Abstract
The collar region plays a crucial role in leaf angle formation and plant architecture, which is important for improving crop yield given the challenges of diminishing arable land and changing environmental conditions. To determine collar region-preferential genes (CRPGs) affecting plant architecture and crop yield, we conducted genome-wide transcriptomic analysis. By integrating our RNA sequencing data with public rice anatomical expression data, we identified 657 CRPGs. Verification involved testing six randomly selected CRPGs, all of which exhibited collar-preferential expression. The functional significance of CRPGs was assessed via Gene Ontology enrichment analysis, utilizing MapMan and KEGG, and literature analysis provided additional information for characterized CRPGs. Our findings revealed links between manipulating leaf angle and phytohormone-related pathways and stress responses. Moreover, based on the CRPGs, five transcription factors downstream of the liguleless 1 (LG1) gene were identified. Overall, the identified CRPGs provide potential targets for further research and breeding applications aimed at improving crop productivity by manipulating leaf architecture.
1. Introduction
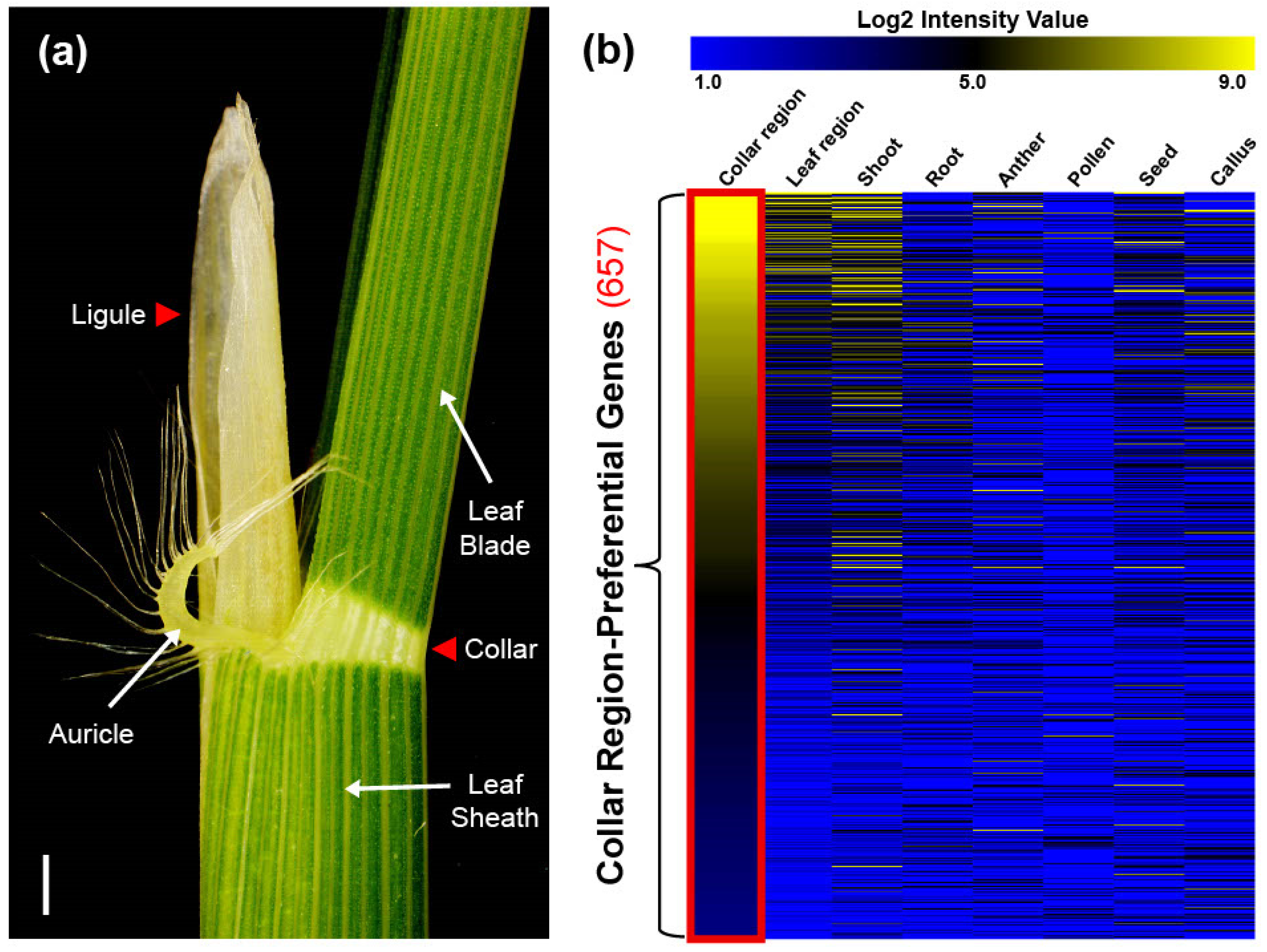
Improving crop yield is an urgent priority owing to complex environmental issues and growing populations [1,2,3]. To meet the global demand for food, efforts to enhance productivity in rice (Oryza sativa L.), one of the world’s most important crops, have focused on manipulating leaf architecture [4]. Rice leaf architecture consists of two distinguishable organs: the vibrant green leaf area and the degenerated white-collar region (Figure 1a). The former facilitates photosynthesis and respiration, whereas the latter shapes the crucial morphological structure known as leaf angle (LA) [5]. Accumulated evidence suggests that the erect leaf phenotype, a key trait in plant leaf architecture, is beneficial for enhancing light capture and photosynthesis efficiency under dense planting, thereby increasing yield [4,6,7]. On the other hand, LA plasticity provides plant flexibility in adapting to rapidly changing environmental conditions [8,9]. Therefore, understanding the collar region comprehensively may contribute to fine-tuning LA for improved breeding applications.
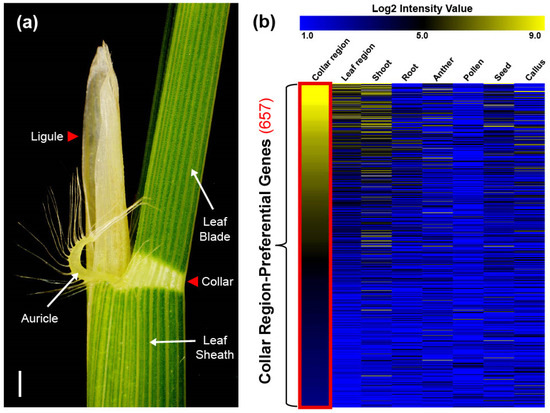
Figure 1.
Illustration of the collar region and heatmap showing the expression of 657 collar region-preferential genes (CRPGs). (a) The collar region adjacent part in 5-week-old rice plants (cv. Dongjin). The collar region is marked by a red triangle. Scale bar = 0.5 cm. (b) Expression of CRPGs in eight anatomical tissues, including the collar region, leaf region, shoot, root, anther, pollen, seed, and callus. The color scheme ranging from blue to black to yellow represents the strength of normalized log2 intensity values. The red rectangle highlights collar region-preferential expression.
Sequential sectioning has been used to investigate morphological changes and the cytological basis of the collar region during its development [10,11]. The cell type divergence and cytological transitions of sclerenchyma and aerenchyma from parenchyma cells and the asymmetric cell constitutions and elongation at the antithetical sides of the cell resulted in LA formation. Dynamic changes in LA are influenced by associated processes, including cell division, cell expansion, and cell wall compositional change [12,13,14]. For instance, overexpression of ILI1 promotes leaf inclination through cell elongation on the adaxial side [15]. Moreover, extensive studies revealed the involvement of multiple factors like various phytohormone-associated genes and transcription factors (TFs) in the leaf angle determination. Intricate phytohormone regulatory networks are an important aspect of collar region development and LA regulation. FISH BONE (FIB) acts as an auxin biosynthesis gene and decreases indole-3-acetic acid content, and thus, enlarged LA was caused by the mutation of FIB [16]. Also, the overexpression of auxin signaling genes, including OsGH3-2 [17], OsIAA1 [18], and OsARF19 [19], indicated that auxin negatively affects leaf inclination. Conversely, the promoting effect of brassinosteroids in leaf angle enlargement and its crosstalk control with other phytohormones, such as auxin, gibberellin, and abscisic acid, have been revealed by functional studies [20,21,22,23,24]. Among many TFs, LIGULELESSs play dominant roles in initiating collar region organogenesis, which is fundamental to LA formation. OsLG2/2L gene-edited plants exhibit localization perturbations in the boundary between blade and sheath, which further disrupt OsLG1-mediated collar differentiation [25,26]. This organogenesis initiation triggers well-ordered cytological changes to occur, leading to leaf bending.
Via systemic dissection with a focus on collar region development, many stage-specific genes have been identified, and they have been proposed to be potential targets for LA manipulation [10,11]. It is helpful for illustrating the complex regulatory mechanisms of collar region development. However, despite a few examples of success in yield enhancement by modulating these genes, the pleiotropic expression still hinders their practical engineering and application. This multi-functional phenomenon of a single gene is often accompanied by negative effects on plant growth, making it difficult to obtain the ideal leaf phenotype without affecting other developmental processes. For example, although RNA interference mutants of OsBUL1 exhibit the erect leaf phenotype, their grain size is significantly reduced, adversely affecting final productivity [27]. To mitigate these adverse impacts, it is important to identify tissue-preferential genes. Modifying these genes may achieve the desired phenotype while mitigating the negative effects.
In the present study, we performed global identification of collar region-preferential genes (CRPGs) by integrating our RNA sequencing (RNA-Seq) data with various anatomical transcriptome datasets. Subsequently, 657 CRPGs were identified, and after validating the collar-preferential expression pattern through qRT-PCR, functional enrichment analysis using Gene Ontology (GO) and MapMan toolkit was conducted. Five regulatory genes identified from the transcription term in GO were suggested as downstream TFs of OsLG1, a key regulatory gene in the initiation of collar development in rice. Based on these findings, a conceptual regulatory model for collar development was proposed. Our findings could accelerate the modification of compacted leaf structure in rice to improve productivity while efficiently using energy, aligning with sustainable agriculture trends.
