Investigation of the Influence of Polyamines on Mature Embryo Culture and DNA Methylation of Wheat (Triticum aestivum L.) Using the Machine Learning Algorithm Method
Abstract
:1. Introduction
2. Results
2.1. In Vitro Parameters
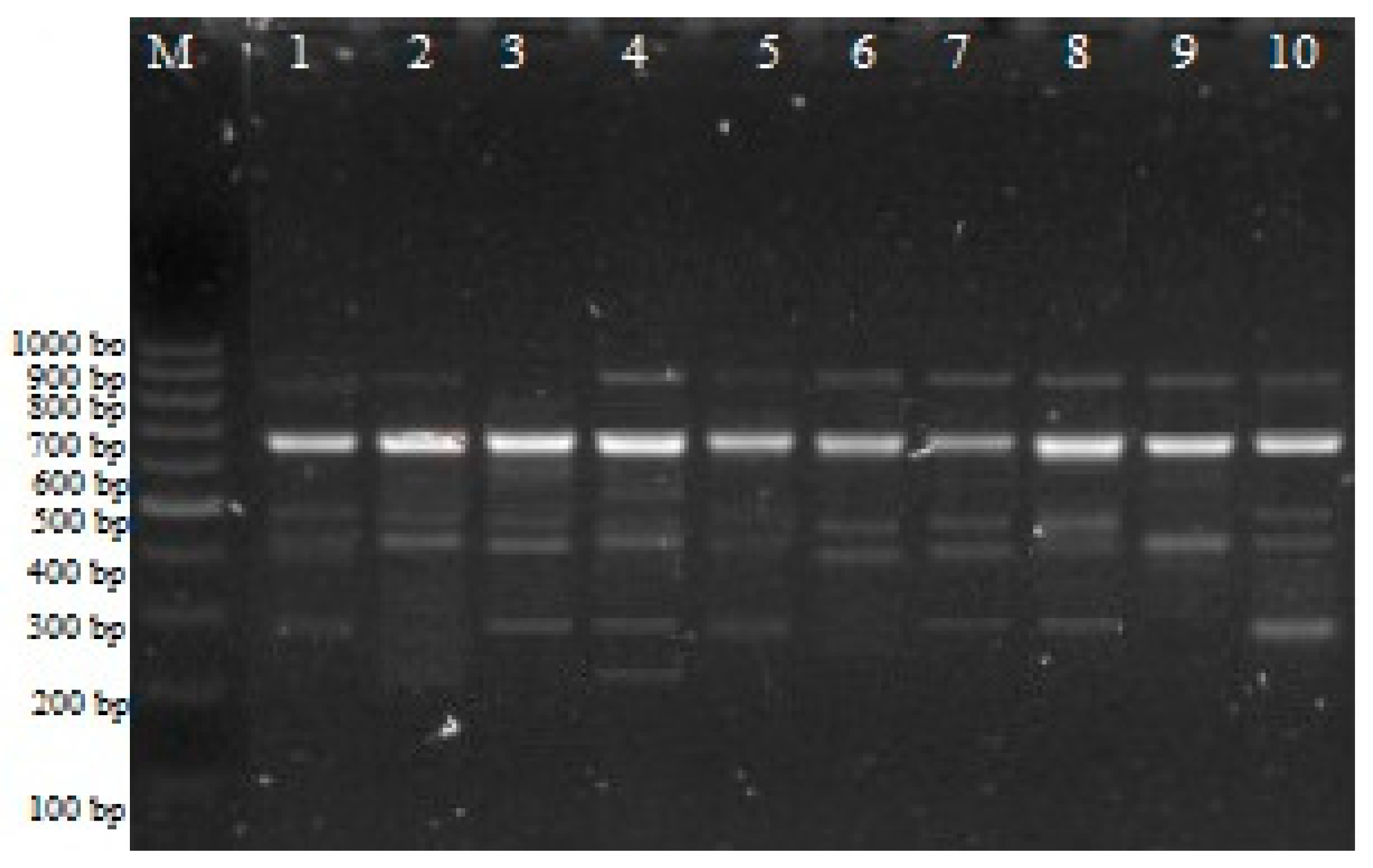
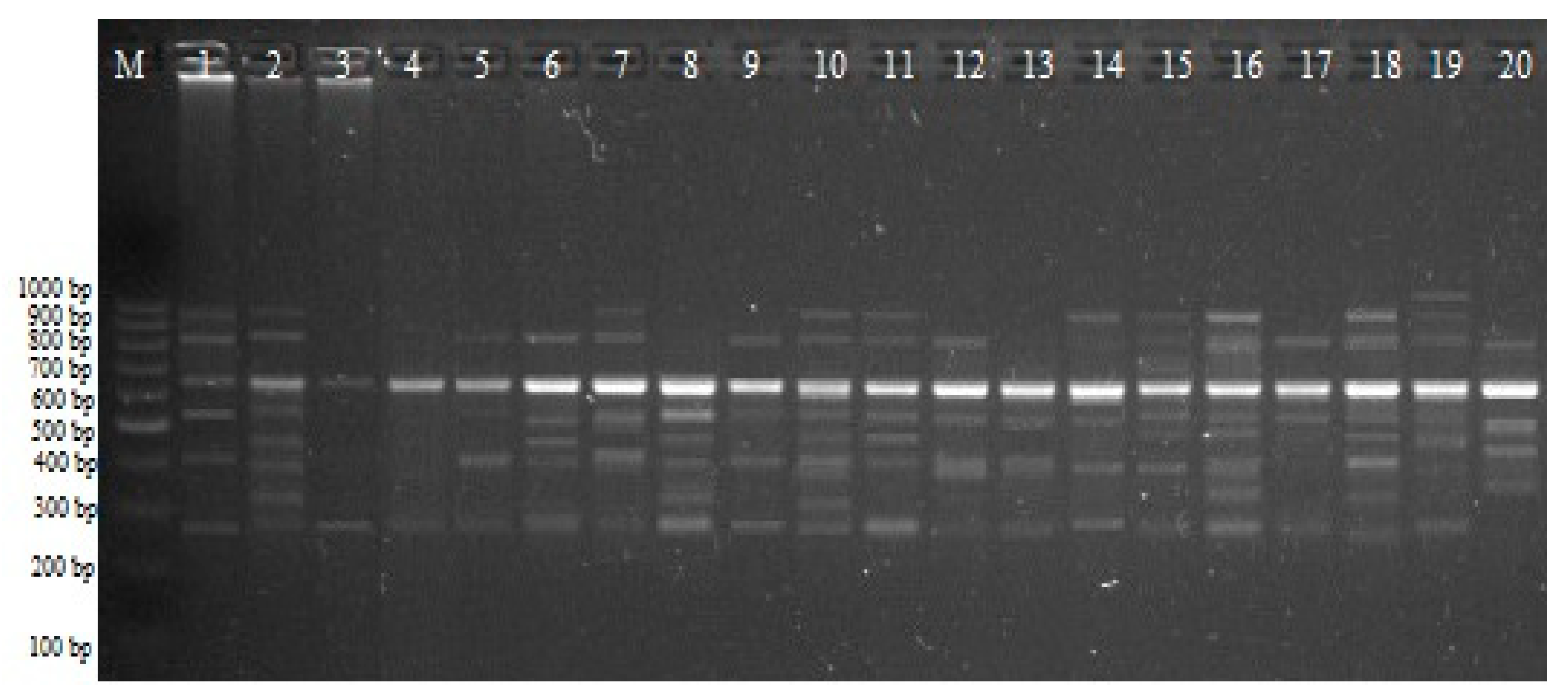
2.2. iPBS Analysis
2.3. Machine Learning (ML) Analysis
3. Discussion
4. Materials and Methods
4.1. In Vitro Conditions
4.1.1. Plant Material, Callus Initiation, Embryogenic Callus Formation, and Plant Regeneration
4.1.2. Statistical Analysis
4.2. Genotypic Assay
4.2.1. Isolation of Genomic DNA, iPBS, and CRED–iPBS PCR Assays and Electrophoresis
4.2.2. Genetics Analysis
4.3. Modeling Using Machine Learning Algorithms
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine function in plants: Metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [PubMed]
- Alcázar, R.; Tiburcio, A.F. Plant polyamines in stress and development: An emerging area of research in plant sciences. Front. Plant Sci. 2014, 5, 319. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Hu, Q.; Pu, Q.; Chen, M.; Zhu, X.; Gao, C.; Cao, Y. Spermidine enhanced free polyamine levels and expression of polyamine biosynthesis enzyme gene in rice spikelets under heat tolerance before heading. Sci. Rep. 2020, 10, 1–9. [Google Scholar]
- Kapoor, R.T. Role of polyamines in plants under abiotic stresses: Regulation of biochemical interactions. Plant Stress Mitigators 2023, 209–220. [Google Scholar] [CrossRef]
- Aydin, M.; Pour, A.H.; Haliloğlu, K.; Tosun, M. Effect of polyamines on somatic embryogenesis via mature embryo in wheat. Turk. J. Biol. 2016, 40, 1178–1184. [Google Scholar] [CrossRef]
- Redha, A.; Suleman, P. Effects of exogenous application of polyamines on wheat anther cultures. Plant Cell Tissue Organ Cult. 2011, 105, 345–353. [Google Scholar]
- Qureshi, M.A.; Shafqat, W.; ud Din, S.; Raza, M.K.; Sharif, N.; Ikram, S.; Rana, H.A.A. Effect of different plant growth regulators on in vitro regeneration of mango: A report. J. Glob. Innov. Agric. Sci. 2021, 9, 95–98. [Google Scholar]
- Miguel, C.; Marum, L. An epigenetic view of plant cells cultured in vitro: Somaclonal variation and beyond. J. Exp. Bot. 2011, 62, 3713–3725. [Google Scholar]
- Shah, A.A.; Ahmed, S.; Ali, A.; Yasin, N.A. 2–Hydroxymelatonin Mitigates Cadmium Stress in Cucumis sativus Seedlings: Modulation of Antioxidant Enzymes and Polyamines. Chemosphere 2020, 243, 125308. [Google Scholar]
