Haploid Induction in Indica Rice: Exploring New Opportunities
Abstract
1. Introduction
2. In Vitro Techniques for Induction of Haploids and Doubled Haploids
2.1. Microspore-Derived Doubled Haploids in Rice Breeding: Standard Protocol
2.2. In Vitro Haploid Induction in Rice: The Current Scenario
2.3. Factors Influencing Androgenic Success: Hereditary Factors
2.3.1. The Genotype
2.3.2. Microspore Maturity
2.4. Factors Influencing Androgenic Success: Environmental Factors
2.4.1. Donor Plant Environment
2.4.2. Anther Pre-Treatment
2.4.3. In Vitro Culture Conditions
2.5. Plants Regenerated through In Vitro Haploid Cell Culture
3. In Vivo Methods of Haploid Induction
3.1. Wide Hybridization and Embryo Rescue
3.2. Use of Haploid Inducer Lines
3.2.1. Paternal Haploids
3.2.2. Maternal Haploids
3.3. CENH3 and MTL Are Two Promising Candidate Genes for Haploid Induction in Rice
4. Conclusions and Future Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Campbell, M.T.; Du, Q.; Liu, K.; Sharma, S.; Zhang, C.; Walia, H. Characterization of the transcriptional divergence between the subspecies of cultivated rice (Oryza sativa). BMC Genom. 2020, 21, 394. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. Statistical Yearbook: World Food and Agriculture; FAO: Rome, Italy, 2021. [Google Scholar] [CrossRef]
- Mohidem, N.A.; Hashim, N.; Shamsudin, R.; Che Man, H. Rice for Food Security: Revisiting Its Production, Diversity, Rice Milling Process and Nutrient Content. Agriculture 2022, 12, 741. [Google Scholar] [CrossRef]
- Valera, H.; Pede, V.; What Do We Know about the Future of Rice in Relation to Food System Transformation. CGIAR Initiative on Foresight. CGIAR News. 2023. Available online: https://www.cgiar.org/news-events/news/what-do-we-know-about-the-future-of-rice-in-relation-to-food-system-transformation/ (accessed on 25 August 2023).
- Mew, T.W.; Hibino, H.; Savary, S.; Vera Cruz, C.M.; Opulencia, R.; Hettel, G.P. (Eds.) Rice Diseases: Biology and Selected Management Practices; International Rice Research Institute: Los Baños, Philippines, 2018; Available online: https://rice-diseases.irri.org (accessed on 25 August 2023).
- Eckardt, N.; A Ainsworth, E.; Bahuguna, R.N.; Broadley, M.R.; Busch, W.; Carpita, N.C.; Castrillo, G.; Chory, J.; DeHaan, L.R.; Duarte, C.M.; et al. Climate change challenges, plant science solutions. Plant Cell 2023, 35, 24–66. [Google Scholar] [CrossRef] [PubMed]
- Kasha, K.J.; Maluszynski, M. Production of doubled haploids in crop plants: An Introduction. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer Science & Business Media: New York, NY, USA, 2003; pp. 1–4. [Google Scholar] [CrossRef]
- Murovec, J.; Bohanec, B. Haploids and doubled haploids in plant breeding. In Plant Breeding; Abdurakhmonov, I., Ed.; IntechOpen: Rijeka, Croatia, 2012; pp. 87–106. [Google Scholar] [CrossRef]
- Samantaray, S.; Ali, J.; Nicolas, K.L.C.; Katara, J.L.; Verma, R.L.; Parameswaran, C.; Devanna, B.N.; Kumar, A.; Dash, B.; Bhuyan, S.S. Doubled Haploids in Rice Improvement: Approaches, Applications, and Future Prospects. In Rice Improvement: Physiological, Molecular Breeding and Genetic Perspectives; Ali, J., Wani, S.H., Eds.; Springer Nature: Cham, Switzerland, 2021; pp. 425–447. [Google Scholar] [CrossRef]
- Lantos, C.; Jancsó, M.; Székely, A.; Nagy, E.; Szalóki, T.; Pauk, J. Improvement of Anther Culture to integrate Doubled Haploid Technology in Temperate Rice (Oryza sativa L.) Breeding. Plants 2022, 11, 3446. [Google Scholar] [CrossRef]
- Blakeslee, A.F.; Belling, J.; Farnham, M.E.; Bergner, A.D. A haploid mutant in the Jimson weed, “Datura stramonium”. Science 1922, 55, 646–647. [Google Scholar] [CrossRef]
- Magoon, M.L.; Khanna, K.R. Haploids. Caryologia 1963, 16, 191–235. [Google Scholar] [CrossRef]
