front. Plant Sci. 2022, 13, 870695. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.J.; Tao, J.J.; Cheng, T.; Bian, X.H.; Wei, W.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Soybean miR172a improves salt tolerance and can function as a long-distance signal. Mol. Plant 2016, 9, 1337–1340. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Guan, R.; Yu, L.; Berkowitz, O.; David, R.; Whelan, J.; Ford, M.; Wege, S.; Qiu, L.; Gilliham, M. Enhanced reactive oxygen detoxification occurs in salt-stressed soybean roots expressing GmSALT3. Physiol. Plant. 2022, 174, e13709. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fu, X.; Zhang, S.; Chen, G.; Li, S.; Shangguan, T.; Zheng, Y.; Xu, F.; Chen, Z.H.; Xu, S. Evolutionary and regulatory pattern analysis of soybean Ca2+ ATPases for abiotic stress tolerance. Front. Plant Sci. 2022, 13, 898256. [Google Scholar] [CrossRef]
- Lu, L.; Wei, W.; Tao, J.J.; Lu, X.; Bian, X.H.; Hu, Y.; Cheng, T.; Yin, C.C.; Zhang, W.K.; Chen, S.Y.; et al. Nuclear factor Y subunit GmNFYA competes with GmHDA13 for interaction with GmFVE to positively regulate salt tolerance in soybean. Plant Biotechnol. J. 2021, 19, 2362–2379. [Google Scholar] [CrossRef]
- Feng, P.; Sun, X.; Liu, X.; Li, Y.; Sun, Q.; Lu, H.; Li, M.; Ding, X.; Dong, Y. Epigenetic regulation of plant tolerance to salt stress by histone acetyltransferase GsMYST1 from wild soybean. Front. Plant Sci. 2022, 13, 860056. [Google Scholar] [CrossRef]
- Yung, W.S.; Wang, Q.; Huang, M.; Wong, F.L.; Liu, A.; Ng, M.S.; Li, K.P.; Sze, C.C.; Li, M.W.; Lam, H.M. Priming-induced alterations in histone modifications modulate transcriptional responses in soybean under salt stress. Plant J. 2022, 109, 1575–1590. [Google Scholar] [CrossRef]
- Li, S.; Wang, N.; Ji, D.; Zhang, W.; Wang, Y.; Yu, Y.; Zhao, S.; Lyu, M.; You, J.; Zhang, Y.; et al. A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell 2019, 31, 2107–2130. [Google Scholar] [CrossRef]
- Jin, T.; Sun, Y.; Zhao, R.; Shan, Z.; Gai, J.; Li, Y. Overexpression of peroxidase gene GsPRX9 confers salt tolerance in soybean. Int. J. Mol. Sci. 2019, 20, 3745. [Google Scholar] [CrossRef]
- Zhao, Y.; Cui, Y.; Huang, S.; Yu, J.; Wang, X.; Xin, D.; Li, X.; Liu, Y.; Dai, Y.; Qi, Z.; et al. Genome-wide analysis of the glucose-6-phosphate dehydrogenase family in soybean and functional identification of GmG6PDH2 involvement in salt stress. Front. Plant Sci. 2020, 11, 214. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.W.; Wong, F.L.; Yung, W.S.; Ku, Y.S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K.; et al. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell. Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.H.; Li, W.; Niu, C.F.; Wei, W.; Hu, Y.; Han, J.Q.; Lu, X.; Tao, J.J.; Jin, M.; Qin, H.; et al. A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 2020, 225, 268–283. [Google Scholar] [CrossRef] [PubMed]
- Pi, E.; Qu, L.; Hu, J.; Huang, Y.; Qiu, L.; Lu, H.; Jiang, B.; Liu, C.; Peng, T.; Zhao, Y.; et al. Mechanisms of soybean roots’ tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol. Cell. Proteom. 2016, 15, 266–288. [Google Scholar] [CrossRef]