- Yu, Y.; Jin, C.; Sun, C.; Wang, J.; Ye, Y.; Zhou, W.; Lu, L.; Lin, X. Inhibition of ethylene production by putrescine alleviates aluminium–induced root inhibition in wheat plants. Sci. Rep. 2016, 6, 18888. [Google Scholar]
- Sharma, S.K.; Hazeldine, S.; Crowley, M.L.; Hanson, A.; Beattie, R.; Varghese, S.; Woster, P.M. Polyamine–based small molecule epigenetic modulators. MedChemComm. 2012, 3, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Madhumati, B. Potential and application of molecular markers techniques for plant genome analysis. Int. J. Pure Appl. Biosci. 2014, 2, 169–188. [Google Scholar]
- Hosseinpour, A.; Aydin, M.; Haliloglu, K. Plant regeneration system in recalcitrant rye (Secale cereale L.). Biologia 2020, 75, 1017–1028. [Google Scholar] [CrossRef]
- Hosseinpour, A.; Ilhan, E.; Özkan, G.; Öztürk, H.İ.; Haliloglu, K.; Cinisli, K.T. Plant growth–promoting bacteria (PGPBs) and copper (II) oxide (CuO) nanoparticle ameliorates DNA damage and DNA Methylation in wheat (Triticum aestivum L.) exposed to NaCl stress. J. Plant Biochem. Biotechnol. 2022, 31, 751–764. [Google Scholar] [CrossRef]
- Turkoglu, A. Effects of mammalian sex hormones on regeneration capacity, retrotransposon polymorphism and genomic instability in wheat (Triticum aestivum L.). Plant Cell Tissue Organ Cult. 2023, 152, 647–659. [Google Scholar] [CrossRef]
- Türkoğlu, A.; Haliloğlu, K.; Tosun, M.; Szulc, P.; Demirel, F.; Eren, B.; Bujak, H.; Karagöz, H.; Selwet, M.; Özkan, G.; et al. Sodium Azide as a Chemical Mutagen in Wheat (Triticum aestivum L.): Patterns of the Genetic and Epigenetic Effects with iPBS and CRED–iPBS Techniques. Agriculture 2023, 13, 1242. [Google Scholar] [CrossRef]
- Türkoglu, A.; Tosun, M.; Haliloğlu, K. Mutagenic effects of sodium azide on in vitro mutagenesis, polymorphism and genomic instability in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2022, 49, 10165–10174. [Google Scholar] [CrossRef]
- Türkoğlu, A.; Tosun, M.; Haliloğlu, K. Evaluation of ethyl methanesulfonate–induced in vitro mutagenesis, polymorphism and genomic instability in wheat (Triticum aestivum L.). J. Crop. Sci. Biotechnol. 2023, 26, 199–213. [Google Scholar] [CrossRef]
- Hosseinpour, A.; Özkan, G.; Nalci, Ö.; Haliloğlu, K. Estimation of genomic instability and DNA methylation due to aluminum (Al) stress in wheat (Triticum aestivum L.) using iPBS and CRED–iPBS analyses. Turk. J. Bot. 2019, 43, 27–37. [Google Scholar] [CrossRef]