- Germana, M.A. Anther culture for haploid and doubled haploid production. Plant Cell Tissue Organ Cult. 2011, 104, 283–300. [Google Scholar] [CrossRef]
- Chaikam, V.; Molenaar, W.; Melchinger, A.E.; Boddupalli, P.M. Doubled haploid technology for line development in maize: Technical advances and prospects. Theor. Appl. Genet. 2019, 132, 3227–3243. [Google Scholar] [CrossRef]
- Guha, S.; Maheshwari, S.C. In vitro production of embryos from anthers of Datura. Nature 1964, 204, 497. [Google Scholar] [CrossRef]
- Afza, R.; Shen, M.; Zapata-Arias, F.J.; Xie, J.; Fundi, H.K.; Lee, K.-S.; Bobadilla-Mucino, E.; Kodym, A. Effect of spikelet position on rice anther culture efficiency. Plant Sci. 2000, 153, 155–159. [Google Scholar] [CrossRef]
- Niroula, R.K.; Bimb, H.P. Effect of genotype and callus induction medium on green plant regeneration from anther of Nepalese rice cultivars. Asian J. Plant Sci. 2009, 8, 368–374. [Google Scholar] [CrossRef]
- Silva, T.D.; Ratnayake, W.J. Anther culture potential of Indica rice varieties, Kuruluthuda and BG 250. Trop. Agric. Res. Ext. 2009, 12, 53–56. [Google Scholar] [CrossRef][Green Version]
- Rukmini, M.; Rao, G.J.N.; Rao, R.N. Effect of cold pretreatment and phytohormones on anther culture efficiency of two Indica rice (Oryza sativa L.) hybrids—Ajay and Rajalaxmi. J. Exp. Biol. Agric. Sci. 2013, 1, 69–76. [Google Scholar]
- Rahman, Z.A.; Seman, Z.A.; Othman, A.N.; Ghaffar, M.B.A.; Razak, A.B.; Yusof, M.F.M. Efficient callus induction and plant regeneration of Malaysian indica rice MR219 using anther culture. Biocatal. Agric. Biotechnol. 2021, 31, 101865. [Google Scholar] [CrossRef]
- Sakina, A.; Mir, S.; Najeeb, S.; Zargar, S.M.; Nehvi, F.A.; Rather, Z.A.; Salgotra, R.K.; Shikari, A.B. Improved protocol for efficacious in vitro androgenesis and development of doubled haploids in temperate japonica rice. PLoS ONE 2020, 15, e0241292. [Google Scholar] [CrossRef]
- Sarao, N.K.; Gosal, S.S. In vitro androgenesis for accelerated breeding in rice. In Biotechnologies of Crop Improvement; Gosal, S., Wani, S., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 1, pp. 407–435. [Google Scholar] [CrossRef]
- Mayakaduwa, D.M.R.G.; Silva, T.D. Flow cytometric detection of haploids, diploids and mixoploids among the anther-derived plants in Indica rice (Oryza sativa L.). J. Anim. Plant Sci. 2019, 29, 1344–1351. [Google Scholar]
- Kumar, A.; Sandhu, N.; Yadav, S.; Pradhan, S.K.; Anandan, A.; Pandit, E. Rice varietal development to meet future challenges. In The future Rice Strategy for India; Mohanty, S., Chengappa, P.G., Mruthyunjaya Ladha, J.K., Baruah, S., Kannan, E., Eds.; Academic Press: London, UK, 2017; pp. 161–220. [Google Scholar]
- Javed, M.A.; Ishii, T.; Kamijima, O.; Misoo, S. The role of alternating culture temperatures and maltose in enhancing the anther culture efficiency of salt tolerant Indica rice (Oryza sativa L.) cultivars, Pokkali and Nona Bokra. Plant Biotechnol. 2007, 24, 283–287. [Google Scholar] [CrossRef]
- Grewal, D.; Manito, C.; Bartolome, V. Doubled haploids generated through anther culture from crosses of elite Indica and Japonica cultivars and/or lines of rice: Large-scale production, agronomic performance, and molecular characterization. Crop Sci. 2011, 51, 2544–2553. [Google Scholar] [CrossRef]
- Tripathy, S.K. Anther culture for double haploid breeding in rice-a way forward. Rice Genom. Genet. 2018, 9, 1–6. [Google Scholar] [CrossRef]
- Zhang, Z.H. The practicability of anther culture breeding. In Review of Advances in Plant Biotechnology 1985-88: 2nd International Symposium on Genetic Manipulation in Crops; Mujeeb-Kazi, A., Sitch, L.A., Eds.; International Maize and Wheat Improvement Center: Méx, Mexico; International Rice Research Institute: Los Baños, Philippines, 1989; pp. 31–42. [Google Scholar]