2. Results
2.1. Genome-Wide Identification of Collar Region-Preferential Genes
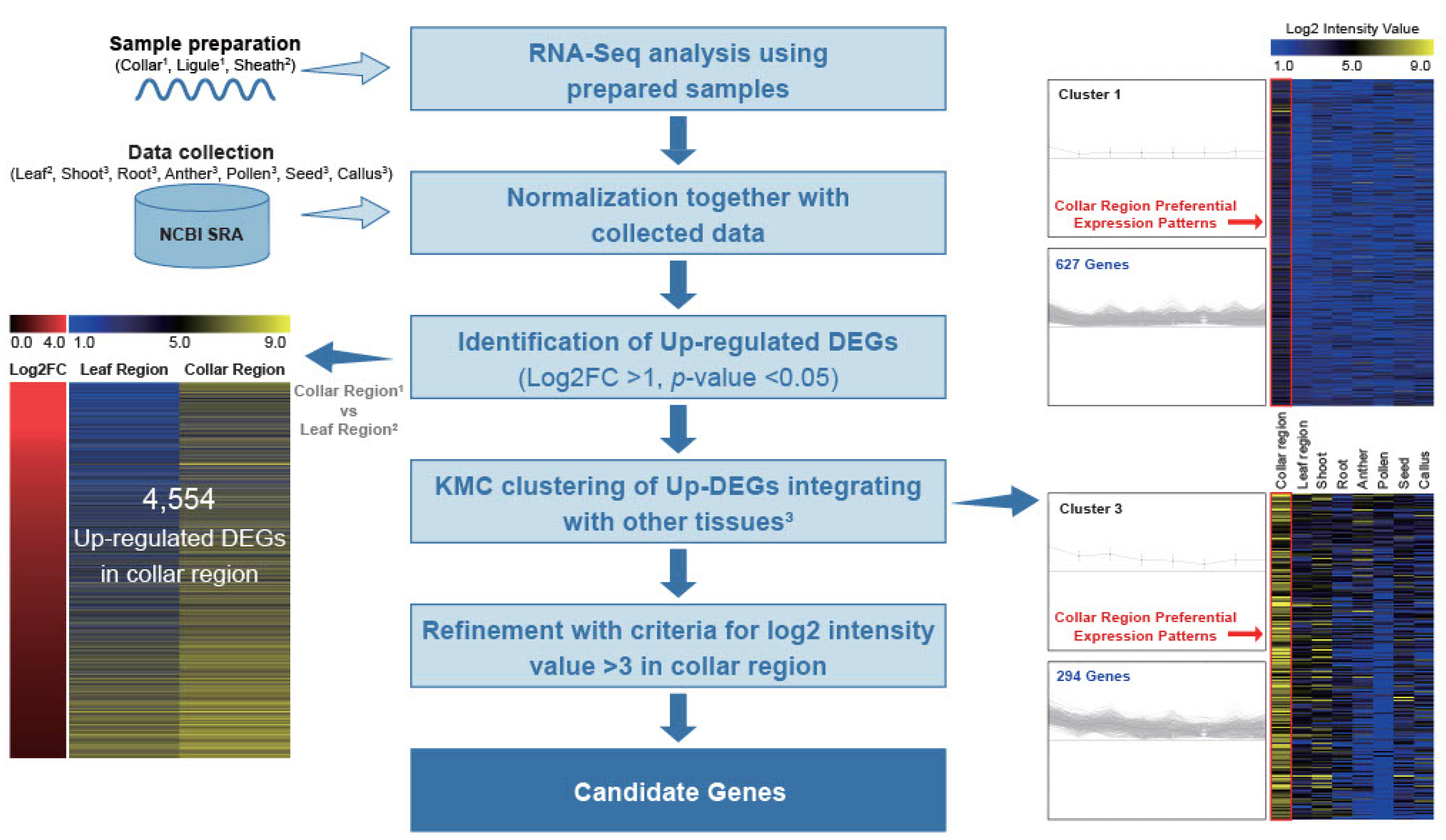
To investigate CRPGs, we grouped tissues according to their properties. The collar and ligule were grouped as the collar region, whereas the leaf blade and sheath were grouped as the leaf region (Figure 1a). RNA samples from collar, ligule, and sheath were collected from 5- to 6-week-old rice plants in the vegetative stage, and the expression raw data of leaf, shoot, root, anther, pollen, seed, and callus were collected from NCBI Sequence Read Archive (SRA; http://ncbi.nlm.nih.gov/sra/, accessed on 28 December 2022). Further, they were normalized using a previously described method all at once [28]. By comparisons of collar and leaf regions, we preliminary identified 4554 differentially upregulated genes in the collar region, with a log2 fold change > 1 and a p-value < 0.05. These genes were further classified into 12 distinct clusters using K-means clustering (KMC) analysis. Two of the twelve clusters showed collar region-preferential expression patterns. After manually filtering genes with a log2 intensity value >3 in the collar region, we finally identified 657 CRPGs (Figure 2). The expression heatmap of the identified CRPGs was reconstructed using MeV software (Figure 1b).
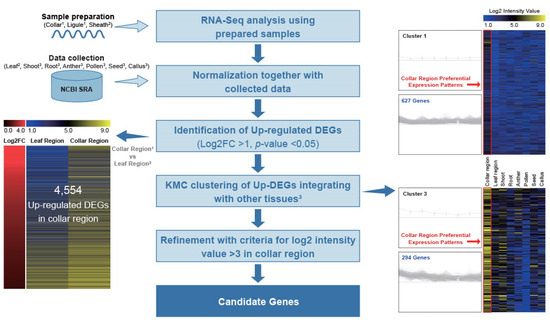
Figure 2.
Workflow for the genome-wide identification of CRPGs. Light and deep blue horizontal arrows represent input data and output results, respectively. Upregulated differentially expressed genes (DEGs) were identified by comparing the collar region with the leaf region. In the left panel, the output shows the log2 fold change combined with anatomical expression data of upregulated DEGs (collar region vs. leaf region), where the red color indicates upregulation in the collar region. The color scheme ranging from blue to black to yellow represents the strength of normalized log2 intensity values. The right panel displays a centroid graph with expression images of collar region-preferential gene clusters obtained from KMC analysis. The number of genes in each cluster is shown, and expression in the collar region is highlighted with a red rectangle. Collar region1: collar1 and ligule1; leaf region2: leaf2 and sheath2; other tissues3: shoot3, root3, anther3, pollen3, seed3, and callus3.
2.2. Authentication of Tissue-Preferential Expression Patterns of Six CRPGs
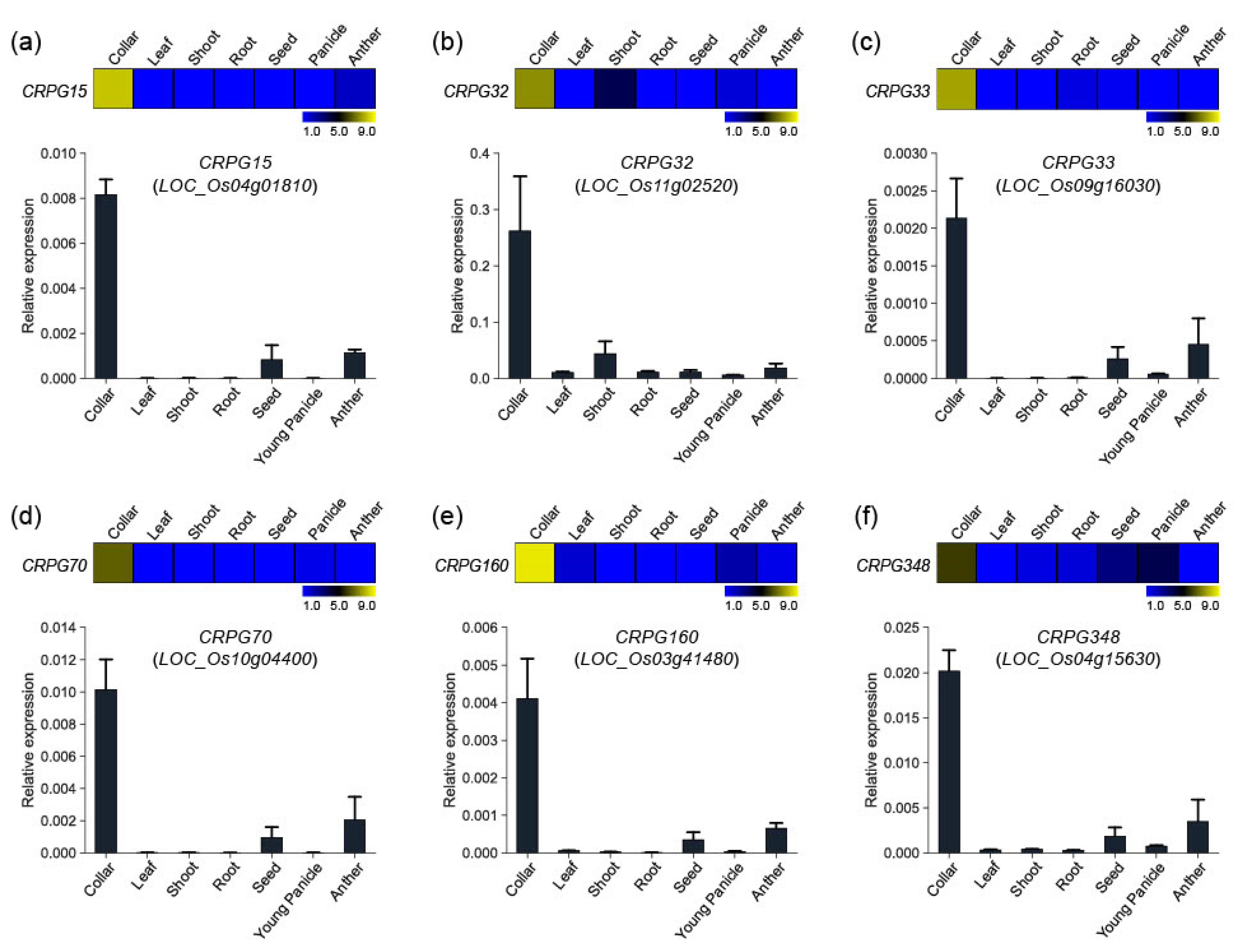
To verify the reliability of the collar region-preferred expression patterns of our candidate genes, we randomly selected six genes and performed in silico expression and quantitative real-time PCR analysis. The genes included one secondary metabolism related gene (CRPG15, Os04g01810), one TF (CRPG32, Os11g02520), one hormone-related gene (CRPG33, Os09g16030), one transferase (CRPG70, Os10g04400), one unannotated gene (CRPG160, Os03g41480), and one protein kinase (CRPG348, Os04g15630). Anatomical expression data were downloaded from the CAFRI-Rice database [28] by entering locus IDs retrieved from the Rice Genome Annotation Project website (RGAP; http://rice.plantbiology.msu.edu/, accessed on 28 December 2022). The data were reconstructed using MeV software. Real-time PCR experiments confirmed the expression of the six genes (Figure 3). As expected, all these showed preferential expression patterns in the collar tissue, corroborating our previous identification results.
