The Endophytic Microbiome of Wild Grapevines Vitis amurensis Rupr. and Vitis coignetiae Pulliat Growing in the Russian Far East
Abstract
:1. Introduction
2. Results
2.1. High-Throughput Sequencing
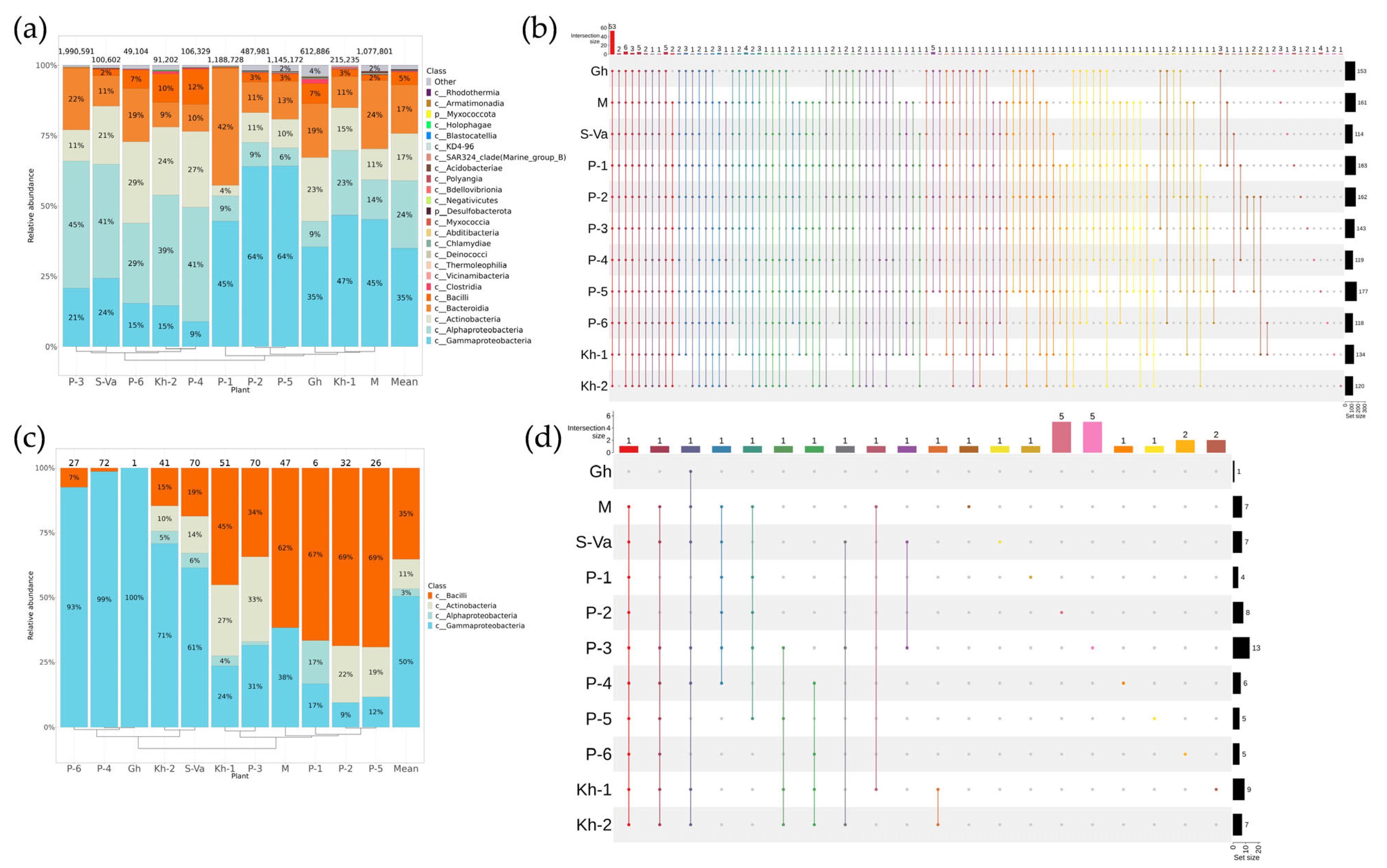
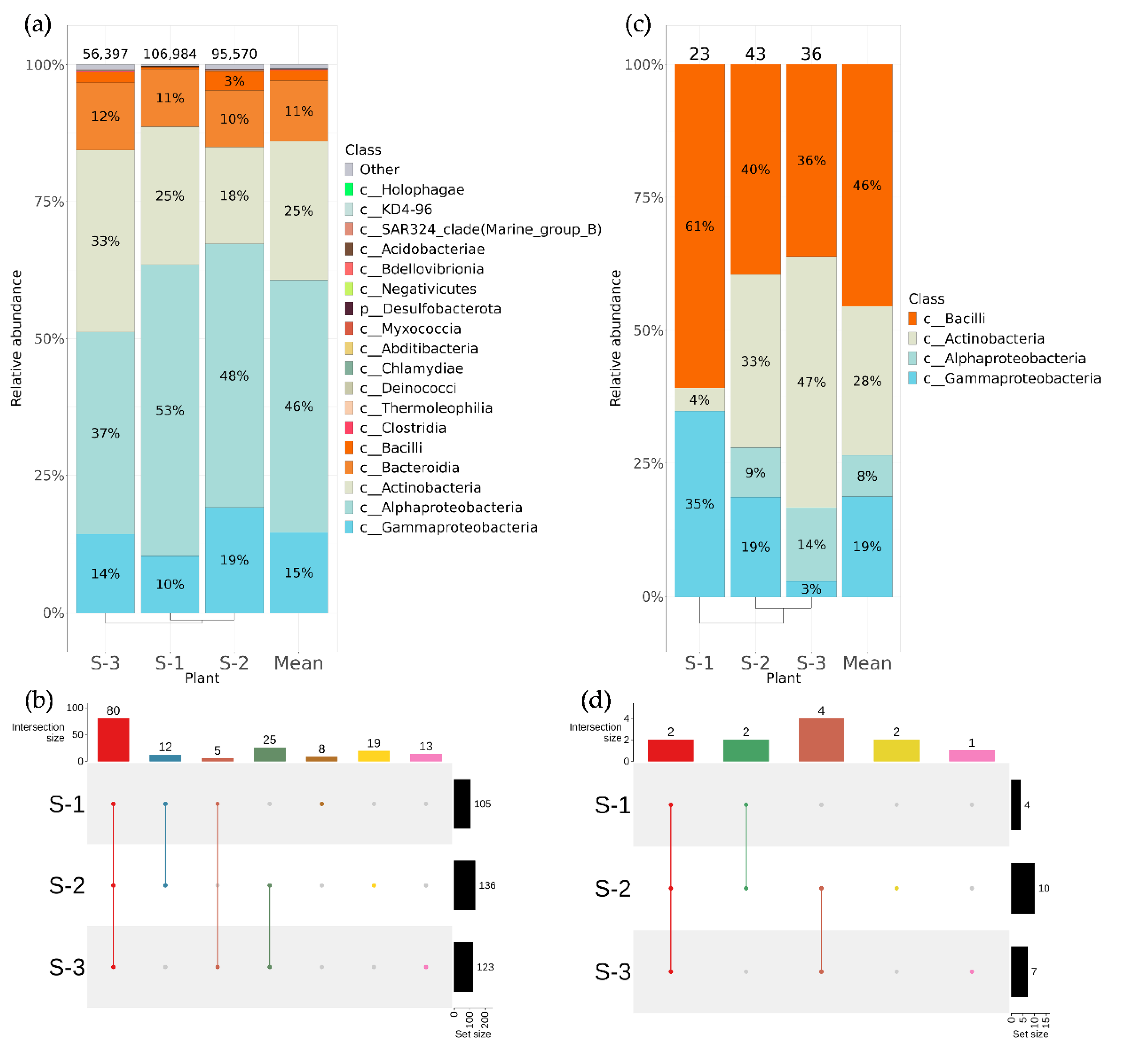
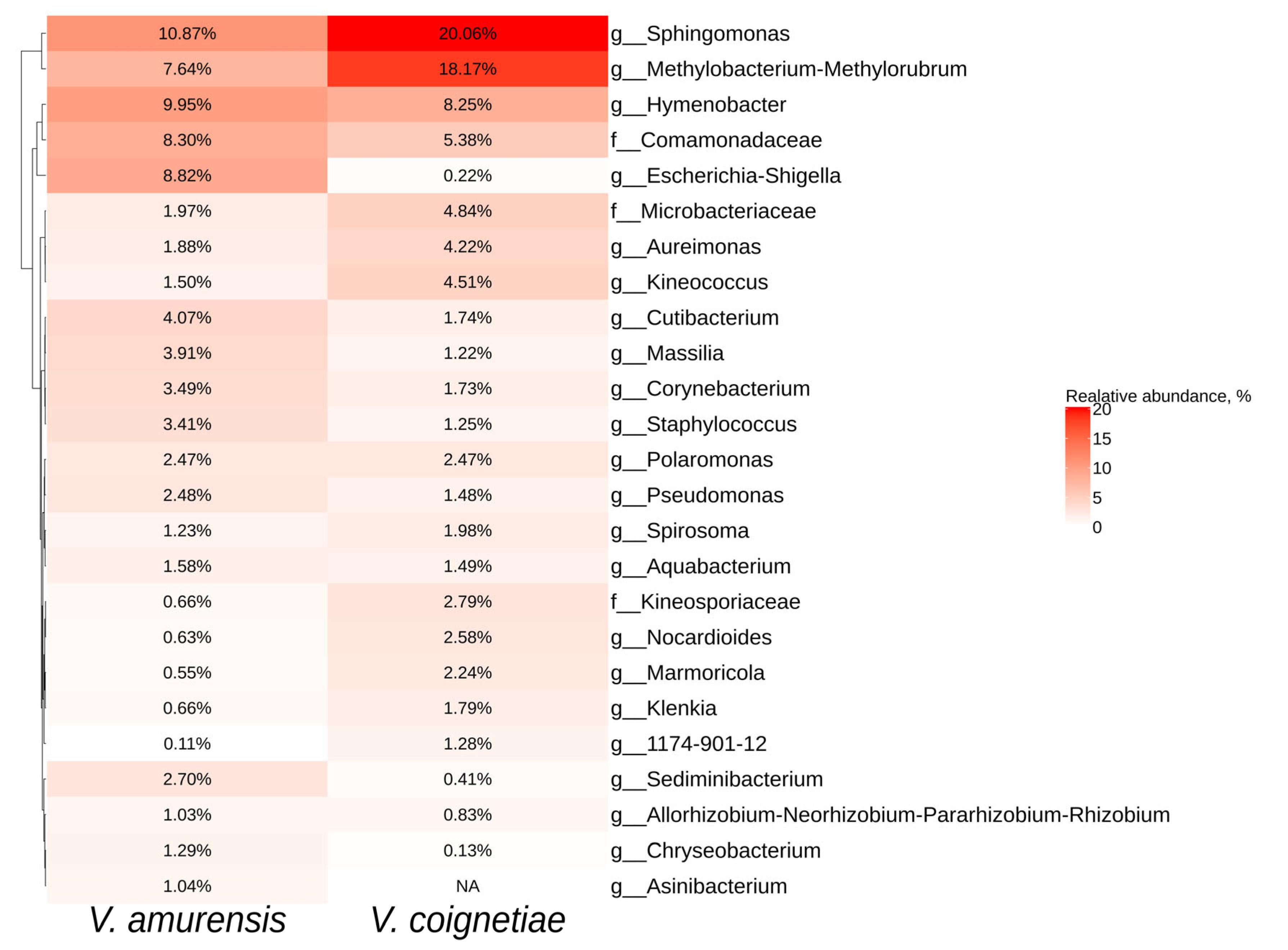
2.2. Biodiversity of Endophytic Bacteria in Vitis amurensis and Vitis coignetiae
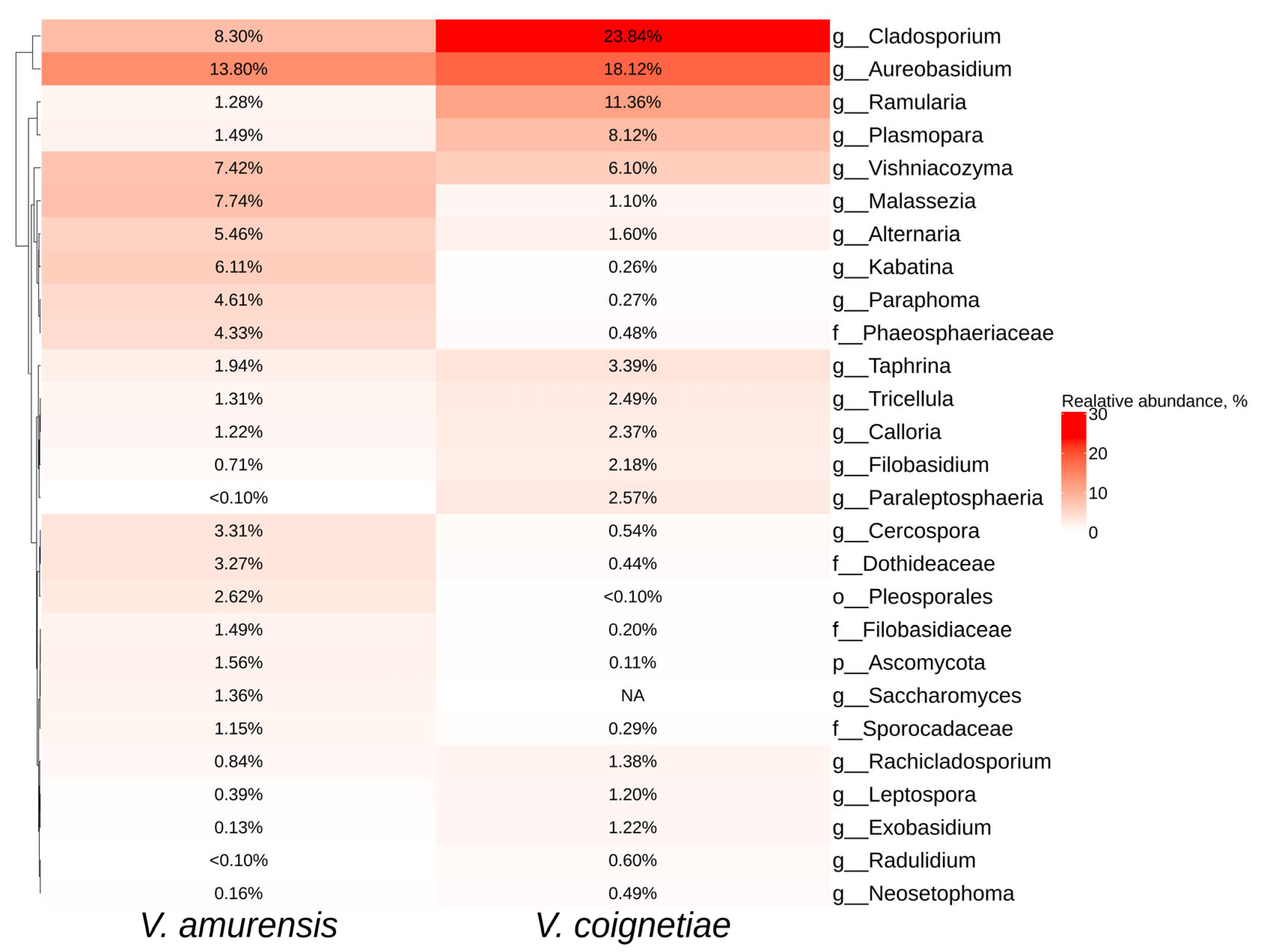
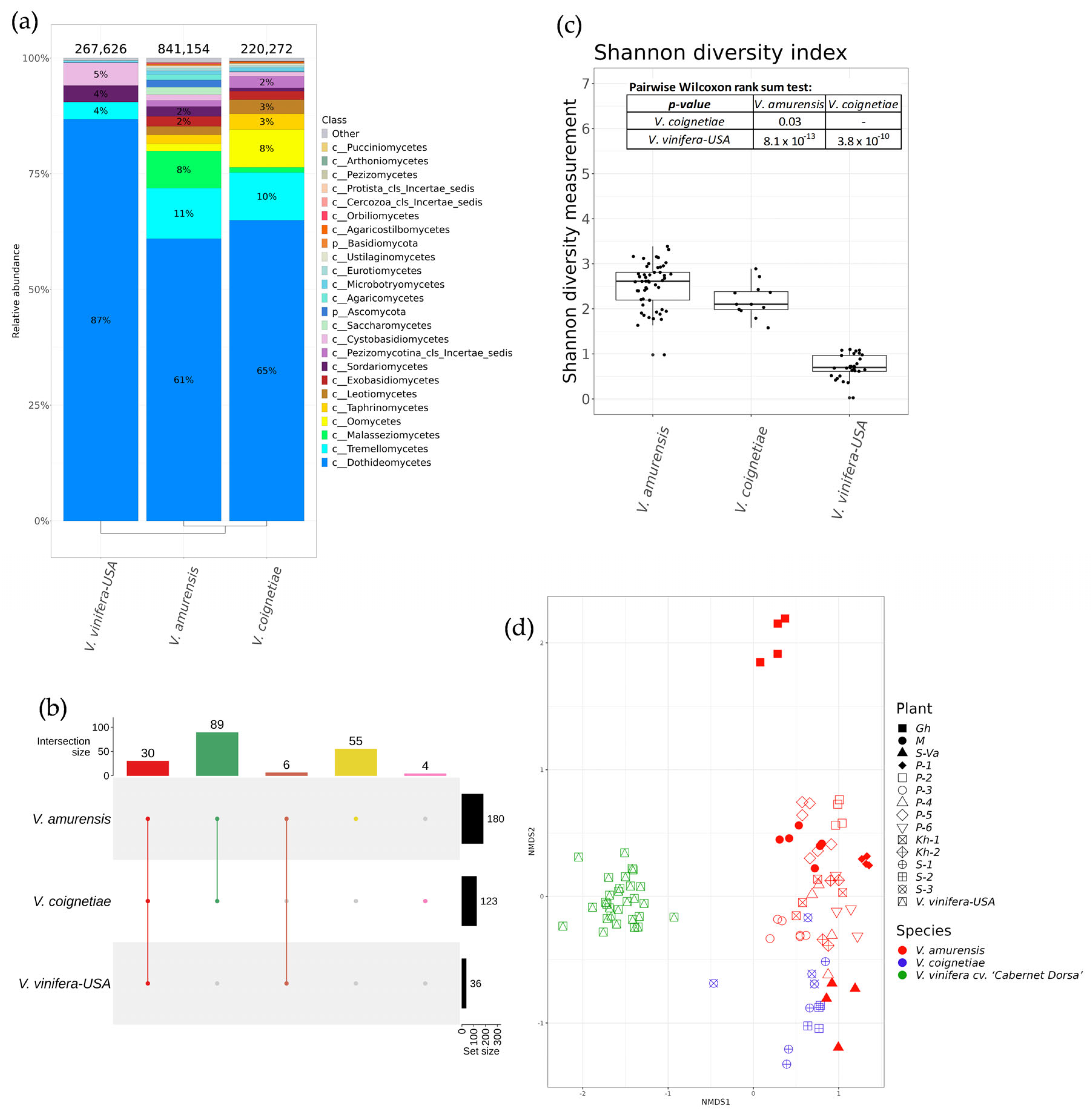
2.3. The Biodiversity of Fungal and Fungus-Like Endophytes of Vitis amurensis and Vitis coignetiae Grapevines
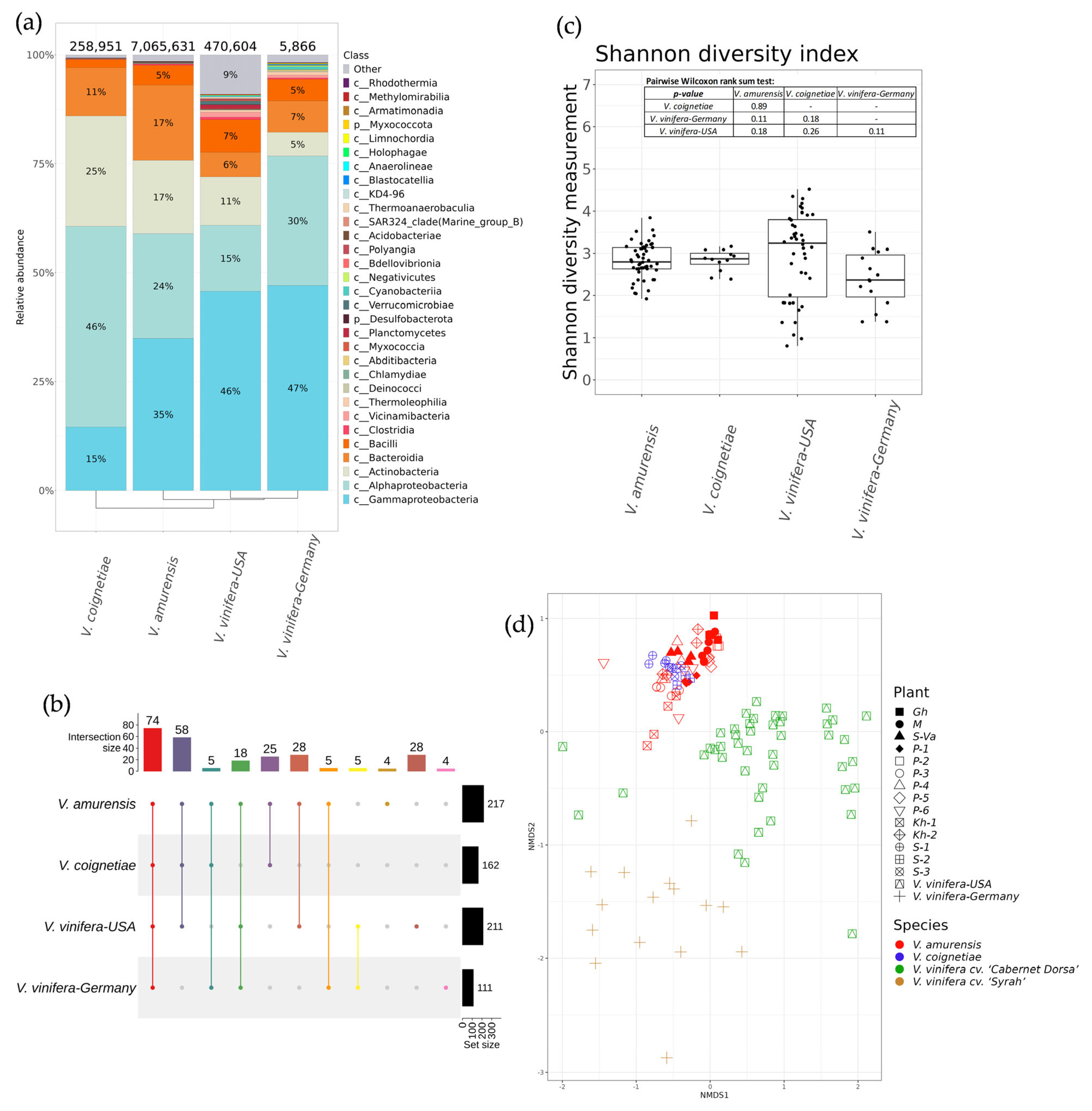
2.4. Comparative Analysis of Endophytic Microbial Communities in Vitis amurensis, Vitis coignetiae, and Vitis vinifera
3. Discussion
4. Materials and Methods
4.1. Plant Material and Sampling
4.2. Isolation and Identification of the Endophytic Bacteria and Fungi
4.3. DNA Extraction, Library Preparation, and Illumina MiSeq Sequencing
4.4. Data Processing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strobel, G.; Daisy, B.; Castillo, U.; Harper, J. Natural Products from Endophytic Microorganisms. J. Nat. Prod. 2004, 67, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Firdous, J.; Lathif, N.A.; Mona, R.; Muhamad, N. Endophytic Bacteria and Their Potential Application in Agriculture: A Review. Indian J. Agric. Res. 2019, 53, 1–7. [Google Scholar]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial Endophyte Colonization and Distribution within Plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felber, A.C.; Orlandelli, R.C.; Rhoden, S.A.; Garcia, A.; Costa, A.T.; Azevedo, J.L.; Pamphile, J.A. Bioprospecting Foliar Endophytic Fungi of Vitis labrusca Linnaeus, Bordô and Concord Cv. Ann. Microbiol. 2016, 66, 765–775. [Google Scholar] [CrossRef]
- Gao, Q.; Zhang, Y.; Gao, C.; Li, H.; Cheng, Y.; Qian, X.; Zhang, L.; Liu, J.; Ogunyemi, S.O.; Guan, J. The Microbial Diversity in Relation to Postharvest Quality and Decay: Organic vs. Conventional Pear Fruit. Foods 2023, 12, 1980. [Google Scholar] [CrossRef]
- Saxena, S. Biologically Active Secondary Metabolites from Endophytic Alternaria Species. In Endophytes: Potential Source of Compounds of Commercial and Therapeutic Applications; Patil, R.H., Maheshwari, V.L., Eds.; Springer: Singapore, 2021; pp. 1–20. ISBN 9789811593710. [Google Scholar]