- Liang, S.; Huang, S. Huayu 15, a high-yielding rice variety bred by anther culture. In Biotechnology in Agriculture and Forestry; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1991; Volume 14, pp. 230–247. [Google Scholar] [CrossRef]
- Wang, P.; Bai, Y.-L.; Wang, M.-X.; Hu, B.-H.; Pu, Z.-G.; Zhang, Z.-Y.; Zhang, Q.; Xu, D.-W.; Luo, W.-L.; Chen, Z.-Q. Breeding of CMS maintainer lines through anther culture assisted by high-resolution melting-based markers. J. Integr. Agric. 2020, 19, 2965–2973. [Google Scholar] [CrossRef]
- He, T.; Yang, Y.; Tu, S.B.; Yu, M.Q.; Li, X.F. Selection of interspecific hybrids for anther culture of Indica rice. Plant Cell Tissue Organ Cult. 2006, 86, 271–277. [Google Scholar] [CrossRef][Green Version]
- Dewi, I.S.; Purwoko, B.S.; Aswidinnoor, H.; Somantri, I.H.; Chozin, M.A. Plant regeneration from anther cultures of several genotypes of Indica rice tolerant to aluminium toxicity. Indones. J. Agric. 2009, 2, 1–5. [Google Scholar]
- Dash, B.; Bhuyan, S.S.; Singh, S.K.; Chandravani, M.; Swain, N.; Rout, P.; Katara, J.L.; Parameswaran, C.; Devanna, B.N.; Samantaray, S. Androgenesis in indica rice: A comparative competency in development of doubled haploids. PLoS ONE 2022, 17, e0267442. [Google Scholar] [CrossRef]
- Senadhira, D.; Zapata-Arias, F.; Gregorio, G.; Alejar, M.; de la Cruz, H.; Padolina, T.; Galvez, A. Development of the first salt-tolerant rice cultivar through indica/indica anther culture. Field Crops Res. 2002, 76, 103–110. [Google Scholar] [CrossRef]
- Ahmadi, B.; Ebrahimzadeh, H. In vitro androgenesis: Spontaneous vs. artificial genome doubling and characterization of regenerants. Plant Cell Rep. 2020, 39, 299–316. [Google Scholar] [CrossRef] [PubMed]
- Mohiuddin, A.K.M.; Karim, N.H.; Shahanaz, S. Development of improved doubled-haploids through anther culture of Indica rice (Oryza sativa L.). Ann. Biol. Res. 2014, 5, 6–13. [Google Scholar]
- Naik, N.; Rout, P.; Umakanta, N.; Verma, R.L.; Katara, J.L.; Sahoo, K.K.; Samantaray, S. Development of doubled haploids from an elite Indica rice hybrid (BS6444G) using anther culture. Plant Cell Tissue Organ Cult. 2017, 128, 679–689. [Google Scholar] [CrossRef]
- Mishra, R.; Rao, G.J.N. In-vitro androgenesis in rice: Advantages, constraints and future prospects. Rice Sci. 2016, 23, 57–68. [Google Scholar] [CrossRef]
- Miah, M.A.A.; Earle, E.D.; Khush, G.S. Inheritance of callus formation ability in anther cultures of rice, Oryza sativa L. Theor. Appl. Genet. 1985, 70, 113–116. [Google Scholar] [CrossRef]
- Silva, T.D. Indica rice anther culture: Can the impasse be surpassed. Plant Cell Tissue Organ Cult. 2010, 100, 1–11. [Google Scholar] [CrossRef]
- Herath, H.M.I.; Bandara, D.C. Anther culture performance in selected high yielding Indica (of Sri Lanka) and Japonica rice varieties and their inter sub-specific hybrids. J. Natl. Sci. Found. Sri Lanka 2011, 39, 149–154. [Google Scholar] [CrossRef]
- Yan, J.; Xue, Q.; Zhu, J. Genetic studies of anther culture ability in rice (Oryza sativa). Plant Cell Tissue Organ Cult. 1996, 45, 253–258. [Google Scholar] [CrossRef]
- Gajecka, M.; Marzec, M.; Chmielewska, B.; Jelonek, J.; Zbieszczyk, J.; Szarejko, I. Changes in plastid biogenesis leading to the formation of albino regenerants in barley microspore culture. BMC Plant Biol. 2021, 21, 22. [Google Scholar] [CrossRef]
- Yamagishi, M. Heterogeneous plastid genomes in anther culture-derived albino rice plants. Euphytica 2002, 123, 67–74. [Google Scholar] [CrossRef]
- Datta, S.K. Androgenic haploids: Factors controlling development and its application in crop improvement. Curr. Sci. 2005, 89, 1870–1878. [Google Scholar]
- Raghavan, V. From microspore to embryoid: Faces of the angiosperm pollen grain. In Progress in Plant Cellular and Molecular Biology; Current Plant Science and Biotechnology in Agriculture; Nijkamp, H.J.J., Van Der Plas, L.H.W., Van Aartrijk, J., Eds.; Springer: Dordrecht, The Netherlands, 1990; pp. 213–221. [Google Scholar] [CrossRef]