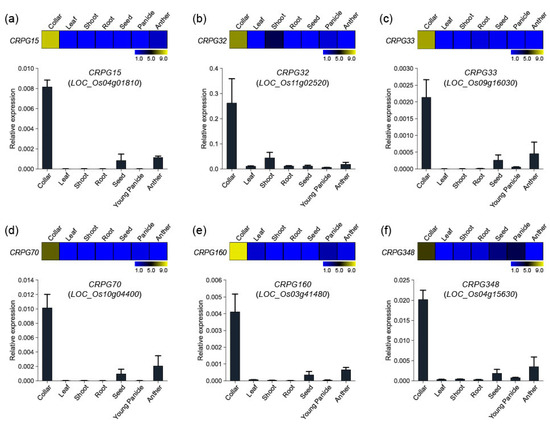
Figure 3.
Expression validation of six CRPGs. (a) CRPG15/Os04g01810, (b) CRPG32/Os11g02520, (c) CRPG33/Os09g16030, (d) CRPG70/Os10g04400, (e) CRPG160/Os03g41480, and (f) CRPG348/Os04g15630. Expression heatmaps were constructed using the MeV program, and numeric values represent the average normalized log2 intensity values obtained from the CAFRI-Rice database. Collar region-preferential expression pattern was examined using real-time PCR analysis in seven distinct tissues (collar, leaf, shoot, root, seed, young panicle, and anther). Ubi5 (Os01g22490) was used as an internal control in real-time PCR experiments.
2.3. Literature Analysis of Functionally Characterized CRPGs
Detailed archives of functionally characterized genes are useful for interpreting the functional significance of genes identified from large-scale datasets, and they provide a valuable resource that may facilitate further application. We used two databases, funRiceGenes (http://funricegenes.github.io/, accessed on 28 December 2022) [29] and Overview of Functionally Characterized Genes in Rice Online (OGRO; http://qtaro.abr.affrc.go.jp/ogro/table, accessed on 28 December 2022) [30] to identify 31 characterized genes among 657 CRPGs (Table 1; Figure S1). In terms of collar region development and LA formation, three CRPGs have been reported previously. Mutations in OsLG1 lead to complete loss of collar, ligule, and auricle tissues, resulting in the erect leaf phenotype [25]. A bHLH transcription factor, namely OsBC1, regulates LA development by forming a trimeric complex [27], whereas its interaction partner, OsBUL1, was excluded from analysis in this study owing to its ubiquitous expression. Epigenetic modification of RAV6 affects LA and grain size in a brassinosteroid-dependent manner [31]. Additionally, OsGA2ox6 [32], OsMPH1 [33], and RePRP2.1 [34] alter cell elongation in other tissues, hinting the possibility that their parallel function in the collar region is to be expected. Further, more than half of the characterized CRPGs were phytohormone-related or stress-responsive genes, emphasizing the intricate internal signaling networks coupled with the development and regulation of collar region plasticity in rice.

Table 1.
Summary of functionally characterized genes among 657 CRPGs in rice.
2.4. Functional Enrichment Analysis and Classification of CRPGs
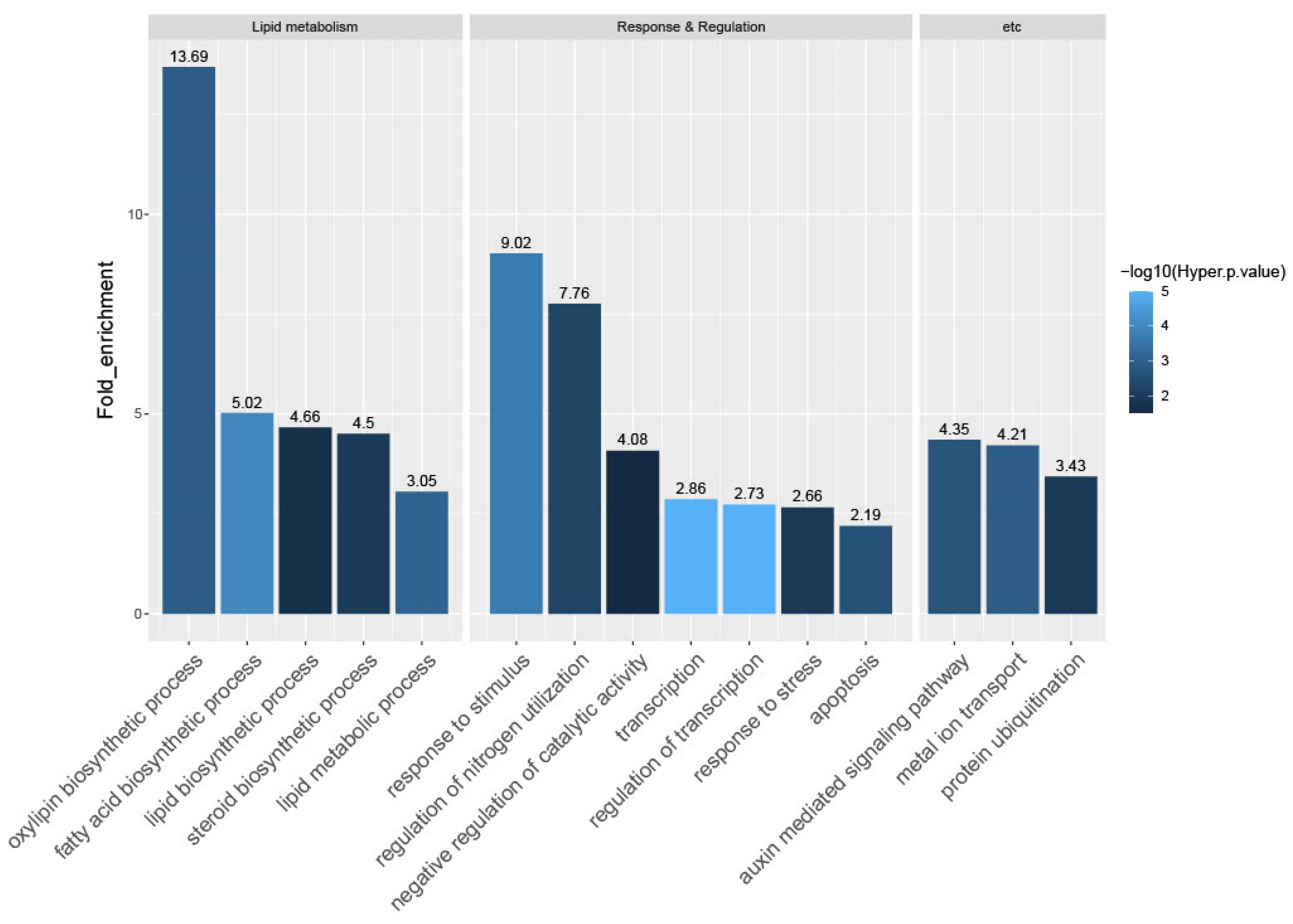
To explore the biological role of the candidate genes, we conducted GO enrichment analysis using the Rice Oligonucleotide Array Database (ROAD; http://www.ricearray.org/, accessed on 28 December 2022) [66]. Based on the criteria query number >2, hypergeometric p-value <0.05, and fold-enrichment value >2, we found that 15 GO terms were over-represented in CRPGs. These included five terms associated with lipid metabolism: oxylipin biosynthetic process (13.69; GO:0031408); fatty acid biosynthetic process (5.02; GO:0006633); lipid biosynthetic process (4.66; GO:0008610); steroid biosynthetic process (4.50; GO:0006694); and lipid metabolic process (3.05; GO:0006629). In addition, seven terms were associated with response and regulation: response to stimulus (9.02; GO:0050896); regulation of nitrogen utilization (7.76; GO:0006808); negative regulation of catalytic activity (4.08; GO:0043086); transcription (2.86; GO:0006350); regulation of transcription (2.73; GO:0045449); response to stress (2.66; GO:0006950); and apoptosis (2.19; GO:0006915). Also, three other terms were enriched: auxin-mediated signaling pathway (4.35; GO:0009734); metal ion transport (4.21; GO:0030001); and protein ubiquitination (3.43; GO:0016567) (Figure 4).
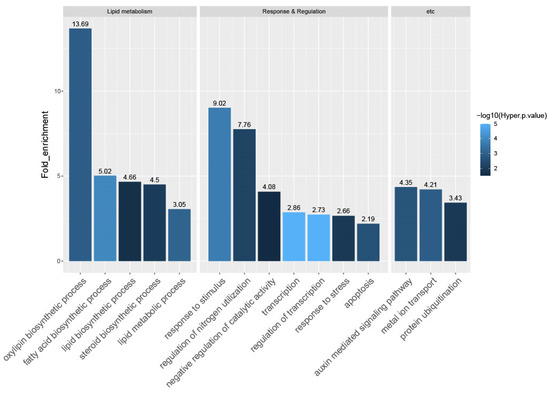
Figure 4.