- D’Agostino, L.; Di Pietro, M.; Di Luccia, A. Nuclear aggregates of polyamines are supramolecular structures that play a crucial role in genomic DNA protection and conformation. FEBS J. 2005, 272, 3777–3787. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J.; Ruiz-Medrano, R.; Dominguez, A. Selective inhibition of cytosine DNA methylases by polyamines. FEBS Lett. 1995, 357, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Jones, A.M.P. Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl. Microbiol. Biotechnol. 2020, 104, 9449–9485. [Google Scholar] [CrossRef] [PubMed]
- Niazian, M.; Niedbała, G. Machine learning for plant breeding and biotechnology. Agriculture 2020, 10, 436. [Google Scholar] [CrossRef]
- Aasim, M.; Katırcı, R.; Akgur, O.; Yildirim, B.; Mustafa, Z.; Nadeem, M.A.; Yılmaz, G. Machine learning (ML) algorithms and artificial neural network for optimizing in vitro germination and growth indices of industrial hemp (Cannabis sativa L.). Ind. Crop. Prod. 2022, 181, 114801. [Google Scholar] [CrossRef]
- Aasim, M.; Ali, S.A.; Bekiş, P.; Nadeem, M.A. Light–emitting diodes induced in vitro regeneration of Alternanthera reineckii mini and validation via machine learning algorithms. Vitr. Cell. Dev. Biol. Plant 2022, 58, 816–825. [Google Scholar] [CrossRef]
- Hesami, M.; Condori-Apfata, J.A.; Valderrama Valencia, M.; Mohammadi, M. Application of artificial neural network for modeling and studying in vitro genotype–independent shoot regeneration in wheat. Appl. Sci. 2020, 10, 5370. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M. Modeling and optimizing in vitro sterilization of chrysanthemum via multilayer perceptron–non–dominated sorting genetic algorithm–II (MLP–NSGAII). Front. Plant Sci. 2019, 10, 282. [Google Scholar] [CrossRef]
- Kirtis, A.; Aasim, M.; Katırcı, R. Application of artificial neural network and machine learning algorithms for modeling the in vitro regeneration of chickpea (Cicer arietinum L.). Plant Cell Tissue Organ Cult. 2022, 150, 141–152. [Google Scholar] [CrossRef]
- Mirza, K.; Aasim, M.; Katırcı, R.; Karataş, M.; Ali, S.A. Machine learning and artificial neural networks–based approach to model and optimize ethyl methanesulfonate and sodium azide induced in vitro regeneration and morphogenic traits of water hyssops (Bacopa monnieri L.). J. Plant Growth Reg. 2022, 42, 3471–3485. [Google Scholar] [CrossRef]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Akar, Ö.; Güngör, O. Classification of multispectral images using Random Forest algorithm. J. Geod. Geoinfor. 2012, 1, 105–112. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 16–17 August 2016; pp. 785–794. [Google Scholar]