- Azar, N.; Liarzi, O.; Zavitan, M.; Samara, M.; Nasser, A.; Ezra, D. Endophytic Penicillium Species Secretes Mycophenolic Acid That Inhibits the Growth of Phytopathogenic Fungi. Microb. Biotechnol. 2022, 16, 1629–1638. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, J.; White, J.F.; Li, C. Epichloë bromicola from Wild Barley Improves Salt-Tolerance of Cultivated Barley by Altering Physiological Responses to Salt Stress. Front. Microbiol. 2022, 13, 1044735. [Google Scholar] [CrossRef]
- Negi, R.; Kaur, T.; Devi, R.; Kour, D.; Yadav, A.N. Assessment of Nitrogen-Fixing Endophytic and Mineral Solubilizing Rhizospheric Bacteria as Multifunctional Microbial Consortium for Growth Promotion of Wheat and Wild Wheat Relative Aegilops kotschyi. Heliyon 2022, 8, e12579. [Google Scholar] [CrossRef]
- Sánchez-Cruz, R.; Tpia Vázquez, I.; Batista-García, R.A.; Méndez-Santiago, E.W.; Sánchez-Carbente, M.d.R.; Leija, A.; Lira-Ruan, V.; Hernández, G.; Wong-Villarreal, A.; Folch-Mallol, J.L. Isolation and Characterization of Endophytes from Nodules of Mimosa pudica with Biotechnological Potential. Microbiol. Res. 2019, 218, 76–86. [Google Scholar] [CrossRef]
- Todasca, M.-C.; Fotescu, L.; Chira, N.-A.; Deleanu, C.; Rosca, S. Composition Changes in Wines Produced by Different Growing Techniques Examined through 1H-NMR Spectroscopy. Rev. Chim. 2011, 62, 131–134. [Google Scholar]
- West, E.R.; Cother, E.J.; Steel, C.C.; Ash, G.J. The Characterization and Diversity of Bacterial Endophytes of Grapevine. Can. J. Microbiol. 2010, 56, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Mitter, B.; Colli-Mull, J.G.; Gangl, H.; Sessitsch, A. Endophytes of Grapevine Flowers, Berries, and Seeds: Identification of Cultivable Bacteria, Comparison with Other Plant Parts, and Visualization of Niches of Colonization. Microb. Ecol. 2011, 62, 188–197. [Google Scholar] [CrossRef]
- Baldan, E.; Nigris, S.; Populin, F.; Zottini, M.; Squartini, A.; Baldan, B. Identification of Culturable Bacterial Endophyte Community Isolated from Tissues of Vitis vinifera “Glera”. Plant Biosyst.—Int. J. Deal. All Asp. Plant Biol. 2014, 148, 508–516. [Google Scholar] [CrossRef]
- Campisano, A.; Antonielli, L.; Pancher, M.; Yousaf, S.; Pindo, M.; Pertot, I. Bacterial Endophytic Communities in the Grapevine Depend on Pest Management. PLoS ONE 2014, 9, e112763. [Google Scholar] [CrossRef]
- Andreolli, M.; Lampis, S.; Zapparoli, G.; Angelini, E.; Vallini, G. Diversity of Bacterial Endophytes in 3 and 15 Year-Old Grapevines of Vitis vinifera cv. Corvina and Their Potential for Plant Growth Promotion and Phytopathogen Control. Microbiol. Res. 2016, 183, 42–52. [Google Scholar] [CrossRef]