Gene Ontology (GO) enrichment analysis of CRPGs. The 657 CRPGs were mapped to GO terms in the biological process category. Enriched GO terms were categorized into three groups, including lipid metabolism, response and regulation, and others. The numbers above the bar represent the fold-enrichment value of the GO terms. The colors of the bars indicate statistical significance [−log10(hyper p-value)].
Terms for oxylipin biosynthetic process were the most enriched, as represented by lipoxygenase (LOX) and the specialized cytochrome P450 enzyme, allene oxide synthase (AOS). Their expression is consistent with the elevated expression of rice jasmonic acid (JA) biosynthetic genes, such as OsLOX2 and OsAOS1, in the collar compared with the adjacent leaf region [10]. The only enriched pathway in our KEGG analysis was alpha-linolenic acid metabolism (Figure S2). Alpha-linolenic acid serves as the substrate for LOX in the biosynthesis of JA, which is the most extensively studied oxylipin in plants [67]. The subsequent process is taken over by AOS [68]. This finding was further supported by the regulation overview visualized through MapMan analysis (Figure S3). The aforementioned genes were classified into functional groups associated with jasmonate synthesis, indicating the active biosynthesis of JA and its crucial biological functions in the collar region. In addition, the identification of auxin-related genes in the MapMan overview analysis was consistent with the terms “response to stimulus” and “auxin-mediated signaling pathway” in the GO analysis results. Other hormone-related genes were also observed in the hormone metabolism category. Additionally, approximately half of the total mapped CRPGs were associated with transcription (GO:0006350) and regulation of transcription (GO:0045449), highlighting the importance of TFs in tissue-preferential regulation. Overall, our functional analysis revealed the tissue-preferential regulation by TFs and phytohormones, particularly JA and auxin, and emphasized their intricate networks in relation to collar region development and LA formation.
2.5. Case Study Using the lg1 Mutant Revealed Downstream Regulatory Elements
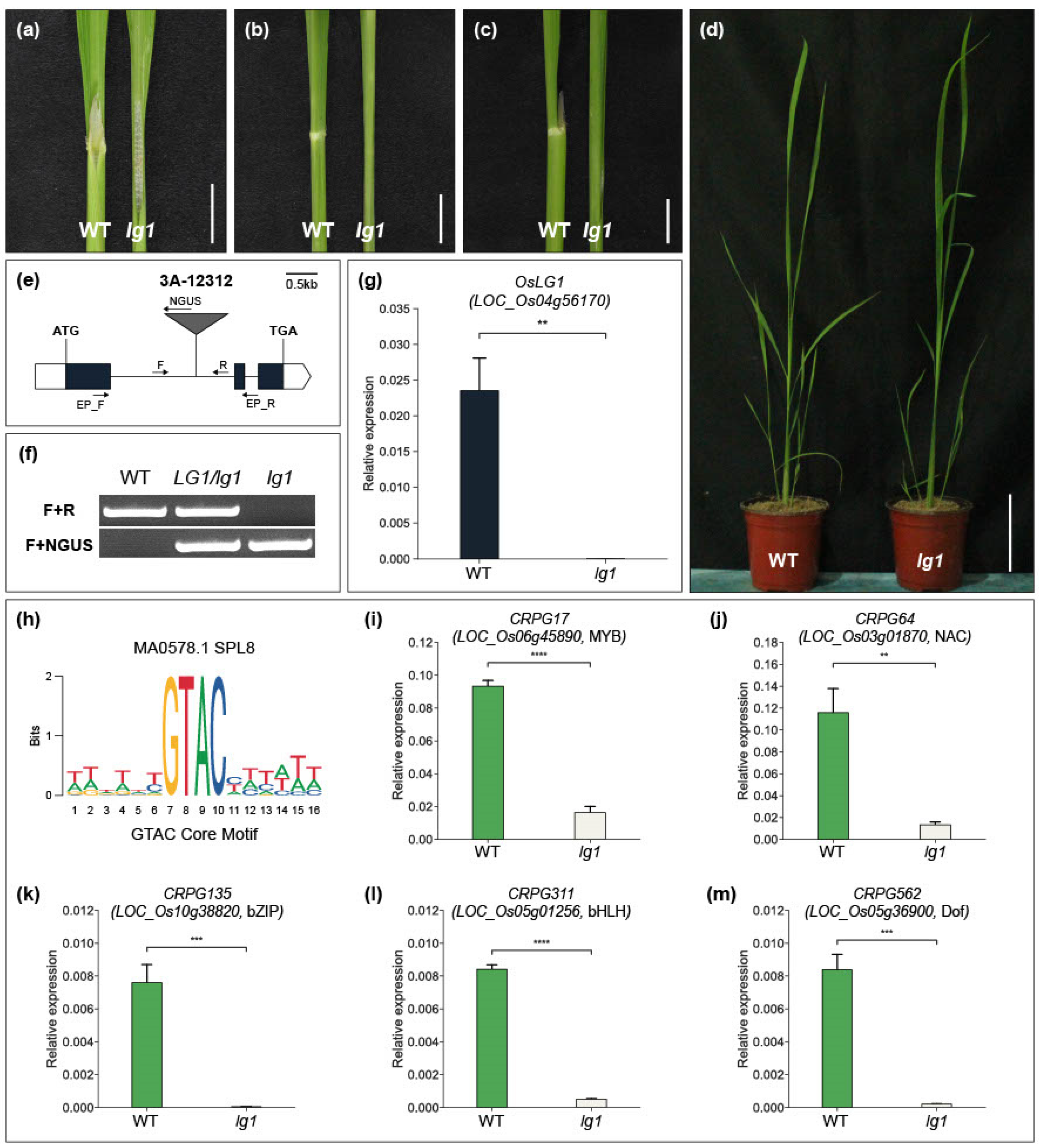
Among the functionally characterized CRPGs, LIGULELESS1 (LG1) is a key gene that initiates collar region organogenesis. Knockout of the LG1 gene leads to the entire loss of the collar region and an erect leaf structure in rice. We identified a T-DNA insertion line (3A-12312) of OsLG1/LOC_Os04g56170 in our T-DNA mutant pool (Figure 5e). T-DNA was inserted into the first exon of OsLG1, and wild-type plants and oslg1 mutants were segregated through genotyping experiments (Figure 5f). Quantitative real-time PCR analysis showed that there was no OsLG1 mRNA detected in 3A-12312 line homozygous plants (Figure 5g). As previously reported, oslg1 plants exhibited a complete absence of the collar region, and 4-week-old lg1 plants showed a more obvious erect leaf structure than wild-type plants (Figure 5a–d). LG1 is a member of the SQUAMOSA PROMOTER BINDING-LIKE (SPL) TF family. In Arabidopsis and wheat, LG1 orthologous genes directly bind to the promoter region containing the GTAC core motif [69,70] (Figure 5h). Considering the conserved function of LG1 in multiple crops (rice, maize, and wheat) [70,71], we hypothesized that CRPGs containing GTAC motifs in their promoters could potentially be regulated by LG1. To identify potential targets, we used Find Individual Motif Occurrences (FIMO), which is embedded in the MEME tool, to screen for TFs among CRPGs that possess the GTAC motif within their 2 kb promoter region. As a result, we identified five TFs as potential downstream regulatory genes of OsLG1. The expression levels of the genes encoding these five TFs were significantly reduced in the oslg1 mutant compared with wild-type plants, supporting our hypothesis regarding the regulatory pathway (Figure 5i–m). However, the direct binding ability of OsLG1 to the GTAC motif of the five CRPGs requires further investigation.
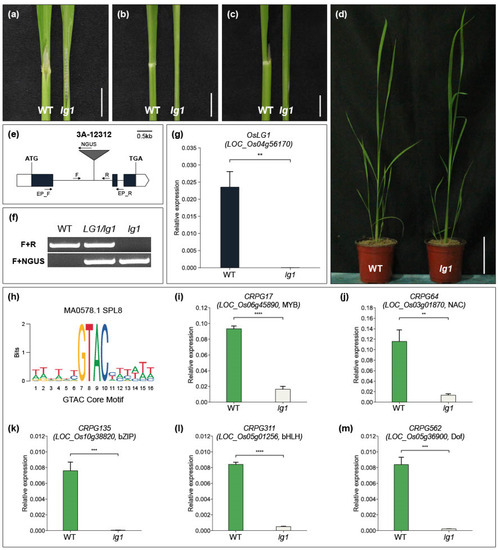
Figure 5.
Phenotype of the rice lg1 mutant and discovery of downstream regulatory factors of OsLG1. (a–d) Comparison of wild-type (WT) plant (left) and lg1 mutant (right) collar region, showing the adaxial side (a), abaxial side (b), lateral side (c), and overall features (d). Scale bars in (a–c) indicate 1 cm, and in (d) indicate 10 cm. (e) Schematic diagram of T-DNA insertional line 3A-12312 for OsLG1. White boxes represent the 5′ and 3′ UTR regions, and black boxes represent the exon region. T-DNA was inserted in the first intron. F, R, and NGUS are primers used for genotyping. EP_F and EP_R are primers for checking expressions. Scale bar: 0.5 kb. (f) Segregation analysis of the lg1 mutant through genotyping. (g) Expression validation of OsLG1 in WT and lg1 mutant. (h) Sequence logo for GTAC core motif. (i–m) Expression of five candidate genes in the WT and lg1 mutant. The internal control used in this study was OsUbi5 (Os01g22490). The experiment included three biological replicates, and the t-test was conducted on independent samples with Bonferroni correction. ** 0.001 < p ≤ 0.01; *** 0.0001 < p ≤ 0.001; **** p ≤ 0.0001.