- Dunwell, J.M. Haploids in flowering plants: Origins and exploitation. Plant Biotechnol. J. 2010, 8, 377–424. [Google Scholar] [CrossRef]
- Mayakaduwa, D.M.R.G.; Silva, T.D. A cytological indicator allows rapid assessment of microspore maturity, leading to improved in vitro anther response in Indica rice (Oryza sativa L.). In Vitro Cell. Dev. Biol. Plant 2017, 53, 591–597. [Google Scholar] [CrossRef]
- Bishnoi, U.; Jain, R.K.; Rohilla, J.S.; Chowdhury, V.K.; Gupta, K.R.; Chowdhury, J.B. Anther culture of recalcitrant Indica× basmati rice hybrids. Euphytica 2000, 114, 93–101. [Google Scholar] [CrossRef]
- Bagheri, N.; Jelodar, N.B.; Ghanbari, A. Evaluation of effective factors in anther culture of Iranian rice (Oryza sativa L.) cultivars. Biharean Biol. 2009, 3, 119–124. [Google Scholar]
- Serrat, X.; Cardona, M.; Gil, J.; Brito, A.M.; Moysset, L.; Nogués, S.; Lalanne, E. A Mediterranean Japonica rice (Oryza sativa) cultivar improvement through anther culture. Euphytica 2014, 195, 31–44. [Google Scholar] [CrossRef][Green Version]
- Cha-um, S.; Srianan, B.; Pichakum, A.; Kirdmanee, C. An efficient procedure for embryogenic callus induction and double haploid plant regeneration through anther culture of Thai aromatic rice (Oryza sativa L. subsp. Indica). In Vitro Cell. Dev. Biol.-Plant 2009, 45, 171–179. [Google Scholar] [CrossRef]
- Rout, P.; Naik, N.; Ngangkham, U.; Verma, R.L.; Katara, J.L.; Singh, O.N.; Samantaray, S. Doubled haploids generated through anther culture from an elite long duration rice hybrid, CRHR32: Method optimization and molecular characterization. Plant Biotechnol. 2016, 33, 177–186. [Google Scholar] [CrossRef]
- Foroughi-Wehr, B.; Wenzel, G. Andro- and parthenogenesis. In Plant Breeding; Hayward, M.D., Bosemark, N.O., Romagosa, I., Cerezo, M., Eds.; Springer: Dordrecht, The Netherlands, 1993; pp. 261–277. [Google Scholar] [CrossRef]
- Heberle-Bors, E. In vitro haploid formation from pollen: A critical review. Theor. Appl. Genet. 1985, 71, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Raina, S.K. Doubled haploid breeding in cereals. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 1997; pp. 141–186. [Google Scholar]
- Tuvesson, S.; von Post, R.; Ljungberg, A. Wheat anther culture. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 71–76. [Google Scholar] [CrossRef]
- Davies, P.A. Barley isolated microspore culture (IMC) method. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 49–52. [Google Scholar] [CrossRef]
- Immonen, S.; Tenhola-Roininen, T. Protocol for rye anther culture. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 141–149. [Google Scholar] [CrossRef]
- Silva, T.D. Microspore embryogenesis. In Embryogenesis; Sato, K.I., Ed.; IntechOpen: Rijeka, Croatia, 2012; pp. 573–596. [Google Scholar] [CrossRef][Green Version]
- Sopory, S.K.; Munshi, M. Anther culture. In In Vitro Haploid Production in Higher Plants; Current Plant Science and Biotechnology in Agriculture; Jain, S.M., Sopory, S.K., Veilleux, R.E., Eds.; Springer: Dordrecht, The Netherlands, 1996; Volume 23, pp. 145–176. [Google Scholar] [CrossRef]
- Smykal, P. Pollen embryogenesis- the stress mediated switch from gametophytic to sporophytic development: Current status and future prospects. Biol. Plant. 2000, 43, 481–489. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Bhojwani, S.S. Enhanced pollen grain embryogenesis and plant regeneration in anther cultures of Brassica juncea cv. PR-45. Euphytica 1993, 70, 191–196. [Google Scholar] [CrossRef]
- Guzman, M.; Arias, F.J.Z. Increasing anther culture efficiency in rice (Oryza sativa L.) using anthers from ratooned plants. Plant Sci. 2000, 151, 107–114. [Google Scholar] [CrossRef]