- Pi, E.; Xu, J.; Li, H.; Fan, W.; Zhu, C.; Zhang, T.; Jiang, J.; He, L.; Lu, H.; Wang, H.; et al. Enhanced salt tolerance of rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis. Mol. Cell. Proteom. 2019, 18, 2225–2243. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Dai, L.; Sun, H.; Chen, M.; Sun, Y. Effects of moderate soil salinity on osmotic adjustment and energy strategy in soybean under drought stress. Plant Physiol. Biochem. 2019, 139, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Yu, T.F.; Fu, J.D.; Su, H.G.; Chen, J.; Zhou, Y.B.; Chen, M.; Guo, J.; Ma, Y.Z.; Wei, W.L.; et al. Genome-wide analysis of the GRAS gene family and functional identification of GmGRAS37 in drought and salt tolerance. Front. Plant Sci. 2020, 11, 604690. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, S.; Wang, G.; Du, Y.; Zhang, Z.; Yu, G.; Ren, C.; Zhang, Y.; Du, J. Exogenous melatonin enhances soybean (Glycine max (L.) Merr.) seedling tolerance to saline-alkali stress by regulating antioxidant response and DNA damage repair. Physiol. Plant. 2022, 174, e13731. [Google Scholar] [CrossRef]
- Staniak, M.; Czopek, K.; Stępień-Warda, A.; Kocira, A.; Przybyś, M. Cold stress during flowering alters plant structure, yield and seed quality of different soybean genotypes. Agronomy 2021, 11, 2059. [Google Scholar] [CrossRef]
- Szczerba, A.; Płażek, A.; Pastuszak, J.; Kopeć, P.; Hornyák, M.; Dubert, F. Effect of low temperature on germination, growth, and seed yield of four soybean (Glycine max L.) cultivars. Agronomy 2021, 11, 800. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Hagihara, S.; Hirai, D. Field assessment of a major QTL associated with tolerance to cold-induced seed coat discoloration in soybean. Breed. Sci. 2019, 69, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Molecular regulation of plant responses to environmental temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, Y.; Randall, S.K. Functionality of soybean CBF/DREB1 transcription factors. Plant Sci. 2016, 246, 80–90. [Google Scholar] [CrossRef]
- Tacarindua, C.R.; Shiraiwa, T.; Homma, K.; Kumagai, E.; Sameshima, R. The effects of increased temperature on crop growth and yield of soybean grown in a temperature gradient chamber. Field Crop. Res. 2013, 154, 74–81. [Google Scholar] [CrossRef]
- Alsajri, F.A.; Wijewardana, C.; Irby, J.T.; Bellaloui, N.; Krutz, L.J.; Golden, B.; Gao, W.; Reddy, K.R. Developing functional relationships between temperature and soybean yield and seed quality. Agron. J. 2020, 112, 194–204. [Google Scholar] [CrossRef]
- Chebrolu, K.K.; Fritschi, F.B.; Ye, S.; Krishnan, H.B.; Smith, J.R.; Gillman, J.D. Impact of heat stress during seed development on soybean seed metabolome. Metabolomics 2016, 12, 1–14. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, Z.; Lu, Q.; Wang, P.; Li, Y.; Lv, Q.; Song, X.; Li, D.; Gu, Y.; Liu, L.; et al. Overexpression of a GmGBP1 ortholog of soybean enhances the responses to flowering, stem elongation and heat tolerance in transgenic tobaccos. Plant Mol. Biol. 2013, 82, 279–299. [Google Scholar] [CrossRef]
- Huang, Y.; Xuan, H.; Yang, C.; Guo, N.; Wang, H.; Zhao, J.; Xing, H. GmHsp90A2 is involved in soybean heat stress as a positive regulator. Plant Sci. 2019, 285, 26–33. [Google Scholar] [CrossRef]
- Valdés-López, O.; Batek, J.; Gomez-Hernandez, N.; Nguyen, C.T.; Isidra-Arellano, M.C.; Zhang, N.; Joshi, T.; Xu, D.; Hixson, K.K.; Weitz, K.K.; et al. Soybean roots grown under heat stress show global changes in their transcriptional and proteomic profiles. Front. Plant Sci. 2016, 7, 517. [Google Scholar] [CrossRef]
- Zinn, K.E.; Tunc-Ozdemir, M.; Harper, J.F. Temperature stress and plant sexual reproduction: Uncovering the weakest links. J. Exp. Bot. 2010, 61, 1959–1968. [Google Scholar] [CrossRef]