- Sundararajan, S.; Sivakumar, H.P.; Nayeem, S.; Rajendran, V.; Subiramani, S.; Ramalingam, S. Influence of exogenous polyamines on somatic embryogenesis and regeneration of fresh and long–term cultures of three elite indica rice cultivars. Cereal Res. Commun. 2021, 49, 245–253. [Google Scholar] [CrossRef]
- Astarita, L.V.; Handro, W.; Floh, E.I.S. Changes in polyamines content associated with zygotic embryogenesis in the Brazilian pine, Araucaria angustifolia (Bert.) O. Ktze. Braz. J. Bot. 2003, 26, 163–168. [Google Scholar] [CrossRef]
- Liu, J.H.; Nada, K.; Honda, C.; Kitashiba, H.; Wen, X.P.; Pang, X.M.; Moriguchi, T. Polyamine biosynthesis of apple callus under salt stress: Importance of the arginine decarboxylase pathway in stress response. J. Exp. Bot. 2006, 57, 2589–2599. [Google Scholar] [CrossRef] [PubMed]
- Martınez, L.E.; Agüero, C.B.; Lopez, M.E.; Galmarini, C.R. Improvement of in vitro gynogenesis induction in onion (Allium cepa L.) using polyamines. Plant Sci. 2000, 156, 221–226. [Google Scholar] [CrossRef]
- Dewi, I.S.; Purwoko, B.S. Role of polyamines in inhibition of ethylene biosynthesis and their effects on rice anther culture development. Ind. J. Agric. Sci. 2008, 9, 60–67. [Google Scholar] [CrossRef]
- Tang, W.; Newton, R.J.; Outhavong, V. Exogenously added polyamines recover browning tissues into normal callus cultures and improve plant regeneration in pine. Physiol. Plant 2004, 122, 386–395. [Google Scholar] [CrossRef]
- Li, J.; Doyle, K.M.; Tatlisumak, T. Polyamines in the brain: Distribution, biological interactions, and their potential therapeutic role in brain ischaemia. Curr. Med. Chem. 2007, 14, 1807–1813. [Google Scholar]
- Purwoko, B.; Dewi, I.; Usman, A. Poliamina meningkatkan regenerasi tanaman hijau pada kultur antera padi cv Taipei 309. Hayati 2001, 8, 117–120. [Google Scholar]
- Paul, A.; Mitter, K.; Raychaudhuri, S.S. Effect of polyamines on in vitro somatic embryogenesis in Momordica charantia L. Plant Cell Tissue Organ Cult. 2009, 97, 303–311. [Google Scholar] [CrossRef]
- Rakesh, B.; Sudheer, W.N.; Nagella, P. Role of polyamines in plant tissue culture: An overview. Plant Cell Tissue Organ Cult. 2021, 145, 487–506. [Google Scholar] [CrossRef]
- Xiong, L.; Liu, C.; Liu, D.; Yan, Z.; Yang, X.; Feng, G. Optimization of an indirect regeneration system for common bean (Phaseolus vulgaris L.). Plant Biotechnol. Rep. 2023, 1–13. [Google Scholar] [CrossRef]
- Liu, Q.; Xu, L.; Li, Y.; Xu, W.; Vetukuri, R.R.; Xu, X. Overexpression of an autophagy–related gene DiATG3 from Davidia involucrata improves plant thermotolerance by enhancing the accumulation of polyamines and regulating genes in calcium and MAPK signaling pathways. Environ. Exp. Bot. 2023, 208, 105235. [Google Scholar] [CrossRef]