- Salvetti, E.; Campanaro, S.; Campedelli, I.; Fracchetti, F.; Gobbi, A.; Tornielli, G.B.; Torriani, S.; Felis, G.E. Whole-Metagenome-Sequencing-Based Community Profiles of Vitis vinifera L. cv. Corvina Berries Withered in Two Post-Harvest Conditions. Front. Microbiol. 2016, 7, 937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deyett, E.; Rolshausen, P.E. Temporal Dynamics of the Sap Microbiome of Grapevine Under High Pierce’s Disease Pressure. Front. Plant Sci. 2019, 10, 1246. [Google Scholar] [CrossRef]
- Gamalero, E.; Bona, E.; Novello, G.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Berta, G.; Lingua, G. Discovering the Bacteriome of Vitis vinifera cv. Pinot Noir in a Conventionally Managed Vineyard. Sci. Rep. 2020, 10, 6453. [Google Scholar] [CrossRef] [Green Version]
- Deyett, E.; Rolshausen, P.E. Endophytic Microbial Assemblage in Grapevine. FEMS Microbiol. Ecol. 2020, 96, fiaa053. [Google Scholar] [CrossRef]
- Csótó, A.; Kovács, C.; Pál, K.; Nagy, A.; Peles, F.; Fekete, E.; Karaffa, L.; Kubicek, C.P.; Sándor, E. The Biocontrol Potential of Endophytic Trichoderma Fungi Isolated from Hungarian Grapevines, Part II, Grapevine Stimulation. Pathogens 2023, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Carro-Huerga, G.; Compant, S.; Gorfer, M.; Cardoza, R.E.; Schmoll, M.; Gutiérrez, S.; Casquero, P.A. Colonization of Vitis vinifera L. by the Endophyte Trichoderma sp. Strain T154: Biocontrol Activity Against Phaeoacremonium minimum. Front. Plant Sci. 2020, 11, 1170. [Google Scholar] [CrossRef] [PubMed]
- Moloinyane, S.; Nchu, F. The Effects of Endophytic Beauveria bassiana Inoculation on Infestation Level of Planococcus ficus, Growth and Volatile Constituents of Potted Greenhouse Grapevine (Vitis vinifera L.). Toxins 2019, 11, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.-Z.; Ma, M.-D.; Yuan, M.-Q.; Huang, Z.-Y.; Yang, W.-X.; Zhang, H.-B.; Huang, L.-H.; Ren, A.-Y.; Shan, H. Fungal Endophytes as a Metabolic Fine-Tuning Regulator for Wine Grape. PLoS ONE 2016, 11, e0163186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griggs, R.G.; Steenwerth, K.L.; Mills, D.A.; Cantu, D.; Bokulich, N.A. Sources and Assembly of Microbial Communities in Vineyards as a Functional Component of Winegrowing. Front. Microbiol. 2021, 12, 673810. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Feng, B.; Yao, Z.; Wei, B.; Zhao, Y.; Shi, S. Antifungal Activity of Endophytic Bacillus K1 Against Botrytis cinerea. Front. Microbiol. 2022, 13, 935675. [Google Scholar] [CrossRef] [PubMed]
- Hamaoka, K.; Aoki, Y.; Suzuki, S. Isolation and Characterization of Endophyte Bacillus velezensis KOF112 from Grapevine Shoot Xylem as Biological Control Agent for Fungal Diseases. Plants 2021, 10, 1815. [Google Scholar] [CrossRef]