- Djanaguiraman, M.; Schapaugh, W.; Fritschi, F.; Nguyen, H.; Prasad, P.V. Reproductive success of soybean (Glycine max L. Merril) cultivars and exotic lines under high daytime temperature. Plant Cell Environ. 2019, 42, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, A.C.; De Smet, I.; Sozzani, R.; Locke, A.M. Field-grown soybean shows genotypic variation in physiological and seed composition responses to heat stress during seed development. Environ. Exp. Bot. 2022, 195, 104768. [Google Scholar] [CrossRef]
- Siebers, M.H.; Yendrek, C.R.; Drag, D.; Locke, A.M.; Rios Acosta, L.; Leakey, A.D.; Ainsworth, E.A.; Bernacchi, C.J.; Ort, D.R. Heat waves imposed during early pod development in soybean (Glycine max) cause significant yield loss despite a rapid recovery from oxidative stress. Glob. Chang. Biol. 2015, 21, 3114–3125. [Google Scholar] [CrossRef]
- Li, J.; Nadeem, M.; Chen, L.; Wang, M.; Wan, M.; Qiu, L.; Wang, X. Differential proteomic analysis of soybean anthers by iTRAQ under high-temperature stress. J. Proteom. 2020, 229, 103968. [Google Scholar] [CrossRef]
- Bureau, M.F.; Mederski, H.J.; Evans, C.E. The effect of phosphatic fertilizer material and soil phosphorus level on the yield and phosphorus uptake of soybeans. Agron. J. 1953, 45, 150–154. [Google Scholar] [CrossRef]
- Ning, L.H.; Du, W.K.; Song, H.N.; Shao, H.B.; Qi, W.C.; Sheteiwy, M.S.A.; Yu, D.Y. Identification of responsive miRNAs involved in combination stresses of phosphate starvation and salt stress in soybean root. Environ. Exp. Bot. 2019, 167, 103823. [Google Scholar] [CrossRef]
- Pascual, M.B.; Echevarria, V.; Gonzalo, M.J.; Hernández-Apaolaza, L. Silicon addition to soybean (Glycine max L.) plants alleviate zinc deficiency. Plant Physiol. Biochem. 2016, 108, 132–138. [Google Scholar] [CrossRef]
- Singh, S.; Husain, T.; Kushwaha, B.K.; Suhel, M.; Fatima, A.; Mishra, V.; Singh, S.K.; Bhatt, J.A.; Rai, M.; Prasad, S.M.; et al. Regulation of ascorbate-glutathione cycle by exogenous nitric oxide and hydrogen peroxide in soybean roots under arsenate stress. J. Hazard. Mater. 2021, 409, 123686. [Google Scholar] [CrossRef]
- Zeng, C.Q.; Liu, W.X.; Hao, J.Y.; Fan, D.N.; Chen, L.M.; Xu, H.N.; Li, K.Z. Measuring the expression and activity of the CAT enzyme to determine Al resistance in soybean. Plant Physiol. Biochem. 2019, 144, 254–263. [Google Scholar] [CrossRef]
- Shu, W.; Zhou, Q.; Xian, P.; Cheng, Y.; Lian, T.; Ma, Q.; Zhou, Y.; Li, H.; Nian, H.; Cai, Z. GmWRKY81 encoding a WRKY transcription factor enhances aluminum tolerance in soybean. Int. J. Mol. Sci. 2022, 23, 6518. [Google Scholar] [CrossRef]
- Bashir, W.; Anwar, S.; Zhao, Q.; Hussain, I.; Xie, F. Interactive effect of drought and cadmium stress on soybean root morphology and gene expression. Ecotoxicol. Environ. Saf. 2019, 175, 90–101. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Komatsu, S.; Kuji, R.; Nanjo, Y.; Hiraga, S.; Furukawa, K. Comprehensive analysis of endoplasmic reticulum-enriched fraction in root tips of soybean under flooding stress using proteomics techniques. J. Proteom. 2012, 77, 531–560. [Google Scholar] [CrossRef]
- Komatsu, S.; Yamamoto, A.; Nakamura, T.; Nouri, M.Z.; Nanjo, Y.; Nishizawa, K.; Furukawa, K. Comprehensive analysis of mitochondria in roots and hypocotyls of soybean under flooding stress using proteomics and metabolomics techniques. J. Proteome Res. 2011, 10, 3993–4004. [Google Scholar] [CrossRef]
- Yin, X.; Komatsu, S. Nuclear proteomics reveals the role of protein synthesis and chromatin structure in root tip of soybean during the initial stage of flooding stress. J. Proteome Res. 2016, 15, 2283–2298. [Google Scholar] [CrossRef]