- Shariatpanahi, M.E.; Bal, U.; Heberle-Bors, E.; Touraev, A. Stresses applied for the re-programming of plant microspores towards in vitro embryogenesis. Physiol. Plant. 2006, 127, 519–534. [Google Scholar] [CrossRef]
- Touraev, A.; Vicente, O.; Heberle-Bors, E. Initiation of microspore embryogenesis by stress. Trends Plant Sci. 1997, 2, 297–302. [Google Scholar] [CrossRef]
- Huang, B. Wheat anther culture: Effect of temperature. In Biotechnology in Agriculture and Forestry: Wheat; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1990; Volume 13, pp. 403–415. [Google Scholar]
- Herath, H.M.I.; Bandara, D.C.; Samarajeewa, P.K.; Wijesundara, D.S.A. Effect of low temperature pre-treatment on anther culture in selected Indica, Japonica rice varieties and their inter sub-specific hybrids. Ceylon J. Sci. (Biol. Sci.) 2009, 38, 11–16. [Google Scholar] [CrossRef]
- Lee, S.T.; Huang, W.L. Osmotic stress stimulates shoot organogenesis in callus of rice (Oryza sativa L.) via auxin signalling and carbohydrate metabolism regulation. Plant Growth Regul. 2014, 73, 193–204. [Google Scholar] [CrossRef]
- Duncan, E.J.; Heberle, E. Effect of temperature shock on nuclear phenomena in microspores of Nicotiana tabacum and consequently on plantlet production. Protoplasma 1976, 90, 173–177. [Google Scholar] [CrossRef]
- Rashid, A. Pollen dimorphism in relation to pollen plant formation. Physiol. Plant. 1983, 58, 544–548. [Google Scholar] [CrossRef]
- Sunderland, N.; Xu, Z.H. Shed pollen culture in Hordeum vulgare. J. Exp. Bot. 1982, 33, 1086–1095. [Google Scholar] [CrossRef]
- Zoriniants, S.; Tashpulatov, A.S.; Heberle-Bors, E.; Touraev, A. The role of stress in the induction of haploid microspore embryogenesis. In Biotechnology in Agriculture and Forestry: Haploids in Crop Improvement II; Palmer, C.E., Keller, W.A., Kasha, K.J., Eds.; Springer: Heidelberg, Germany, 2005; Volume 56, pp. 35–52. [Google Scholar] [CrossRef]
- Nayyar, H.; Bains, T.; Kumar, S. Low temperature induced floral abortion in chickpea: Relationship to abscisic acid and cryoprotectants in reproductive organs. Environ. Exp. Bot. 2005, 53, 39–47. [Google Scholar] [CrossRef]
- Oliver, S.N.; van Dongen, J.T.; Alfred, S.C.; Mamun, E.A.; Zhao, X.; Saini, H.S.; Fernandes, S.F.; Blanchard, C.L.; Sutton, B.G.; Geigenberger, P.; et al. Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ. 2005, 28, 1534–1551. [Google Scholar] [CrossRef]
- Huang, W.L.; Liu, L.F. Carbohydrate metabolism in rice during callus induction and shoot regeneration induced by osmotic stress. Bot. Bull. Acad. Sin. 2002, 43, 107–113. [Google Scholar]
- Wetherell, D.F. Enhanced adventive embryogenesis resulting from plasmolysis of cultured wild carrot cells. Plant Cell Tissue Organ Cult. 1984, 3, 221–227. [Google Scholar] [CrossRef]
- Garrido, D.; Vicente, O.; Heberle-Bors, E.; Rodriguez-Garcia, M.I. Cellular changes during the acquisition of embryogenic potential in isolated pollen grains of Nicotiana tabacum. Protoplasma 1995, 186, 220–230. [Google Scholar] [CrossRef]
- Kyo, M.; Harada, H. Specific phosphoproteins in the initial period of tobacco pollen embryogenesis. Planta 1990, 182, 58–63. [Google Scholar] [CrossRef]
- Heberle-Bors, E. Isolated pollen culture in tobacco: Plant reproductive development in a nutshell. Sex. Plant Reprod. 1989, 2, 1–10. [Google Scholar] [CrossRef]
- Maraschin, S.D.F.; De Priester, W.; Spaink, H.P.; Wang, M. Androgenic switch: An example of plant embryogenesis from the male gametophyte perspective. J. Exp. Bot. 2005, 56, 1711–1726. [Google Scholar] [CrossRef] [PubMed]
- Mayakaduwa, D.M.R.G.; Silva, T.D. In vitro response of Indica rice microspores subjected to cold stress: A cytological and histological perspective. In Vitro Cell. Dev. Biol. Plant 2021, 57, 843–855. [Google Scholar] [CrossRef]