3. Discussion
Manipulating LA is an important trait for the efficient use of limited land and energy resources. Due to its importance, several studies have illuminated the organogenesis process of the collar region, which is a determinant organ of LA, and the involving genetic factors in different perspectives [10,11]. Despite the collar region mRNA profiling in prior studies, it is necessary to identify additional candidate genes with organ-preferential expression owing to the developmental and positional dynamics of the collar region. Also, previous studies have predominantly focused on stage-specific genes during collar region development, but the pleiotropic expression of these genes poses a major challenge for practical applications. To overcome this challenge and offer a more comprehensive set of potential candidates, we performed genome-wide identification and functional analysis of CRPGs in rice.
Through the integration of our RNA-Seq analysis with NCBI SRA datasets, we successfully identified 657 CRPGs. To validate the reliability of our in silico expression analysis data, we conducted expression testing on a randomly selected subset of six CRPGs (Figure 1, Figure 2 and Figure 3). Functional enrichment analyses using GO and KEGG provided valuable insights into the biological implications of these genes (Figure 4). Notably, the most prominent terms among genes exhibiting preferential expression in the collar region were phytohormone-related, particularly JA biosynthesis and auxin signaling. Previous studies have shown that auxin has a negative effect on collar region development and regulation, including biosynthesis and signaling, while also interacting with other phytohormones or nutrients [72]. Given the complexity of this regulatory system, it may be more effective to maximize the responsiveness of tissue-specific signaling genes rather than fine-tuning hormone levels to control leaf inclination. Regarding JA, except for MeJA, which represses leaf inclination through a brassinosteroid-dependent mechanism [73], there is limited information on the action of JA-related genes, specifically in the collar region, while the involvement of JA in collar region development has been hinted. In line with this notion, although no previously functionally characterized genes related to programmed cell death (PCD) have been identified, the term “apoptosis” was enriched in our study. Notably, Zhou et al. (2017) [10] highlighted the crucial role of PCD in the overall development of the collar region, yet PCD-related terms were not specifically identified in their study. This discrepancy may be attributed to the unspecified expression of the genes analyzed. In summary, the functional assessment of CRPGs in relation to these relevant terms may provide insights into the molecular mechanisms underlying JA or PCD in the collar region that are yet to be elucidated.
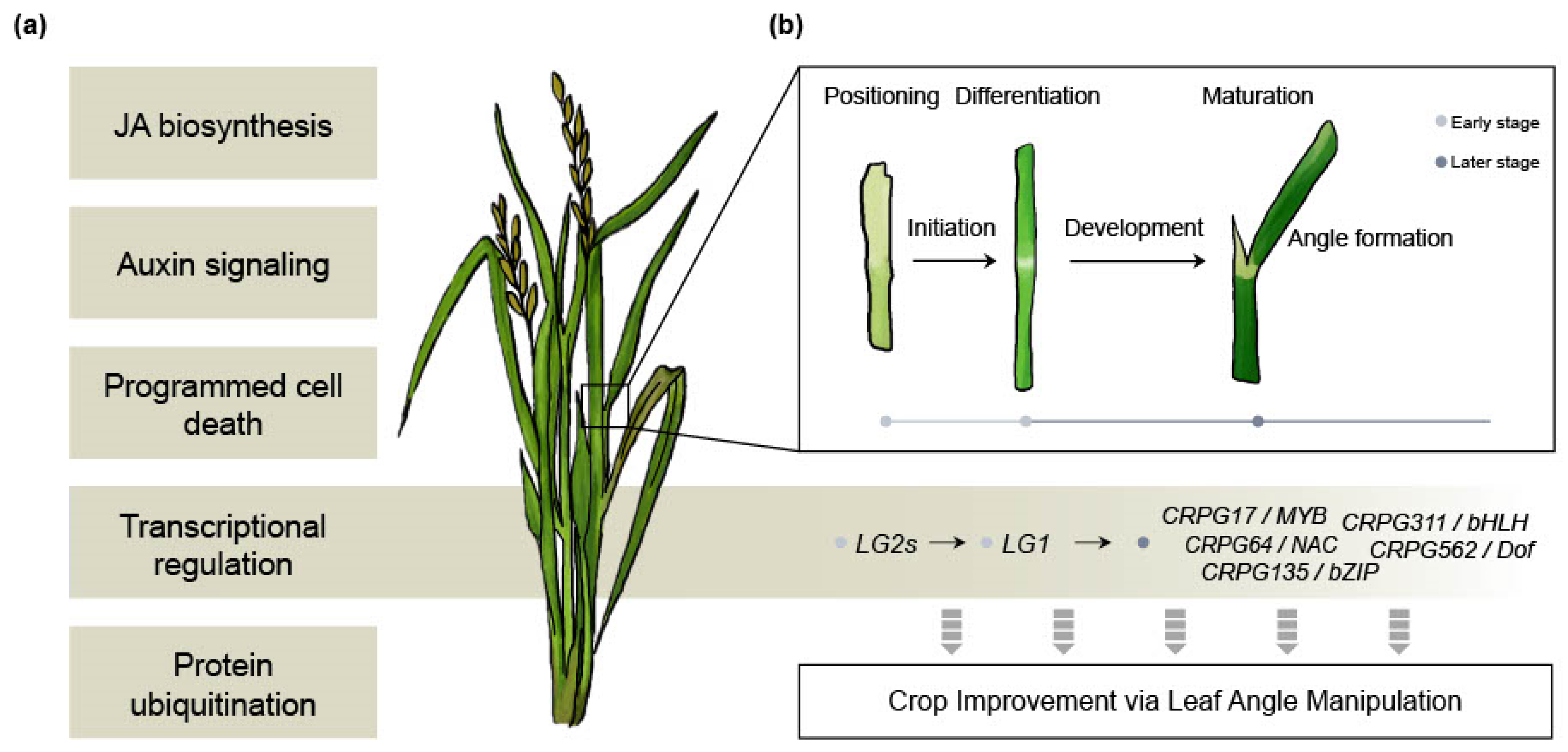
The LG1 gene (Table 1) is renowned for its involvement in the erect leaf phenotype in crops, making it a promising target for crop improvement [26]. However, the research on the downstream mechanisms of LG1 has not advanced proportionally to its significance. In our study, we identified five downstream regulatory genes of OsLG1 among the CRPGs (one each from the MYB, NAC, bZIP, bHLH, and Dof TF families) (Figure 5). Previous studies have documented the role of these TF families in governing LA regulation. For example, OsMYB7 contributes to leaf inclination by modulating auxin levels and promoting cell elongation on the adaxial side [74]. Although the erect leaf phenotype offers advantages in terms of enhanced yield under certain conditions, for the adaptation of crops to diverse geographical environments, it is considered more desirable to have erect leaves while retaining some degree of plasticity rather than a complete loss of structural flexibility. From this perspective, the five CRPGs downstream of OsLG1 hold potential as promising candidates for exhibiting the desired phenotype without negatively impacting plant growth. Although further investigation into the underlying molecular mechanisms is necessary, we present an overall hypothetical model that will be valuable for future studies (Figure 6).
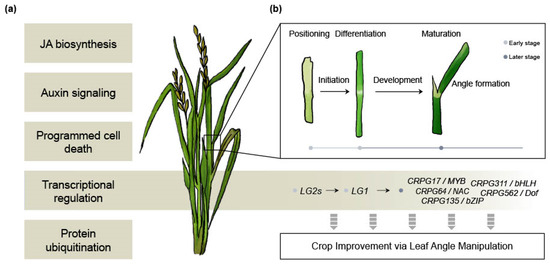
Figure 6.
Overall hypothetical model. (a) Biological processes involved in collar region development. (b) Detailed developmental processes in the sequence of leaf angle formation, along with potential molecular regulators that could serve as candidates for crop improvement.
Direct-sowing cultivation has garnered considerable attention as a strategy to reduce carbon emissions. However, direct-sowing often involves dense planting, which presents challenges in terms of energy utilization efficiency and disease management [75]. As a consequence, the stress-related CRPGs identified in our study, along with the aforementioned candidates, hold potential not only for uncovering molecular mechanisms but also for practical applications. Moreover, the development of the collar region exhibits a certain level of conservation across various crops, providing a valuable foundation for expanding research to other crops that have received comparatively less attention than rice. Collectively, comprehensive investigations and dissections of currently identified CRPGs will be the cornerstone of biotechnological applications and crop breeding to achieve the so-called geo-adapted crops with optimal plant ideotype.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Dongjin rice (O. sativa ssp. japonica) and T-DNA insertional line (3A-12312, cv. Dongjin) seeds were respectively sterilized with 50% sodium hypochlorite and germinated on half-strength Murashige and Skoog medium for one week in an incubator at 28 °C/22 °C (day/night) under continuous light conditions. Seedling plants (7 days old) were transferred to the soil condition in a greenhouse or artificial growth chamber (28 °C/25 °C day/night, 14/10 h light/dark, and 80% relative humidity) at the Kyung Hee University, Yongin, Republic of Korea, for further development.
4.2. RNA-Seq Analysis and Data Collection
The collar, ligule, and sheath from 5- to 6-week-old WT vegetative-stage plants (cv. Dongjin) cultivated under greenhouse conditions were sampled for RNA-Seq analysis. We used a RNeasy Plant Mini Kit from Qiagen to extract RNA from samples and then a TruSeq Stranded mRNA LT Sample Prep Kit for library construction. The libraries were sequenced on an Illumina platform (Illumina NovaSeq 6000) [76], with two biological replicates for each sample. Raw expression datasets for leaf, shoot, root, anther, pollen, seed, and callus tissues were downloaded from NCBI SRA (http://ncbi.nlm.nih.gov/sra/, accessed on 28 December 2022). An identical pipeline was used for preprocessing and reference genome alignment of the sequenced raw data of the abovementioned tissues, followed by normalization, as described previously [28]. The accession number for the collar, ligule, and sheath is E-MTAB-11005 in ArrayExpress; that for leaf, shoot, root, anther, pollen, seed, and callus tissues is DRP000391 in NCBI SRA.