- Yin, X.; Komatsu, S. Quantitative proteomics of nuclear phosphoproteins in the root tip of soybean during the initial stages of flooding stress. J. Proteom. 2015, 119, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Rehman, H.M.; Chen, S.; Zhang, S.; Khalid, M.; Uzair, M.; Wilmarth, P.A.; Ahmad, S.; Lam, H.M. Membrane proteomic profiling of soybean leaf and root tissues uncovers salt-stress-responsive membrane proteins. Int. J. Mol. Sci. 2022, 23, 13270. [Google Scholar] [CrossRef]
- Wei, Y.; Jiang, C.; Han, R.; Xie, Y.; Liu, L.; Yu, Y. Plasma membrane proteomic analysis by TMT-PRM provides insight into mechanisms of aluminum resistance in tamba black soybean roots tips. PeerJ 2020, 8, e9312. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Kobayashi, Y.; Nishizawa, K.; Nanjo, Y.; Furukawa, K. Comparative proteomics analysis of differentially expressed proteins in soybean cell wall during flooding stress. Amino Acids 2010, 39, 1435–1449. [Google Scholar] [CrossRef] [PubMed]
- Arai, Y.; Hayashi, M.; Nishimura, M. Proteomic analysis of highly purified peroxisomes from etiolated soybean cotyledons. Plant Cell Physiol. 2008, 49, 526–539. [Google Scholar] [CrossRef][Green Version]
- Arai, Y.; Hayashi, M.; Nishimura, M. Proteomic identification and characterization of a novel peroxisomal adenine nucleotide transporter supplying ATP for fatty acid beta-oxidation in soybean and Arabidopsis. Plant Cell 2008, 20, 3227–3240. [Google Scholar] [CrossRef] [PubMed]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef]
- He, Y.; Yu, C.; Zhou, L.; Chen, Y.; Liu, A.; Jin, J.; Qi, Y.; Jiang, D. Rubisco decrease is involved in chloroplast protrusion and Rubisco-containing body formation in soybean (Glycine max.) under salt stress. Plant Physiol. Biochem. 2014, 74, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Li, H.; Zhang, X.; Yu, K.; Chao, M.; Han, S.; Zhang, D. Physiological and proteomics analyses reveal low-phosphorus stress affected the regulation of photosynthesis in soybean. Int. J. Mol. Sci. 2018, 19, 1688. [Google Scholar]
- Fan, Y.; Chen, J.; Wang, Z.; Tan, T.; Li, S.; Li, J.; Wang, B.; Zhang, J.; Cheng, Y.; Wu, X.; et al. Soybean (Glycine max L. Merr.) seedlings response to shading: Leaf structure, photosynthesis and proteomic analysis. BMC Plant Biol. 2019, 19, 34. [Google Scholar] [CrossRef] [PubMed]
- Shelke, D.B.; Pandey, M.; Nikalje, G.C.; Zaware, B.N.; Suprasanna, P.; Nikam, T.D. Salt responsive physiological, photosynthetic and biochemical attributes at early seedling stage for screening soybean genotypes. Plant Physiol. Biochem. 2017, 118, 519–528. [Google Scholar]
- Zhou, H.; Liang, X.; Feng, N.; Zheng, D.; Qi, D. Effect of uniconazole to soybean seed priming treatment under drought stress at VC stage. Ecotoxicol. Environ. Saf. 2021, 224, 112619. [Google Scholar]
- Fayazipour, D.; Deckert, J.; Akbari, G.; Soltani, E.; Chmielowska-Bąk, J. Mitochondria specific antioxidant, MitoTEMPO, modulates Cd uptake and oxidative response of soybean seedlings. Antioxidants 2022, 11, 2099. [Google Scholar]
- Chen, Z.; Yan, W.; Sun, L.; Tian, J.; Liao, H. Proteomic analysis reveals growth inhibition of soybean roots by manganese toxicity is associated with alteration of cell wall structure and lignification. J. Proteom. 2016, 143, 151–160. [Google Scholar]