- Chu, C.C. The N6 medium and its applications to anther culture of cereal crops. In Proceedings of the Symposium on Plant Tissue Culture; Science Press: Beijing, China, 1978; pp. 45–50. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioasays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Ali, J.; Nicolas, K.L.C.; Akther, S.; Torabi, A.; Ebadi, A.A.; Marfori-Nazarea, C.M.; Mahender, A. Improved Anther Culture Media for Enhanced Callus Formation and Plant Regeneration in Rice (Oryza sativa L.). Plants 2021, 10, 839. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.; Bhavya, C.; Ashok, T.H. Effect of genotypes and different concentration of growth regulator on callus induction and plant regeneration through anther culture of rice. J. Pharmacogn. Phytochem. 2017, 6, 1354–1358. [Google Scholar]
- Lantos, C.; Jancsó, M.; Székely, Á.; Szalóki, T.; Venkatanagappa, S.; Pauk, J. Development of In Vitro Anther Culture for Doubled Haploid Plant Production in Indica Rice (Oryza sativa L.) Genotypes. Plants 2023, 12, 1774. [Google Scholar] [CrossRef]
- Hooghvorst, I.; Ribas, P.; Nogués, S. Chromosome doubling of androgenic haploid plantlets of rice (Oryza sativa) using antimitotic compounds. Plant Breed. 2020, 139, 754–761. [Google Scholar] [CrossRef]
- Devaux, P. The Hordeum bulbosum (L.) method. In Doubled Haploid Production in Crop Plants; Maluszynski, M., Kasha, K.J., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 15–19. [Google Scholar] [CrossRef]
- Kumlehn, J. Haploid technology. In Biotechnological Approaches to Barley Improvement. Biotechnology in Agriculture and Forestry; Kumlehn, J., Stein, N., Eds.; Springer: Heidelberg, Germany, 2014; Volume 69, pp. 379–392. [Google Scholar] [CrossRef]
- Laurie, D.A. The frequency of fertilization in wheat × pearl millet crosses. Genome 1989, 32, 1063–1067. [Google Scholar] [CrossRef]
- Laurie, D.A.; Bennett, M.D. Wheat × maize hybridization. Can. J. Genet. Cytol. 1986, 28, 313–316. [Google Scholar] [CrossRef]
- Rines, H.W. Oat haploids from wide hybridization. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 155–159. [Google Scholar] [CrossRef]
- Wedzony, M. Protocol for doubled haploid production in hexaploid triticale (x Triticosecale Wittm.) by crosses with maize. In Doubled Haploid Production in Crop Plants: A Manual; Maluszynski, M., Kasha, K., Forster, B.P., Szarejko, I., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 135–140. [Google Scholar] [CrossRef]
- Kisana, N.S.; Nkongolo, K.K.; Quick, J.S.; Johnson, D.L. Production of Doubled Haploids by Anther Culture and Wheat X Maize Method in a Wheat Breeding Programme. Plant Breed. 1993, 110, 96–102. [Google Scholar] [CrossRef]
- Kyum, M.; Kaur, H.; Kamboj, A.; Goyal, L.; Bhatia, D. Strategies and prospects of haploid induction in rice (Oryza sativa). Plant Breed. 2022, 141, 1–11. [Google Scholar] [CrossRef]
- Bennett, M.D.; Finch, R.A.; Barclay, I.R. The time rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma 1976, 54, 175–200. [Google Scholar] [CrossRef]
- Finch, R.A. Tissue-specific elimination of alternative whole parental genomes in one barley hybrid. Chromosoma 1983, 88, 386–393. [Google Scholar] [CrossRef]
- Ravi, M.; Chan, S. Haploid plants produced by centromere-mediated genome elimination. Nature 2010, 464, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Ashtiyani, R.; Ishii, T.; Niessen, M.; Stein, N.; Heckmann, S.; Gurushidze, M.; Banaei-Moghaddam, A.M.; Fuchs, J.; Schubert, V.; Koch, K.; et al. Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. USA 2015, 112, 11211–11216. [Google Scholar] [CrossRef] [PubMed]
- Kuppu, S.; Tan, E.H.; Nguyen, H.; Rodgers, A.; Comai, L.; Chan, S.W.L.; Britt, A.B. Point Mutations in Centromeric Histone Induce Post-zygotic Incompatibility and Uniparental Inheritance. PLoS Genet. 2015, 11, e1005494. [Google Scholar] [CrossRef]