4.3. Identification of Candidates Coupled with Heatmap Analysis
Using the DESeq2 package, we conducted statistical testing based on normalized read counts to identify differentially expressed genes (DEGs) upregulated in the collar region compared with the leaf region. We selected genes with a log2 fold change >1 and a p-value < 0.05. The integrated log2 intensity values of the resulting upregulated DEGs were then loaded into the MeV program (version 4.9.0), where KMC analysis was performed using the Euclidean distance algorithm [77]. This clustering analysis grouped the upregulated DEGs into 12 distinct clusters, from which two that exhibited a preferential expression pattern in the collar region were selected. Subsequently, we applied an additional filter requiring a log2 intensity value >3 in the collar region. Finally, a heatmap showing the expression patterns of these final candidates was generated using the MeV program (version 4.9.0) and employing the single-color array method. Information regarding the 657 CRPGs identified is listed in Table S1.
4.4. Literature Analysis
The OGRO (http://qtaro.abr.affrc.go.jp/ogro/table, accessed on 28 December 2022) and funRiceGenes (http://funricegenes.github.io/, accessed on 28 December 2022) databases were searched to identify functionally characterized CRPGs. Detailed information regarding these functionally characterized CRPGs is provided in Table 1.
4.5. Enrichment Analysis Via GO, KEGG, and MapMan
The Gene IDs of the 657 CRPGs were used as entries for query mapping in ROAD (http://ricephylogenomics-khu.org/road/home.php, accessed on 28 December 2022). To identify significant GO terms (GO type: biological process), certain criteria were applied: query number >2; fold-enrichment value >2; and hypergeometric p-value < 0.05 [78]. The fold-enrichment values were obtained by dividing the query number by the query-expected value. To conduct KEGG enrichment analysis, we used R studio (2023.06.0+421) along with the clusterProfiler package version 4.8.1. The input data included cluster information and Rice Annotation Project Database IDs (http://rapdb.dna.affrc.go.jp, accessed on 28 December 2022), with the organism code for rice specified as “dosa”. The results were filtered by employing an adjusted p-value threshold of <0.05 [79]. For visualization, R studio version 4.3.0 and the ggplot2 package version 3.4.2 were used [80]. MapMan version 3.6.0RC1 was used for visualizing CRPGs mapped to various pathways or processes [81]. An overview of metabolism and regulation was analyzed. Detailed information regarding GO and MapMan analysis is presented in Table S2 and Table S3, respectively.
4.6. RNA Extraction and qRT-PCR
Wild-type plants (cv. Dongjin) and lg1 mutants were grown under growth chamber conditions prior to RNA extraction. Subsequently, 1 cm of the collar region was collected and immediately frozen in liquid nitrogen. Total RNA was isolated manually (RNAiso; Takara Bio, Shiga, Japan) and quantified using a NanoDrop Spectrophotometer ND-2000 [82]. After synthesizing cDNAs using SuPrimeScript RT Premix [with oligo (dT), 2×] (GeNet Bio, Daegu, Republic of Korea), qRT-PCR was performed on a Rotor Gene Q instrument system (Qiagen, Hidden, Germany) using SYBR Green I (GeNet Bio, Republic of Korea). All reactions were conducted in three biological replications, and data analysis was performed using the 2−ΔΔCt method, as previously described [83]. For other tissues, samples were collected as previously reported [84]. All the primers used in this study are listed in Table S4.
4.7. Motif Scanning
The motif matrix profile of the GTAC core motif (MA0578.1) was downloaded from JASPAR 2022 in MEME format [85]. The function FIMO (Find Individual Motif Occurrences) in MEME suite (version 5.5.3) was used for scanning the 2 kb upstream promoter sequence of TFs [86] among CRPGs that matched with the GTAC motif [87]. The resulting information is in Table S5.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12162959/s1, Figure S1. Venn diagram of functionally characterized CRPGs. Figure S2. KEGG enrichment analysis for 657 CRPGs. Figure S3. MapMan analysis for 657 CRPGs. Results of mapping 657 CRPGs to (a) metabolism overview and (b) regulation overview. Table S1. Detailed information of 657 collar region-preferential genes. Table S2. Summary of enriched GO terms for 657 CRPGs. Table S3. Summary of functional classification of 657 CRPGs using MapMan analysis. Table S4. Summary of primers used in this study. Table S5. Motif scanning result for TFs among CRPGs.
Author Contributions
K.-H.J. designed this work; X.J. and W.-J.H. performed experiments and analyzed the data; X.J. and S.-K.L. generated figures and tables; X.J. and K.-H.J. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01703502 to K.-H.J.)” Rural Development Administration, Republic of Korea, and the Framework of International Cooperation Program managed by the National Research Foundation of Korea (NRF-2021K1A3A1A61002988 to K.-H.J.).
Data Availability Statement
The expression data presented in this study are available in the Supplementary Table S1.
Acknowledgments
We thank Gynheung An (Kyung Hee University) for providing T-DNA lines and thank the Chinese Government Scholarship (CSC NO. 202208260079 to X.J.) under State Scholarship Fund from China Scholarship Council.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812. [Google Scholar] [CrossRef]
- Roychowdhury, R. Crop Improvement in the Era of Climate Change, 1st ed.; I.K. International Publishing: New Delhi, India, 2014; ISBN 978-93-82332-61-9. [Google Scholar]
- Hasanuzzaman, M.; Roychowdhury, R.; Karmakar, J.; Dey, N.; Nahar, K.; Fujita, M. Recent advances in biotechnology and genomic approaches for abiotic stress tolerance in crop plants. In Genomics and Proteomics; Apple Academic Press: New York, NY, USA, 2015; pp. 333–366. ISBN 978-1-77188-114-2. [Google Scholar]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S.; et al. Erect Leaves Caused by Brassinosteroid Deficiency Increase Biomass Production and Grain Yield in Rice. Nat. Biotechnol. 2006, 24, 105–109. [Google Scholar] [CrossRef]
- Hoshikawa, K. The Growing Rice Plant. An Anatomical Monograph; Nosan Gyoson Bunka: Tokyo, Japan, 1989; pp. 199–205. [Google Scholar]
- Donald, C.M. The Breeding of Crop Ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
- Sinclair, T.R.; Sheehy, J.E. Erect Leaves and Photosynthesis in Rice. Science 1999, 283, 1455. [Google Scholar] [CrossRef]
- Zhang, S.-W.; Li, C.-H.; Cao, J.; Zhang, Y.-C.; Zhang, S.-Q.; Xia, Y.-F.; Sun, D.-Y.; Sun, Y. Altered Architecture and Enhanced Drought Tolerance in Rice via the Down-Regulation of Indole-3-Acetic Acid by TLD1/OsGH3.13 Activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Ou, X.; Tang, H.; Wang, R.; Wu, P.; Jia, Y.; Wei, X.; Xu, X.; Kang, S.-H.; Kim, S.-K.; et al. Rice MicroRNA Osa-MiR1848 Targets the Obtusifoliol 14α-Demethylase Gene OsCYP51G3 and Mediates the Biosynthesis of Phytosterols and Brassinosteroids during Development and in Response to Stress. New Phytol. 2015, 208, 790–802. [Google Scholar] [CrossRef]
- Zhou, L.-J.; Xiao, L.-T.; Xue, H.-W. Dynamic Cytology and Transcriptional Regulation of Rice Lamina Joint Development. Plant Physiol. 2017, 174, 1728–1746. [Google Scholar] [CrossRef]
- Wang, R.; Liu, C.; Li, Q.; Chen, Z.; Sun, S.; Wang, X. Spatiotemporal Resolved Leaf Angle Establishment Improves Rice Grain Yield via Controlling Population Density. iScience 2020, 23, 101489. [Google Scholar] [CrossRef]
- Zhao, S.-Q.; Hu, J.; Guo, L.-B.; Qian, Q.; Xue, H.-W. Rice Leaf Inclination2, a VIN3-like Protein, Regulates Leaf Angle through Modulating Cell Division of the Collar. Cell Res. 2010, 20, 935–947. [Google Scholar] [CrossRef]
- Ning, J.; Zhang, B.; Wang, N.; Zhou, Y.; Xiong, L. Increased Leaf Angle1, a Raf-like MAPKKK That Interacts with a Nuclear Protein Family, Regulates Mechanical Tissue Formation in the Lamina Joint of Rice. Plant Cell 2011, 23, 4334–4347. [Google Scholar] [CrossRef]