- Hossain, Z.; Nouri, M.Z.; Komatsu, S. Plant cell organelle proteomics in response to abiotic stress. J. Proteome Res. 2012, 11, 37–48. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, J.; Fang, Q.; Chang, X.; Sun, M.; Li, W.; Li, Y. GmAKT1 is involved in K+ uptake and Na+/K+ homeostasis in Arabidopsis and soybean plants. Plant Sci. 2021, 304, 110736. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, B. Effect of exogenous calcium on growth, nutrients uptake and plasma membrane H+-ATPase and Ca2+-ATPase activities in soybean (Glycine max) seedlings under simulated acid rain stress. Ecotoxicol. Environ. Saf. 2018, 165, 261–269. [Google Scholar] [CrossRef]
- An, P.; Li, X.; Zheng, Y.; Matsuura, A.; Abe, J.; Eneji, A.E.; Tanimoto, E.; Inanaga, S. Effects of NaCl on root growth and cell wall composition of two soya bean cultivars with contrasting salt tolerance. J. Agron. Crop Sci. 2014, 200, 212–218. [Google Scholar] [CrossRef]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Yan, X.; Liao, H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef]
- Yang, Z.; Gao, Z.; Zhou, H.; He, Y.; Liu, Y.; Lai, Y.; Zheng, J.; Li, X.; Liao, H. GmPTF1 modifies root architecture responses to phosphate starvation primarily through regulating GmEXPB2 expression in soybean. Plant J. 2021, 107, 525–543. [Google Scholar] [CrossRef]
- Del Río, L.A.; López-Huertas, E. ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 2016, 57, 1364–1376. [Google Scholar] [CrossRef]
- Wang, N.; Liu, Y.; Cai, Y.; Tang, J.; Li, Y.; Gai, J. The soybean U-box gene GmPUB6 regulates drought tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 155, 284–296. [Google Scholar] [CrossRef]
- Shimada, T.; Takagi, J.; Ichino, T.; Shirakawa, M.; Hara-Nishimura, I. Plant Vacuoles. Annu. Rev. Plant Biol. 2018, 69, 123–145. [Google Scholar] [CrossRef]
- Vorster, B.J.; Cullis, C.A.; Kunert, K.J. Plant vacuolar processing enzymes. Front. Plant Sci. 2019, 10, 479. [Google Scholar] [CrossRef]
- Krishnan, H.B.; Kim, W.S.; Oehrle, N.W.; Smith, J.R.; Gillman, J.D. Effect of heat stress on seed protein composition and ultrastructure of protein storage vacuoles in the cotyledonary parenchyma cells of soybean genotypes that are either tolerant or sensitive to elevated temperatures. Int. J. Mol. Sci. 2020, 21, 4775. [Google Scholar] [CrossRef]
- Cilliers, M.; van Wyk, S.G.; van Heerden, P.D.R.; Kunert, K.J.; Vorster, B.J. Identification and changes of the drought-induced cysteine protease transcriptome in soybean (Glycine max) root nodules. Environ. Exp. Bot. 2018, 148, 59–69. [Google Scholar] [CrossRef]
- Komatsu, S.; Wada, T.; Abaléa, Y.; Nouri, M.Z.; Nanjo, Y.; Nakayama, N.; Shimamura, S.; Yamamoto, R.; Nakamura, T.; Furukawa, K. Analysis of plasma membrane proteome in soybean and application to flooding stress response. J. Proteome Res. 2009, 8, 4487–4499. [Google Scholar] [CrossRef]
- Yin, G.; Sun, H.; Xin, X.; Qin, G.; Liang, Z.; Jing, X. Mitochondrial damage in the soybean seed axis during imbibition at chilling temperatures. Plant Cell Physiol. 2009, 50, 1305–1318. [Google Scholar] [CrossRef]
- Kamal, A.H.; Komatsu, S. Involvement of reactive oxygen species and mitochondrial proteins in biophoton emission in roots of soybean plants under flooding stress. J. Proteome Res. 2015, 14, 2219–2236. [Google Scholar] [CrossRef]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Priya, S.; Sharma, S.K.; Goloubinoff, P. Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides. FEBS Lett. 2013, 587, 1981–1987. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, H.K.; Dong, Q.L.; Zhang, Y.Y.; Wang, Y.M.; Li, H.Y.; Xing, G.J.; Li, Q.Y.; Dong, Y.S. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 2015, 6, 773. [Google Scholar] [CrossRef]