- Li, Z.; Chang, P.; Gao, L.; Wang, X. The Endophytic Fungus Albifimbria verrucaria from Wild Grape as an Antagonist of Botrytis cinerea and Other Grape Pathogens. Phytopathology 2020, 110, 843–850. [Google Scholar] [CrossRef]
- Xie, J.; Guo, X.; Wang, R.; Liu, J.; Liu, Y.; Qiao, G.; Xu, M. The Complete Chloroplast Genome of Vitis amurensis Rupr., an Economic Plant to China. Mitochondrial DNA Part B 2019, 4, 3683–3684. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Y.; Zhang, H.; Huang, H.; Folta, K.M.; Lu, J. Whole Genome Wide Expression Profiles of Vitis amurensis Grape Responding to Downy Mildew by Using Solexa Sequencing Technology. BMC Plant Biol. 2010, 10, 234. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Li, H. Review: Research Progress in Amur Grape, Vitis amurensis Rupr. Can. J. Plant Sci. 2013, 93, 565–575. [Google Scholar] [CrossRef] [Green Version]
- Park, C.; Lee, W.S.; Go, S.-I.; Jeong, S.-H.; Yoo, J.; Cha, H.-J.; Lee, Y.-J.; Kim, H.-S.; Leem, S.-H.; Kim, H.J.; et al. Apoptotic Effects of Anthocyanins from Vitis coignetiae Pulliat Are Enhanced by Augmented Enhancer of the Rudimentary Homolog (ERH) in Human Gastric Carcinoma MKN28 Cells. Int. J. Mol. Sci. 2021, 22, 3030. [Google Scholar] [CrossRef] [PubMed]
- Oshima, Y.; Namao, K.; Kamijou, A.; Matsuoka, S.; Nakano, M.; Terao, K.; Ohizumi, Y. Powerful Hepatoprotective and Hepatotoxic Plant Oligostilbenes, Isolated from the Oriental Medicinal Plant Vitis coignetiae (Vitaceae). Experientia 1995, 51, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Pak, W.; Takayama, F.; Hasegawa, A.; Mankura, M.; Egashira, T.; Ueki, K.; Nakamoto, K.; Kawasaki, H.; Mori, A. Water Extract of Vitis coignetiae Pulliat Leaves Attenuates Oxidative Stress and Inflammation in Progressive NASH Rats. Acta Med. Okayama 2012, 66, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.K.; Park, H.-Y.; Ryu, M.-R.; Kim, Y.; Park, Y. Endothelium-Dependent Vasorelaxant Effects of Dealcoholized Wine Powder of Wild Grape Vitis coignetiae in the Rat Thoracic Aorta. Evid.-Based Complement. Altern. Med. 2016, 2016, e6846084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weidner, S.; Rybarczyk, A.; Karamać, M.; Król, A.; Mostek, A.; Grębosz, J.; Amarowicz, R. Differences in the Phenolic Composition and Antioxidant Properties between Vitis coignetiae and Vitis vinifera Seeds Extracts. Molecules 2013, 18, 3410–3426. [Google Scholar] [CrossRef] [PubMed]