- Chen, J.F.; Cui, L.; Malik, A.A.; Mbira, K.G. In vitro haploid and dihaploid production via unfertilized ovule culture. Plant Cell Tissue Organ Cult. 2011, 104, 311–319. [Google Scholar] [CrossRef]
- Grafi, G.; Ben-Meir, H.; Avivi, Y.; Moshe, M.; Dahan, Y.; Zemach, A. Histone methylation controls telomerase–independent telomere lengthening in cells undergoing dedifferentiation. Dev. Biol. 2007, 306, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Crisp, P.A.; Stelpflug, S.; Kaeppler, S.M.; Li, Q.; Springer, N.M. Heritable epigenomic changes to the maize methylome resulting from tissue culture. Genetics 2018, 209, 983–995. [Google Scholar] [CrossRef]
- Kabita, K.C.; Sharma, S.K.; Sanatombi, K. Analysis of capsaicinoid biosynthesis pathway genes expression in callus cultures of Capsicum chinense Jacq. Cv. ‘Umorok’. Plant Cell Tissue Organ Cult. 2019, 137, 565–573. [Google Scholar] [CrossRef]
- Li, H.; Zhao, X.; Dai, H.; Wu, W.; Mao, W.; Zhang, Z. Tissue culture responsive microRNAs in strawberry. Plant Mol. Biol. Rep. 2012, 30, 1047–1054. [Google Scholar] [CrossRef]
- Temel, A.; Kartal, G.; Gozukirmizi, N. Genetic and epigenetic variations in barley calli cultures. Biotechnol. Biotechnol. Equip. 2008, 22, 911–914. [Google Scholar] [CrossRef]
- Erturk, F.A.; Agar, G.; Arslan, E.; Nardemir, G. Analysis of genetic and epigenetic effects of maize seeds in response to heavy metal (Zn) stress. Environ. Sci. Pollut. Res. 2015, 22, 10291–10297. [Google Scholar] [CrossRef]
- Valero, D.; Perez-Vicente, A.; Martinez-Romero, D.; Castillo, S.; Guillen, F.; Serrano, M. Plum storability improved after calcium and heat postharvest treatments: Role of polyamines. J. Food Sci. 2002, 67, 2571–2575. [Google Scholar] [CrossRef]
- Taspinar, M.S.; Aydin, M.; Sigmaz, B.; Yagci, S.; Arslan, E.; Agar, G. Aluminum–induced changes on DNA damage, DNA methylation and LTR retrotransposon polymorphism in maize. Arab. J. Sci. Eng. 2018, 43, 123–131. [Google Scholar] [CrossRef]
- Tanee, T.; Chadmuk, P.; Sudmoon, R.; Chaveerach, A.; Noikotr, K. Genetic analysis for identification, genomic template stability in hybrids and barcodes of the Vanda species (Orchidaceae) of Thailand. Afr. J. Biotech. 2012, 11, 11772–11781. [Google Scholar]
- Steward, N.; Ito, M.; Yamaguchi, Y.; Koizumi, N.; Sano, H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 2002, 277, 37741–37746. [Google Scholar] [CrossRef] [PubMed]