- Lu, L.; Dong, C.; Liu, R.; Zhou, B.; Wang, C.; Shou, H. Roles of soybean plasma membrane intrinsic protein GmPIP2;9 in drought tolerance and seed development. Front. Plant Sci. 2018, 9, 530. [Google Scholar] [CrossRef]
- Xiao, Y.; Karikari, B.; Wang, L.; Chang, F.; Zhao, T. Structure characterization and potential role of soybean phospholipases A multigene family in response to multiple abiotic stress uncovered by CRISPR/Cas9 technology. Environ. Exp. Bot. 2021, 188, 104521. [Google Scholar] [CrossRef]
- Shimamura, S.; Yoshioka, T.; Yamamoto, R.; Hiraga, S.; Nakamura, T.; Shimada, S.; Komatsu, S. Role of abscisic acid in flood-induced secondary aerenchyma formation in soybean (Glycine max) hypocotyls. Plant Prod. Sci. 2014, 17, 131–137. [Google Scholar] [CrossRef]
- Komatsu, S.; Han, C.; Nanjo, Y.; Altaf-Un-Nahar, M.; Wang, K.; He, D.; Yang, P. Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteome Res. 2013, 12, 4769–4784. [Google Scholar] [CrossRef]
- Yin, X.; Nishimura, M.; Hajika, M.; Komatsu, S. Quantitative proteomics reveals the flooding-tolerance mechanism in mutant and abscisic acid-treated soybean. J. Proteome Res. 2016, 15, 2008–2025. [Google Scholar] [CrossRef]
- Ni, Z.; Hu, Z.; Jiang, Q.; Zhang, H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 2013, 82, 113–129. [Google Scholar] [CrossRef]
- Tamang, B.G.; Li, S.; Rajasundaram, D.; Lamichhane, S.; Fukao, T. Overlapping and stress-specific transcriptomic and hormonal responses to flooding and drought in soybean. Plant J. 2021, 107, 100–117. [Google Scholar] [CrossRef]
- Wei, Y.; Jin, J.; Jiang, S.; Ning, S.; Liu, L. Quantitative response of soybean development and yield to drought stress during different growth stages in the Huaibei Plain, China. Agronomy 2018, 8, 97. [Google Scholar] [CrossRef]
- Papiernik, S.K.; Grieve, C.M.; Lesch, S.M.; Yates, S.R. Effects of salinity, imazethapyr, and chlorimuron application on soybean growth and yield. Commun. Soil. Sci. Plan. 2005, 36, 951–967. [Google Scholar] [CrossRef]
- Luan, X.; Bommarco, R.; Scaini, A.; Vico, G. Combined heat and drought suppress rainfed maize and soybean yields and modify irrigation benefits in the USA. Environ. Res. Lett. 2021, 16, 064023. [Google Scholar] [CrossRef]
- Lee, H.J.; Seo, P.J. Ca2+ talyzing initial responses to environmental stresses. Trends Plant Sci. 2021, 26, 849–870. [Google Scholar] [CrossRef]
- Ambrosino, L.; Colantuono, C.; Diretto, G.; Fiore, A.; Chiusano, M.L. Bioinformatics resources for plant abiotic stress responses: State of the art and opportunities in the fast evolving-omics era. Plants 2020, 9, 591. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of multi-omics technologies for crop improvement. Front. Plant Sci. 2021, 12, 563953. [Google Scholar] [CrossRef]
- Weckwerth, W.; Ghatak, A.; Bellaire, A.; Chaturvedi, P.; Varshney, R.K. PANOMICS meets germplasm. Plant Biotechnol. J. 2020, 10, 1111. [Google Scholar] [CrossRef]

| Subcellular | Organ | Plant | Stress | Technique | Findings | Ref. |
|---|---|---|---|---|---|---|
| ER | root tip | 2-day-old | flood/2 days | 1DE LC–MS/ MS, gel-free LC–MS/MS | Under flood conditions, 117 and 212 proteins increased and decreased, and 111 proteins were enriched in post-translational modification, protein synthesis, folding, degradation, and activation. | [114] |
| ER | root tip | 2-day-old | flood/2 days, drought/2 days | gel-free/label-free LC–MS/MS | A number of 368 and 103 proteins were identified under flood conditions and drought, and proteins enriched in protein glycosylation and signal responded to both stresses. | [27] |