- Sun, S.; Chen, D.; Li, X.; Qiao, S.; Shi, C.; Li, C.; Shen, H.; Wang, X. Brassinosteroid Signaling Regulates Leaf Erectness in Oryza Sativa via the Control of a Specific U-Type Cyclin and Cell Proliferation. Dev. Cell 2015, 34, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-Y.; Bai, M.-Y.; Wu, J.; Zhu, J.-Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/BHLH Transcription Factors Mediate Brassinosteroid Regulation of Cell Elongation and Plant Development in Rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Ito, M.; Sumikura, T.; Nakayama, A.; Nishimura, T.; Kitano, H.; Yamaguchi, I.; Koshiba, T.; Hibara, K.-I.; Nagato, Y.; et al. The Rice FISH BONE Gene Encodes a Tryptophan Aminotransferase, Which Affects Pleiotropic Auxin-Related Processes. Plant J. 2014, 78, 927–936. [Google Scholar] [CrossRef]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 Family Member, OsGH3-2, Modulates Auxin and Abscisic Acid Levels and Differentially Affects Drought and Cold Tolerance in Rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; You, J.; Xiong, L. Characterization of OsIAA1 Gene, a Member of Rice Aux/IAA Family Involved in Auxin and Brassinosteroid Hormone Responses and Plant Morphogenesis. Plant Mol. Biol. 2009, 70, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, S.; Xu, Y.; Yu, C.; Shen, C.; Qian, Q.; Geisler, M.; Jiang, D.A.; Qi, Y. The Auxin Response Factor, OsARF19, Controls Rice Leaf Angles through Positively Regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015, 38, 638–654. [Google Scholar] [CrossRef]
- Yamamuro, C.; Ihara, Y.; Wu, X.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. Loss of Function of a Rice Brassinosteroid Insensitive1 Homolog Prevents Internode Elongation and Bending of the Lamina Joint. Plant Cell 2000, 12, 1591–1606. [Google Scholar] [CrossRef]
- Li, H.; Jiang, L.; Youn, J.-H.; Sun, W.; Cheng, Z.; Jin, T.; Ma, X.; Guo, X.; Wang, J.; Zhang, X.; et al. A Comprehensive Genetic Study Reveals a Crucial Role of CYP90D2/D2 in Regulating Plant Architecture in Rice (Oryza Sativa). New Phytol. 2013, 200, 1076–1088. [Google Scholar] [CrossRef]
- Liu, X.; Yang, C.Y.; Miao, R.; Zhou, C.L.; Cao, P.H.; Lan, J.; Zhu, X.J.; Mou, C.L.; Huang, Y.S.; Liu, S.J.; et al. DS1/OsEMF1 Interacts with OsARF11 to Control Rice Architecture by Regulation of Brassinosteroid Signaling. Rice 2018, 11, 46. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Z.; Xu, Y.; Joo, S.-H.; Kim, S.-K.; Xue, Z.; Xu, Z.; Wang, Z.; Chong, K. OsGSR1 Is Involved in Crosstalk between Gibberellins and Brassinosteroids in Rice. Plant J. 2009, 57, 498–510. [Google Scholar] [CrossRef]
- Han, K.-S.; Ko, K.-W.; Nam, S.-J.; Park, S.-H.; Kim, S.-K. Optimization of a Rice Lamina Inclination Assay for Detection of Brassinosteroids: I. Effect of Phytohormones on the Inclination Activity. J. Plant Biol. 1997, 40, 240–244. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.-J.; Kim, S.L.; Yim, J.; An, G. Mutations in the Rice Liguleless Gene Result in a Complete Loss of the Auricle, Ligule, and Laminar Joint. Plant Mol. Biol. 2007, 65, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liu, C.; Chen, Z.; Sun, S.; Wang, X. Oryza Sativa LIGULELESS 2s Determine Lamina Joint Positioning and Differentiation by Inhibiting Auxin Signaling. New Phytol. 2021, 229, 1832–1839. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; An, G.; Li, H.-Y. Rice Leaf Angle and Grain Size Are Affected by the OsBUL1 Transcriptional Activator Complex. Plant Physiol. 2017, 173, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.-J.; Kim, Y.-J.; Kim, E.-J.; Kumar Nalini Chandran, A.; Moon, S.; Gho, Y.-S.; Yoou, M.-H.; Kim, S.T.; Jung, K.-H. CAFRI-Rice: CRISPR Applicable Functional Redundancy Inspector to Accelerate Functional Genomics in Rice. Plant J. 2020, 104, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Jiang, Y.; Chen, T.; Li, H.; Fu, M.; Wang, Y.; Xu, Y.; Li, Y.; Zhou, Z.; Jia, L.; et al. New Data and New Features of the FunRiceGenes (Functionally Characterized Rice Genes) Database: 2021 Update. Rice 2022, 15, 23. [Google Scholar] [CrossRef]
- Yamamoto, E.; Yonemaru, J.-I.; Yamamoto, T.; Yano, M. OGRO: The Overview of Functionally Characterized Genes in Rice Online Database. Rice 2012, 5, 26. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, J.; Cao, X.; Song, X. Epigenetic Mutation of RAV6 Affects Leaf Angle and Seed Size in Rice. Plant Physiol. 2015, 169, 2118–2128. [Google Scholar] [CrossRef]
- Huang, J.; Tang, D.; Shen, Y.; Qin, B.; Hong, L.; You, A.; Li, M.; Wang, X.; Yu, H.; Gu, M.; et al. Activation of Gibberellin 2-Oxidase 6 Decreases Active Gibberellin Levels and Creates a Dominant Semi-Dwarf Phenotype in Rice (Oryza Sativa L.). J. Genet. Genom. 2010, 37, 23–36. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, C.; Lin, J.; Liu, J.; Liu, B.; Wang, J.; Huang, A.; Li, H.; Zhao, T. OsMPH1 Regulates Plant Height and Improves Grain Yield in Rice. PLoS ONE 2017, 12, e0180825. [Google Scholar] [CrossRef]
- Tseng, I.-C.; Hong, C.-Y.; Yu, S.-M.; Ho, T.-H.D. Abscisic Acid- and Stress-Induced Highly Proline-Rich Glycoproteins Regulate Root Growth in Rice. Plant Physiol. 2013, 163, 118–134. [Google Scholar] [CrossRef]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced Heat and Drought Tolerance in Transgenic Rice Seedlings Overexpressing OsWRKY11 under the Control of HSP101 Promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef]
- Lee, H.; Cha, J.; Choi, C.; Choi, N.; Ji, H.-S.; Park, S.R.; Lee, S.; Hwang, D.-J. Rice WRKY11 Plays a Role in Pathogen Defense and Drought Tolerance. Rice 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, X.; Yin, D.; Yuan, H.; Xie, Q.; Zhao, X.; Li, X.; Zhu, L.; Li, S.; Li, D. Constitutive Expression of OsDof4, Encoding a C2-C2 Zinc Finger Transcription Factor, Confesses Its Distinct Flowering Effects under Long- and Short-Day Photoperiods in Rice (Oryza Sativa L.). BMC Plant Biol. 2017, 17, 166. [Google Scholar] [CrossRef]
- Taniguchi, S.; Hosokawa-Shinonaga, Y.; Tamaoki, D.; Yamada, S.; Akimitsu, K.; Gomi, K. Jasmonate Induction of the Monoterpene Linalool Confers Resistance to Rice Bacterial Blight and Its Biosynthesis Is Regulated by JAZ Protein in Rice. Plant Cell Environ. 2014, 37, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Qin, F.; Huang, L.; Sun, Q.; Li, C.; Zhao, Y.; Zhou, D.-X. Rice Histone Deacetylase Genes Display Specific Expression Patterns and Developmental Functions. Biochem. Biophys. Res. Commun. 2009, 388, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Gomi, K.; Satoh, M.; Ozawa, R.; Shinonaga, Y.; Sanada, S.; Sasaki, K.; Matsumura, M.; Ohashi, Y.; Kanno, H.; Akimitsu, K.; et al. Role of Hydroperoxide Lyase in White-Backed Planthopper (Sogatella Furcifera Horváth)-Induced Resistance to Bacterial Blight in Rice, Oryza Sativa L. Plant J. 2010, 61, 46–57. [Google Scholar] [CrossRef]
- Han, M.; Kim, C.-Y.; Lee, J.; Lee, S.-K.; Jeon, J.-S. OsWRKY42 Represses OsMT1d and Induces Reactive Oxygen Species and Leaf Senescence in Rice. Mol. Cells 2014, 37, 532–539. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, H.; Deng, Y.; Xiao, J.; Li, X.; Wang, S. The WRKY45-2 WRKY13 WRKY42 Transcriptional Regulatory Cascade Is Required for Rice Resistance to Fungal Pathogen. Plant Physiol. 2015, 167, 1087–1099. [Google Scholar] [CrossRef]
- Haga, K.; Takano, M.; Neumann, R.; Iino, M. The Rice COLEOPTILE PHOTOTROPISM1 Gene Encoding an Ortholog of Arabidopsis NPH3 Is Required for Phototropism of Coleoptiles and Lateral Translocation of Auxin. Plant Cell 2005, 17, 103–115. [Google Scholar] [CrossRef]
- Lo, S.-F.; Yang, S.-Y.; Chen, K.-T.; Hsing, Y.-I.; Zeevaart, J.A.D.; Chen, L.-J.; Yu, S.-M. A Novel Class of Gibberellin 2-Oxidases Control Semidwarfism, Tillering, and Root Development in Rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef]
- Huang, X.; Duan, M.; Liao, J.; Yuan, X.; Chen, H.; Feng, J.; Huang, J.; Zhang, H.-S. OsSLI1, a Homeodomain Containing Transcription Activator, Involves Abscisic Acid Related Stress Response in Rice (Oryza sativa L.). Sci. World J. 2014, 2014, 809353. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Sharma, R.; Jain, M. Over-Expression of OsHOX24 Confers Enhanced Susceptibility to Abiotic Stresses in Transgenic Rice via Modulating Stress-Responsive Gene Expression. Front. Plant Sci. 2017, 8, 628. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Qi, M.; Sheng, G.; Yang, Y. Inducible Overexpression of a Rice Allene Oxide Synthase Gene Increases the Endogenous Jasmonic Acid Level, PR Gene Expression, and Host Resistance to Fungal Infection. MPMI 2006, 19, 1127–1137. [Google Scholar] [CrossRef]
- Lin, Z.; Li, X.; Shannon, L.M.; Yeh, C.-T.; Wang, M.L.; Bai, G.; Peng, Z.; Li, J.; Trick, H.N.; Clemente, T.E.; et al. Parallel Domestication of the Shattering1 Genes in Cereals. Nat. Genet. 2012, 44, 720–724. [Google Scholar] [CrossRef]
- Ray, S.; Kapoor, S.; Tyagi, A.K. Analysis of Transcriptional and Upstream Regulatory Sequence Activity of Two Environmental Stress-Inducible Genes, NBS-Str1 and BLEC-Str8, of Rice. Transgenic Res. 2012, 21, 351–366. [Google Scholar] [CrossRef]