- Kuppu, S.; Ron, M.; Marimuthu, M.P.; Li, G.; Huddleson, A.; Siddeek, M.H.; Terry, J.; Buchner, R.; Shabek, N.; Comai, L.; et al. A variety of changes, including CRISPR/Cas9-mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol. J. 2020, 18, 2068–2080. [Google Scholar] [CrossRef]
- Sanei, M.; Pickering, R.; Kumke, K.; Nasuda, S.; Houben, A. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Natl. Acad. Sci. USA 2011, 108, E498–E505. [Google Scholar] [CrossRef]
- Kalinowska, K.; Chamas, S.; Unkel, K.; Demidov, D.; Lermontova, I.; Dresselhaus, T.; Kumlehn, J.; Dunemann, F.; Houben, A. State-of-the-art novel developments of in vivo haploid technologies. Theor. Appl. Genet. 2019, 132, 593–605. [Google Scholar] [CrossRef]
- Kaur, K.; Neelam, K.; Singh, J.; Malik, P.; Singh, K. Uncovering natural allelic and structural variants of OsCENH3 gene by targeted resequencing and in silico mining in genus Oryza. Sci. Rep. 2023, 13, 830. [Google Scholar] [CrossRef]
- Husbands, A.; Bell, E.M.; Shuai, B.; Smith, H.M.S.; Springer, P.S. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007, 35, 6663–6671. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.M.S. The indeterminate gametophyte1 Gene of Maize Encodes a LOB Domain Protein Required for Embryo Sac and Leaf Development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tang, W.; Huang, Y.; Niu, X.; Zhao, Y.; Han, Y.; Liu, Y. Down-regulation of a LBD-like gene, OSIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophytes in rice. J. Exp. Bot. 2015, 66, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, K.R.; Coe, E.H., Jr. A Genetic analysis of the origin of maternal haploids in maize. Genetics 1966, 54, 453–464. [Google Scholar] [CrossRef]
- Zhao, X.; Xu, X.; Xie, H.; Chen, S.; Jin, W. Fertilization and uniparental chromosome elimination during crosses with maize haploid inducers. Plant Physiol. 2013, 163, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Prigge, V.; Xu, X.; Li, L.; Babu, R.; Chen, S.; Atlin, G.N.; Melchinger, A.E. New insights into genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics 2012, 190, 781–793. [Google Scholar] [CrossRef]
- Li, Y.; Lin, Z.; Yue, Y.; Zhao, H.; Fei, X.; Lizhu, E.; Liu, C.; Chen, S.; Lai, J.; Song, W. Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat. Plants 2021, 7, 1579–1588. [Google Scholar] [CrossRef]
- Bhowmik, P.; Bilichak, A. Advances in Gene Editing of Haploid Tissues in Crops. Genes 2021, 12, 1410. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Liu, C.; Li, X.; Meng, D.; Zhong, Y.; Chen, C.; Dong, X.; Xu, X.; Chen, B.; Li, W.; Li, L.; et al. A 4-bp Insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 2017, 10, 520–522. [Google Scholar] [CrossRef]
- Gilles, L.M.; Khaled, A.; Laffaire, J.B.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Widiez, T. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017, 36, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Coe, E.H., Jr. A Line of Maize with High Haploid Frequency. Am. Nat. 1959, 93, 381–382. [Google Scholar] [CrossRef]
- Kermicle, J.L. Androgenesis Conditioned by a Mutation in Maize. Science 1969, 166, 1422–1424. [Google Scholar] [CrossRef]
- Singh, A.; Baranwal, V.; Shankar, A.; Kanwar, P.; Ranjan, R.; Yadav, S.; Pandey, A.; Kapoor, S.; Pandey, G.K. Rice Phospholipase A Superfamily: Organization, Phylogenetic and Expression Analysis during Abiotic Stresses and Development. PLoS ONE 2012, 7, e30947. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Liang, D.; Liu, J.; Sahoo, G.; Kelliher, T. OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 2018, 4, 530–533. [Google Scholar] [CrossRef]