| Chloroplast | leaf | 10-day-old | O3 (120 ppb)/3 days | 2DE MALDI–TOF/MS | A total of 32 proteins were responsive to ozone stress, and proteins associated with photosynthesis mostly decreased. | [23] |
| Mitochondrion | root and hypocotyl | 2-day-old | flood/2 days | 2DE, BN-PAGE LC–MS/MS | In mitochondria, 34 matrix proteins and 16 membrane proteins changed by flood conditions, and proteins related to the tricarboxylic acid cycle increased, while inner membrane carriers and proteins related to complexes III, IV, and V of electron transport chains decreased. | [115] |
| Mitochondrion | root tip | 2-day-old | flood with A12O3 (5, 30–60, 135 nm at 50 ppm)/2 days | gel-free/ label-free LC–MS/MS | Under flood conditions, six, five, and six proteins were specifically changed in plants exposed to 5, 30–60, and 135 nm A12O3, indicating various sizes of nanoparticles affected membrane permeability and the tricarboxylic acid cycle activity through mediating mitochondrial proteins. | [25] |
| Nucleus | root tip | 2-day-old | flood/3, 6, 24 h | gel-free/label-free LC–MS/MS | A total of 237 proteins were altered by flood conditions in a time–course manner, while 365 proteins were changed by the initial 3 h flood and mapped to pre-mRNA processing and pre-ribosome biogenesis. | [116] |
| Nucleus | root tip | 2-day-old | flood/3 h | gel-free/label-free LC–MS/MS | Under flood conditions, 14 nuclear phosphoproteins were changed, and phosphorylation of zinc finger/BTB domain-containing protein 47 was responsive to activated ABA signal for stress tolerance. | [117] |
| Plasma membrane | leaf and root | 7-day-old | NaCl (50, 100 mM)/7 days | gel-free/label-free LC–MS/MS | Under the presence of salt, 140 and 57 proteins were specific to the root and leaf, and they were mainly involved in transport inside the cell. | [118] |
| Plasma membrane | root tip | 14-day-old | AlCl3 (50 μM in 0.5 mM CaCl2)/3 days | TMT LC–MS/MS | A total of 90 proteins were differentially accumulated and enriched in membrane trafficking/transporters, cell wall modification, defense response, and signal transduction. | [119] |
| Cell wall | root and hypocotyl | 2-day-old | flood/2 days | 2DE MALDI–TOF/MS | Under flood conditions, 16 proteins responded with decreased stem 28/31 kDa glycoprotein precursors, germin-like protein precursors, and increased methionine synthases. | [120] |
| Peroxisome | cotyledon | seed | dark/7 day | 2DE MALDI–TOF/MS | Identified peroxisomal proteins harboring peroxisomal targeting signal sequences were associated with the beta-oxidation cycle, glyoxylate cycle, and stress response. | [121] |
| Peroxisome | cotyledon | seed | dark/7 day | BN-PAGE MALDI–TOF/MS | GmPNC1 was identified, and PNC1 contributed to the transport of adenine nucleotides consumed during post-germinative growth. | [122] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Komatsu, S. Subcellular Proteomics to Elucidate Soybean Response to Abiotic Stress. Plants 2023, 12, 2865. https://doi.org/10.3390/plants12152865
Wang X, Komatsu S. Subcellular Proteomics to Elucidate Soybean Response to Abiotic Stress. Plants. 2023; 12(15):2865. https://doi.org/10.3390/plants12152865
Chicago/Turabian StyleWang, Xin, and Setsuko Komatsu. 2023. "Subcellular Proteomics to Elucidate Soybean Response to Abiotic Stress" Plants 12, no. 15: 2865. https://doi.org/10.3390/plants12152865
APA StyleWang, X., & Komatsu, S. (2023). Subcellular Proteomics to Elucidate Soybean Response to Abiotic Stress. Plants, 12(15), 2865. https://doi.org/10.3390/plants12152865