- Yano, A.; Kikuchi, S.; Takahashi, T.; Kohama, K.; Yoshida, Y. Inhibitory Effects of the Phenolic Fraction from the Pomace of Vitis coignetiae on Biofilm Formation by Streptococcus Mutans. Arch. Oral Biol. 2012, 57, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Faist, H.; Keller, A.; Hentschel, U.; Deeken, R. Grapevine (Vitis vinifera) Crown Galls Host Distinct Microbiota. Appl. Environ. Microbiol. 2016, 82, 5542–5552. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Aghdam, S.A.; Lahowetz, R.M.; Brown, A.M.V. Metapangenomics of Wild and Cultivated Banana Microbiome Reveals a Plethora of Host-Associated Protective Functions. Environ. Microbiome 2023, 18, 36. [Google Scholar] [CrossRef]
- Wang, G.; Ren, Y.; Bai, X.; Su, Y.; Han, J. Contributions of Beneficial Microorganisms in Soil Remediation and Quality Improvement of Medicinal Plants. Plants 2022, 11, 3200. [Google Scholar] [CrossRef]
- Amjad, S.F.; Mansoora, N.; Yaseen, S.; Kamal, A.; Butt, B.; Matloob, H.; Alamri, S.A.M.; Alrumman, S.A.; Eid, E.M.; Shahbaz, M. Combined Use of Endophytic Bacteria and Pre-Sowing Treatment of Thiamine Mitigates the Adverse Effects of Drought Stress in Wheat (Triticum aestivum L.) Cultivars. Sustainability 2021, 13, 6582. [Google Scholar] [CrossRef]
- Anwar, Y.; Khan, S.U.; Ullah, I.; Hemeg, H.A.; Ashamrani, R.; Al-sulami, N.; Ghazi Alniami, E.; Hashem Alqethami, M.; Ullah, A. Molecular Identification of Endophytic Bacteria from Silybum marianum and Their Effect on Brassica napus Growth under Heavy Metal Stress. Sustainability 2023, 15, 3126. [Google Scholar] [CrossRef]
- Iqbal, S.; Iqbal, M.A.; Li, C.; Iqbal, A.; Abbas, R.N. Overviewing Drought and Heat Stress Amelioration—From Plant Responses to Microbe-Mediated Mitigation. Sustainability 2023, 15, 1671. [Google Scholar] [CrossRef]
- Mili, C.; Roy, S.; Tayung, K. Endophytic Fungi of Wild and Domesticated Crop Plants and Their Prospect for Applications in Sustainable Agriculture. In Endophytes: Potential Source of Compounds of Commercial and Therapeutic Applications; Patil, R.H., Maheshwari, V.L., Eds.; Springer: Singapore, 2021; pp. 21–35. ISBN 978-981-15-9371-0. [Google Scholar]
- Hamed, K.E. Isolation of Endophytic Bacteria from Wild Plants in Dry Regions and Investigation of Their Ability to Promote Plant Growth and Inhibit Pathogenic Fungi. Aust. J. Crop Sci. 2022, 16, 143–151. [Google Scholar] [CrossRef]
- Borah, M.; Das, S.; Baruah, H.; Boro, R.C.; Barooah, M. Diversity of Culturable Endophytic Bacteria from Wild and Cultivated Rice Showed Potential Plant Growth Promoting Activities. bioRxiv 2018, 310797. [Google Scholar] [CrossRef] [Green Version]