- Shang, X.-L.; Xie, R.-R.; Tian, H.; Wang, Q.-L.; Guo, F.-Q. Putative Zeatin O-Glucosyltransferase OscZOG1 Regulates Root and Shoot Development and Formation of Agronomic Traits in Rice. J. Integr. Plant Biol. 2016, 58, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-Q.; Meng, X.-P.; Zhang, Y.; Xia, M.; Wang, X.-P. Over-Expression of OsDREB Genes Lead to Enhanced Drought Tolerance in Rice. Biotechnol. Lett. 2008, 30, 2191–2198. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yamaji, N.; Mitani-Ueno, N.; Kashino, M.; Ma, J.F. A Node-Localized Transporter OsZIP3 Is Responsible for the Preferential Distribution of Zn to Developing Tissues in Rice. Plant J. 2015, 84, 374–384. [Google Scholar] [CrossRef]
- Ishii, T.; Numaguchi, K.; Miura, K.; Yoshida, K.; Thanh, P.T.; Htun, T.M.; Yamasaki, M.; Komeda, N.; Matsumoto, T.; Terauchi, R.; et al. OsLG1 Regulates a Closed Panicle Trait in Domesticated Rice. Nat. Genet. 2013, 45, 462–465. [Google Scholar] [CrossRef]
- Qi, J.; Li, J.; Han, X.; Li, R.; Wu, J.; Yu, H.; Hu, L.; Xiao, Y.; Lu, J.; Lou, Y. Jasmonic Acid Carboxyl Methyltransferase Regulates Development and Herbivory-Induced Defense Response in Rice. J. Integr. Plant Biol. 2016, 58, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of Rice DREB1/CBF-Type Transcription Factors Involved in Cold-Responsive Gene Expression in Transgenic Rice. Plant Cell Physiol. 2006, 47, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yu, Y.; Chen, Q.; Mu, G.; Shen, Z.; Zheng, L. OsMYB45 Plays an Important Role in Rice Resistance to Cadmium Stress. Plant Sci. 2017, 264, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.-G.; Lee, C.-Y.; Tseng, C.-S. Heterologous Expression of Rice 9-Cis-Epoxycarotenoid Dioxygenase 4 (OsNCED4) in Arabidopsis Confers Sugar Oversensitivity and Drought Tolerance. Bot. Stud. 2018, 59, 2. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural Variation at the DEP1 Locus Enhances Grain Yield in Rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, J.; Li, Z.; Yi, C.; Liu, J.; Zhang, H.; Tang, S.; Gu, M.; Liang, G. Deletion in a Quantitative Trait Gene QPE9-1 Associated With Panicle Erectness Improves Plant Architecture During Rice Domestication. Genetics 2009, 183, 315–324. [Google Scholar] [CrossRef]
- Qi, W.; Sun, F.; Wang, Q.; Chen, M.; Huang, Y.; Feng, Y.-Q.; Luo, X.; Yang, J. Rice Ethylene-Response AP2/ERF Factor OsEATB Restricts Internode Elongation by Down-Regulating a Gibberellin Biosynthetic Gene. Plant Physiol. 2011, 157, 216–228. [Google Scholar] [CrossRef]
- Xu, Y.-X.; Xiao, M.-Z.; Liu, Y.; Fu, J.-L.; He, Y.; Jiang, D.-A. The Small Auxin-up RNA OsSAUR45 Affects Auxin Synthesis and Transport in Rice. Plant Mol. Biol. 2017, 94, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; Nakamura, H.; Ichikawa, H.; Miyao, A.; Hirochika, H.; Kobayashi, K.; Yamaoka, N.; Nishiguchi, M. Response of an Aspartic Protease Gene OsAP77 to Fungal, Bacterial and Viral Infections in Rice. Rice 2014, 7, 9. [Google Scholar] [CrossRef]
- Wang, H.; Hao, J.; Chen, X.; Hao, Z.; Wang, X.; Lou, Y.; Peng, Y.; Guo, Z. Overexpression of Rice WRKY89 Enhances Ultraviolet B Tolerance and Disease Resistance in Rice Plants. Plant Mol. Biol. 2007, 65, 799–815. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Qian, Q.; Fu, Z.; Wang, M.; Zeng, D.; Li, B.; Wang, X.; Li, J. LAZY1 Controls Rice Shoot Gravitropism through Regulating Polar Auxin Transport. Cell Res. 2007, 17, 402–410. [Google Scholar] [CrossRef]
- Kusano, H.; Asano, T.; Shimada, H.; Kadowaki, K. Molecular Characterization of ONAC300, a Novel NAC Gene Specifically Expressed at Early Stages in Various Developing Tissues of Rice. Mol. Genet. Genom. 2005, 272, 616–626. [Google Scholar] [CrossRef]
- Cao, P.; Jung, K.-H.; Choi, D.; Hwang, D.; Zhu, J.; Ronald, P.C. The Rice Oligonucleotide Array Database: An Atlas of Rice Gene Expression. Rice 2012, 5, 17. [Google Scholar] [CrossRef]
- Wasternack, C.; Feussner, I. The Oxylipin Pathways: Biochemistry and Function. Annu. Rev. Plant Biol. 2018, 69, 363–386. [Google Scholar] [CrossRef]
- Farmer, E.E.; Goossens, A. Jasmonates: What ALLENE OXIDE SYNTHASE Does for Plants. J. Exp. Bot. 2019, 70, 3373–3378. [Google Scholar] [CrossRef]
- Kropat, J.; Tottey, S.; Birkenbihl, R.P.; Depège, N.; Huijser, P.; Merchant, S. A Regulator of Nutritional Copper Signaling in Chlamydomonas Is an SBP Domain Protein That Recognizes the GTAC Core of Copper Response Element. Proc. Natl. Acad. Sci. USA 2005, 102, 18730–18735. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Cao, J.; Yu, K.; Liu, X.; Gao, Y.; Chen, Q.; Zhang, W.; Peng, H.; Du, J.; Xin, M.; et al. Wheat TaSPL8 Modulates Leaf Angle Through Auxin and Brassinosteroid Signaling. Plant Physiol. 2019, 181, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.A.; Harper, L.C.; Krueger, R.W.; Dellaporta, S.L.; Freeling, M. Liguleless1 Encodes a Nuclear-Localized Protein Required for Induction of Ligules and Auricles during Maize Leaf Organogenesis. Genes Dev. 1997, 11, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, J.-J.; Xue, H.-W.; Zhang, G.-H. Leaf Direction: Lamina Joint Development and Environmental Responses. Plant Cell Environ. 2021, 44, 2441–2454. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Wu, H.; Wu, D.; Zhang, Z.; Guo, Z.; Yang, N.; Xia, K.; Zhou, X.; Oh, K.; Matsuoka, M.; et al. Methyl Jasmonate Inhibits Lamina Joint Inclination by Repressing Brassinosteroid Biosynthesis and Signaling in Rice. Plant Sci. 2015, 241, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Yoon, J.; Kim, H.; Lee, S.-J.; Kim, T.; Kang, K.; Paek, N.-C. OsMYB7 Determines Leaf Angle at the Late Developmental Stage of Lamina Joints in Rice. Front. Plant Sci. 2023, 14, 1167202. [Google Scholar] [CrossRef]
- Senapati, M.; Tiwari, A.; Sharma, N.; Chandra, P.; Bashyal, B.M.; Ellur, R.K.; Bhowmick, P.K.; Bollinedi, H.; Vinod, K.K.; Singh, A.K.; et al. Rhizoctonia Solani Kühn Pathophysiology: Status and Prospects of Sheath Blight Disease Management in Rice. Front. Plant Sci. 2022, 13, 881116. [Google Scholar] [CrossRef]
- Hong, W.-J.; Lee, S.K.; Kim, S.-H.; Kim, Y.-J.; Moon, S.; Kim, E.-J.; Silva, J.; Jung, K.-H. Comparative Transcriptome Analysis of Pollen and Anther Wall Reveals Novel Insights into the Regulatory Mechanisms Underlying Anther Wall Development and Its Dehiscence in Rice. Plant Cell Rep. 2022, 41, 1229–1242. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Ochs, M.F., Casagrande, J.T., Davuluri, R.V., Eds.; Springer: Boston, MA, USA, 2010; pp. 267–277. ISBN 978-1-4419-5714-6. [Google Scholar]
- Hong, W.-J.; Jiang, X.; Choi, S.-H.; Kim, Y.-J.; Kim, S.-T.; Jeon, J.-S.; Jung, K.-H. A systemic view of carbohydrate metabolism in rice to facilitate productivity. Plants 2021, 10, 1690. [Google Scholar] [CrossRef]
- Hong, W.-J.; Jiang, X.; Ahn, H.R.; Choi, J.; Kim, S.-R.; Jung, K.-H. Systematic analysis of cold stress response and diurnal rhythm using transcriptome data in rice reveals the molecular networks related to various biological processes. Int. J. Mol. Sci. 2020, 21, 6872. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.S.; Kim, J.K.; Baek, S.-A.; Lee, J.-Y.; Lee, D.; Ha, S.-H. Reciprocal Crosses Between Astaxanthin and Capsanthin Rice Unravel Effects of Metabolic Gene Efficacy in Rice Endosperm. J. Plant Biol. 2021, 64, 371–377. [Google Scholar] [CrossRef]
- Gong, H.; You, J.; Zhang, X.; Liu, Y.; Zhao, F.; Cui, X.; Zhang, Y. Genome-Wide Identification and Functional Analysis of Long Non-Coding RNAs in Sesame Response to Salt Stress. J. Plant Biol. 2021, 64, 555–565. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Kim, M.-H.; Hong, W.-J.; Moon, S.; Kim, E.-J.; Silva, J.; Lee, J.; Lee, S.; Kim, S.T.; Park, S.K.; et al. GORI, Encoding the WD40 Domain Protein, Is Required for Pollen Tube Germination and Elongation in Rice. Plant J. 2021, 105, 1645–1664. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Berhanu Lemma, R.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Pérez, N.; et al. JASPAR 2022: The 9th Release of the Open-Access Database of Transcription Factor Binding Profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Rahman, M.; Rahman, M.M.; Eom, J.-S.; Jeon, J.-S. Genome-Wide Identification, Expression Profiling and Promoter Analysis of Trehalose-6-Phosphate Phosphatase Gene Family in Rice. J. Plant Biol. 2020, 64, 55–71. [Google Scholar] [CrossRef]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for Occurrences of a given Motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).