- Liu, J.; Liang, D.; Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Zhou, H.; Kelliher, T.; Zhang, X.; et al. Rice Haploid Inducer Development by Genome Editing. In Rice Genome Engineering and Gene Editing: Methods in Molecular Biology; Bandyopadhyay, A., Thilmony, R., Eds.; Humana: New York, NY, USA, 2021; Volume 2238, pp. 221–230. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Q.; Shen, Y.; Hua, Y.; Wang, J.; Lin, J.; Wu, M.; Sun, T.; Cheng, Z.; Mercier, R.; et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 2019, 37, 283–286. [Google Scholar] [CrossRef]
- Wang, J.; Cao, Y.; Wang, K.; Liu, C. Development of multiple-heading-date mtl haploid inducer lines in rice. Agriculture 2022, 12, 806. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, D.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Liu, C.; Li, W.; Zhong, Y.; Dong, X.; Hu, H.; Tian, X.; Wang, L.; Chen, B.; Chen, C.; Melchinger, A.E.; et al. Fine mapping of qhir8 affecting in vivo haploid induction in maize. Theor. Appl. Genet. 2015, 128, 2507–2515. [Google Scholar] [CrossRef]
- Wang, S.; Jin, W.; Wang, K. Centromere histone H3- and phospholipase-mediated haploid induction in plants. Plant Methods 2019, 15, 42. [Google Scholar] [CrossRef]
- Hooghvorst, I.; Nogués, S. Chromosome doubling methods in doubled haploid and haploid inducer-mediated genome-editing systems in major crops. Plant Cell Rep. 2021, 40, 255–270. [Google Scholar] [CrossRef] [PubMed]
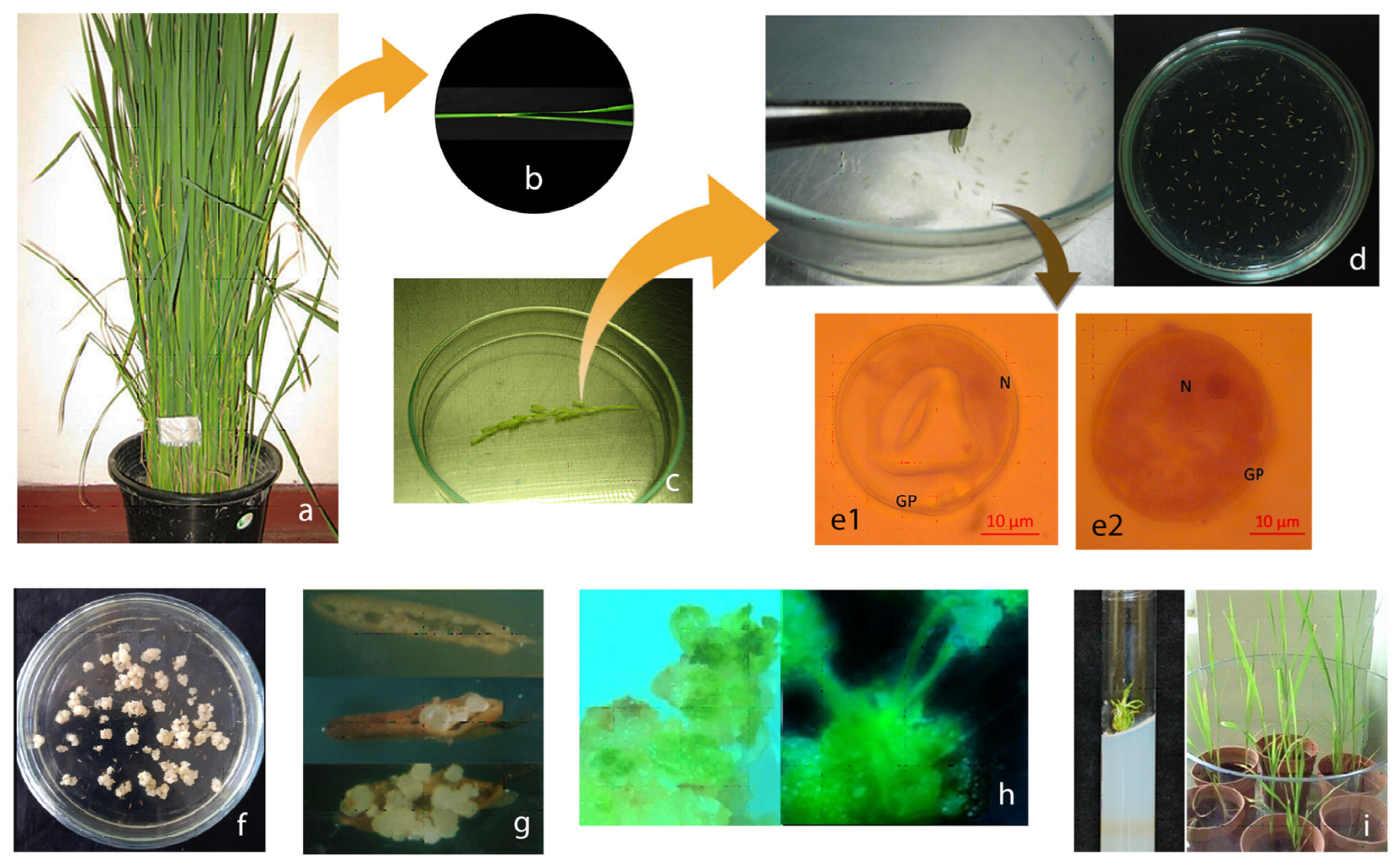
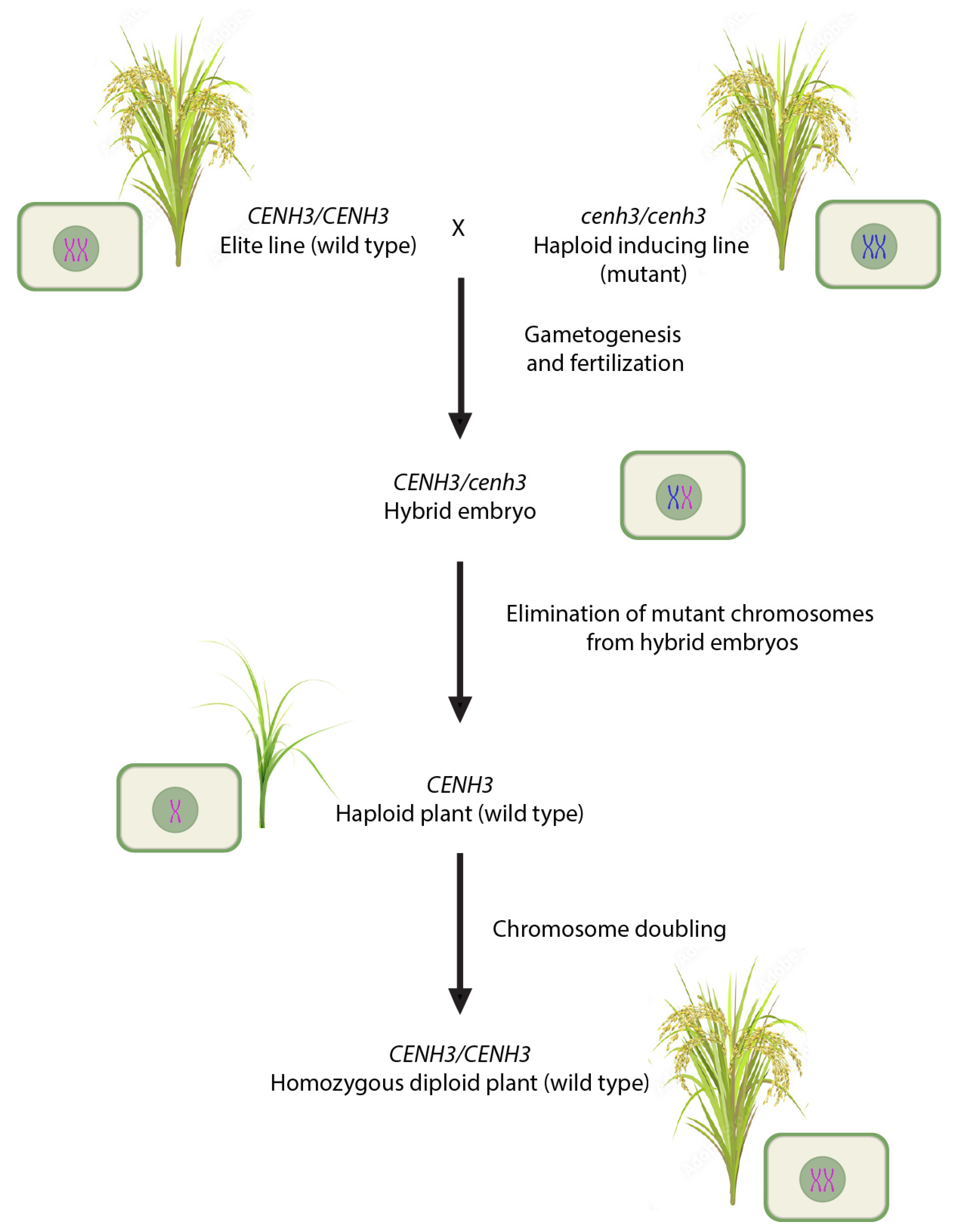
| Gene | Wild Type (WT) Gene Product | WT Expression Analysis | Mutagenic Sites | Mutant Phenotype | Rice Ortholog/Potential for Rice Haploid Induction |
|---|---|---|---|---|---|
| CENH3 | Centromere-specific CENH3 protein (histone variant) consisting of two domains: conserved C-terminal HFD and highly variable N-terminal tail | Kinetochore-loading of CENH3 protein leading to precise chromosome segregation during cell division. | Both domains are targets. | Impaired chromosome segregation during cell division and uniparental chromosome elimination in young embryos. | OsCENH3/ Limited studies in rice have shown an HIR of approximately 1% [104]. |
| IG-1 | LOB-domain protein (Transcription Factor) | Normal female gametogenesis leading to an 8-celled embryo sac (and the development of lateral organs). | Second exon of the gene, upstream of the encoded LOB domain. | Delayed cellularization of the embryo sac leading to extra gametes and abnormal fertilization. | OsIGI/No reports of haploid induction in rice. |
| MTL | Pollen-specific patatin-like phospholipase protein | Localization of proteins in mature pollen membranes leads to normal fertilization of the egg cell. | Exons 1 and 4 (CRISPR/Cas 9 knock-outs) | Mis-localization of the defective protein affects normal fertilization leading to maternal haploid induction. | OsMTL/ HIR of 6% when selfed and 2–5% when out-crossed in rice [120]. |
| DMP | Domain of unknown function 679 Membrane Protein | Expressed in mature pollen and involved in gamete fusion (similar to MTL) | Coding region | Not clearly defined | OsDMP/No reports of haploid induction in rice |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayakaduwa, R.; Silva, T. Haploid Induction in Indica Rice: Exploring New Opportunities. Plants 2023, 12, 3118. https://doi.org/10.3390/plants12173118
Mayakaduwa R, Silva T. Haploid Induction in Indica Rice: Exploring New Opportunities. Plants. 2023; 12(17):3118. https://doi.org/10.3390/plants12173118
Chicago/Turabian StyleMayakaduwa, Ruwani, and Tara Silva. 2023. "Haploid Induction in Indica Rice: Exploring New Opportunities" Plants 12, no. 17: 3118. https://doi.org/10.3390/plants12173118
APA StyleMayakaduwa, R., & Silva, T. (2023). Haploid Induction in Indica Rice: Exploring New Opportunities. Plants, 12(17), 3118. https://doi.org/10.3390/plants12173118