- Toghueo, R.M.K.; Zabalgogeazcoa, I.; Pereira, E.C.; Vazquez de Aldana, B.R. A Diaporthe Fungal Endophyte From a Wild Grass Improves Growth and Salinity Tolerance of Tritordeum and Perennial Ryegrass. Front. Plant Sci. 2022, 13, 896755. [Google Scholar] [CrossRef] [PubMed]
- Tidke, S.; Kl, R.K.; Devappa, R.; Vasist, K.S.; Kosturkova, G.; Gokare, R. Current Understanding of Endophytes: Their Relevance, Importance, and Industrial Potentials. IOSR J. Biotechnol. Biochem. 2017, 03, 43–59. [Google Scholar] [CrossRef]
- Fan, Y.; Gao, L.; Chang, P.; Li, Z. Endophytic Fungal Community in Grape Is Correlated to Foliar Age and Domestication. Ann. Microbiol. 2020, 70, 30. [Google Scholar] [CrossRef]
- Abd-Alla, M.H.; Omar, S.A.; Karanxha, S. The Impact of Pesticides on Arbuscular Mycorrhizal and Nitrogen-Fixing Symbioses in Legumes. Appl. Soil Ecol. 2000, 14, 191–200. [Google Scholar] [CrossRef]
- Mazen, M.B.; Ramadan, T.; Nafady, N.A.; Zaghlol, A.; Hasan, S.H. Comparative Study on the Effect of Chemical Fertilizers, Bio-Fertilizers and Arbuscular Mycorrhizal Fungi on Maize Growth. Biol. For. Int. J. 2018, 10, 182–194. [Google Scholar]
- Bruisson, S.; Zufferey, M.; L’Haridon, F.; Trutmann, E.; Anand, A.; Dutartre, A.; De Vrieze, M.; Weisskopf, L. Endophytes and Epiphytes From the Grapevine Leaf Microbiome as Potential Biocontrol Agents Against Phytopathogens. Front. Microbiol. 2019, 10, 2726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzoska, R.M.; Huntemann, M.; Clum, A.; Chen, A.; Kyrpides, N.; Palaniappan, K.; Ivanova, N.; Mikhailova, N.; Ovchinnikova, G.; Varghese, N.; et al. Complete Genome Sequence for Asinibacterium sp. Strain OR53 and Draft Genome Sequence for Asinibacterium sp. Strain OR43, Two Bacteria Tolerant to Uranium. Microbiol. Resour. Announc. 2019, 8, e01701-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vion, C.; Muro, M.; Bernard, M.; Richard, B.; Valentine, F.; Yeramian, N.; Masneuf-Pomarède, I.; Tempère, S.; Marullo, P. New Malic Acid Producer Strains of Saccharomyces cerevisiae for Preserving Wine Acidity during Alcoholic Fermentation. Food Microbiol. 2023, 112, 104209. [Google Scholar] [CrossRef] [PubMed]
- Hanganu, A.; Todasca, M.-C.; Chira, N.-A.; Rosca, S. Influence of Common and Selected Yeasts on Wine Composition Studied Using 1H-NMR Spectroscopy. Rev. Chim. 2011, 62, 689–692. [Google Scholar]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic Species in the Modulation of Gut Microbiota: An Overview. BioMed Res. Int. 2018, 2018, e9478630. [Google Scholar] [CrossRef] [Green Version]
- Aleynova, O.A.; Nityagovsky, N.N.; Dubrovina, A.S.; Kiselev, K.V. The Biodiversity of Grapevine Bacterial Endophytes of Vitis amurensis Rupr. Plants 2022, 11, 1128. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Nityagovsky, N.N.; Suprun, A.R.; Ananev, A.A.; Dubrovina, A.S.; Kiselev, K.V. The Diversity of Fungal Endophytes from Wild Grape Vitis amurensis Rupr. Plants 2022, 11, 2897. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Dubrovina, A.S.; Tyunin, A.P. The Methylation Status of Plant Genomic DNA Influences PCR Efficiency. J. Plant Physiol. 2015, 175, 59–67. [Google Scholar] [CrossRef]
- Lane, D. 16S/23S RRNA Sequencing. Nucleic Acid. Tech. Bact. Syst. 1991, 115–175. [Google Scholar]
- White, T.J.; Bruns, T.; Lee SJ, W.T.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Aleynova, O.A.; Suprun, A.R.; Nityagovsky, N.N.; Dubrovina, A.S.; Kiselev, K.V. The Influence of the Grapevine Bacterial and Fungal Endophytes on Biomass Accumulation and Stilbene Production by the in vitro Cultivated Cells of Vitis amurensis Rupr. Plants 2021, 10, 1276. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, K.V.; Nityagovsky, N.N.; Aleynova, O.A. A Method of DNA Extraction from Plants for Metagenomic Analysis Based on the Example of Grape Vitis amurensis Rupr. Appl. Biochem. Microbiol. 2023, 59, 361–367. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME Release for Eukaryotes; Version 16.10.2022; UNITE Community: London, UK, 2022. [Google Scholar] [CrossRef]
- Bisanz, J. Qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions. 2018. Available online: https://github.com/jbisanz/qiime2R (accessed on 19 July 2023).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galili, T.; Benjamini, Y.; Simpson, G.; Gregory, J.; Gallotta, M.; Renaudie, J.; Hornik, K.; Ligges, U.; Spiess, A.-N.; Horvath, S.; et al. Dendextend: Extending “dendrogram” Functionality in R. 2023. Available online: https://cran.r-project.org/web/packages/dendextend/ (accessed on 19 July 2023).
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes. 2022. Available online: https://cran.r-project.org/web/packages/RColorBrewer/ (accessed on 19 July 2023).
- Gu, Z. Circlize: Circular Visualization. 2022. Available online: https://cran.r-project.org/web/packages/circlize/ (accessed on 19 July 2023).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 19 July 2023).
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]


| No | Abb. | Location | Map |
|---|---|---|---|
| Vitis amurensis | |||
| 1 | Gh | Greenhouse at the Laboratory of Biotechnology, Federal Scientific Center of the East Asia Terrestrial Biodiversity, Far Eastern Branch of the Russian Academy of Sciences, Vladivostok, Russia | 43.192929, 131.921064 |
| 2 | M | The commercial vineyard “Makarevich” | 43.718600, 132.11040 |
| 3 | S-Va | The botanical garden on Sakhalin Island | 46.943386, 142.758662 |
| 4 | P-1 | Vladivostok, the southern Primorsky Territory of the Russian Far East | 43.2242327, 131.991123 |
| 5 | P-2 | Vladivostok, the southern Primorsky Territory of the Russian Far East | 43.2242327, 131.991123 |
| 6 | P-3 | Russky Island, the southern Primorsky Territory of the Russian Far East | 42.964116, 131.881337 |
| 7 | P-4 | Rikord Island, the southern Primorsky Territory of the Russian Far East | 42.870564,131.655779 |
| 8 | P-5 | Ivanovka village, the center of Primorsky Territory of the Russian Far East | 43.975818, 132.481537 |
| 9 | P-6 | The Verkhne–Ussuriysky Research Station (SSA) of the Federal Scientific Center of the East Asia Terrestrial Biodiversity, Far Eastern Branch of the Russian Academy of Sciences, the center of Primorsky Territory of the Russian Far East | 44.034842, 134.202996 |
| 10 | Kh-1 | Litovko village, the southern Khabarovsk region of the Russian Far East | 49.2540051, 135.1847371 |
| 11 | Kh-2 | Silinsky forest, the southern Khabarovsk region of the Russian Far East | 50.586259, 137.0172029 |
| Vitis coignetiae | |||
| 1 | S-1 | The botanical garden on Sakhalin Island | 46.943386, 142.758662 |
| 2 | S-2 | Near the city of Kholmsk on Sakhalin Island | 46.982750, 142.117957 |
| 3 | S-3 | Near the city of Nevelsk on Sakhalin Island | 46.658645, 141.888784 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aleynova, O.A.; Nityagovsky, N.N.; Ananev, A.A.; Suprun, A.R.; Ogneva, Z.V.; Dneprovskaya, A.A.; Beresh, A.A.; Tyunin, A.P.; Dubrovina, A.S.; Kiselev, K.V. The Endophytic Microbiome of Wild Grapevines Vitis amurensis Rupr. and Vitis coignetiae Pulliat Growing in the Russian Far East. Plants 2023, 12, 2952. https://doi.org/10.3390/plants12162952
Aleynova OA, Nityagovsky NN, Ananev AA, Suprun AR, Ogneva ZV, Dneprovskaya AA, Beresh AA, Tyunin AP, Dubrovina AS, Kiselev KV. The Endophytic Microbiome of Wild Grapevines Vitis amurensis Rupr. and Vitis coignetiae Pulliat Growing in the Russian Far East. Plants. 2023; 12(16):2952. https://doi.org/10.3390/plants12162952
Chicago/Turabian StyleAleynova, Olga A., Nikolay N. Nityagovsky, Alexey A. Ananev, Andrey R. Suprun, Zlata V. Ogneva, Alina A. Dneprovskaya, Alina A. Beresh, Alexey P. Tyunin, Alexandra S. Dubrovina, and Konstantin V. Kiselev. 2023. "The Endophytic Microbiome of Wild Grapevines Vitis amurensis Rupr. and Vitis coignetiae Pulliat Growing in the Russian Far East" Plants 12, no. 16: 2952. https://doi.org/10.3390/plants12162952