- Chi, W.; Feng, P.; Ma, J.; Zhang, L. Metabolites and chloroplast retrograde signaling. Curr. Opin. Plant Biol. 2015, 25, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Seo, P.J. Dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 2018, 23, 235–247. [Google Scholar] [CrossRef]
- Wibowo, A.; Becker, C.; Durr, J.; Price, J.; Spaepen, S.; Hilton, S.; Putra, H.; Papareddy, R.; Saintain, Q.; Harvey, S. Partial maintenance of organ–specific epigenetic marks during plant asexual reproduction leads to heritable phenotypic variation. Proc. Natl. Acad. Sci. USA 2018, 115, E9145–E9152. [Google Scholar] [CrossRef]
- Aasim, M.; Akin, F.; Ali, S.A.; Taskin, M.B.; Colak, M.S.; Khawar, K.M. Artificial neural network modeling for deciphering the in vitro induced salt stress tolerance in chickpea (Cicer arietinum L). Physiol. Mol. Biol. Plants 2023, 29, 289–304. [Google Scholar] [CrossRef]
- Aasim, M.; Ali, S.A.; Altaf, M.T.; Ali, A.; Nadeem, M.A.; Baloch, F.S. Artificial neural network and decision tree facilitated prediction and validation of cytokinin–auxin induced in vitro organogenesis of sorghum (Sorghum bicolor L.). Plant Cell Tissue Organ Cult. 2023, 153, 611–624. [Google Scholar] [CrossRef]
- Niazian, M.; Noori, S.A.S.; Galuszka, P.; Tohidfar, M.; Mortazavian, S.M.M. Genetic stability of regenerated plants via indirect somatic embryogenesis and indirect shoot regeneration of Carum copticum L. Ind. Crop. Prod. 2017, 97, 330–337. [Google Scholar] [CrossRef]
- Arab, M.M.; Yadollahi, A.; Shojaeiyan, A.; Ahmadi, H. Artificial neural network genetic algorithm as powerful tool to predict and optimize in vitro proliferation mineral medium for G× N15 rootstock. Front. Plant Sci. 2016, 7, 1526. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- VSN International Genstat for Windows, 23rd ed.; VSN International: Hemel Hempstead, UK, 2023.
- Zeinalzadehtabrizi, H.; Hosseinpour, A.; Aydin, M.; Haliloglu, K. A modified genomic DNA extraction method from leaves of sunflower for PCR based analyzes. J. Biol. Environ. Sci. 2015, 7, 222–225. [Google Scholar]
- Haliloğlu, K.; Türkoğlu, A.; Balpınar, Ö.; Nadaroğlu, H.; Alaylı, A.; Poczai, P. Effects of zinc, copper and iron oxide nanoparticles on induced DNA methylation, genomic instability and LTR retrotransposon polymorphism in wheat (Triticum aestivum L.). Plants 2022, 11, 2193. [Google Scholar] [CrossRef] [PubMed]
- Sammut, C.; Webb, G.I. Leave-one-out cross-validation. In Encyclopedia of Machine Learning; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010; pp. 600–601. [Google Scholar]
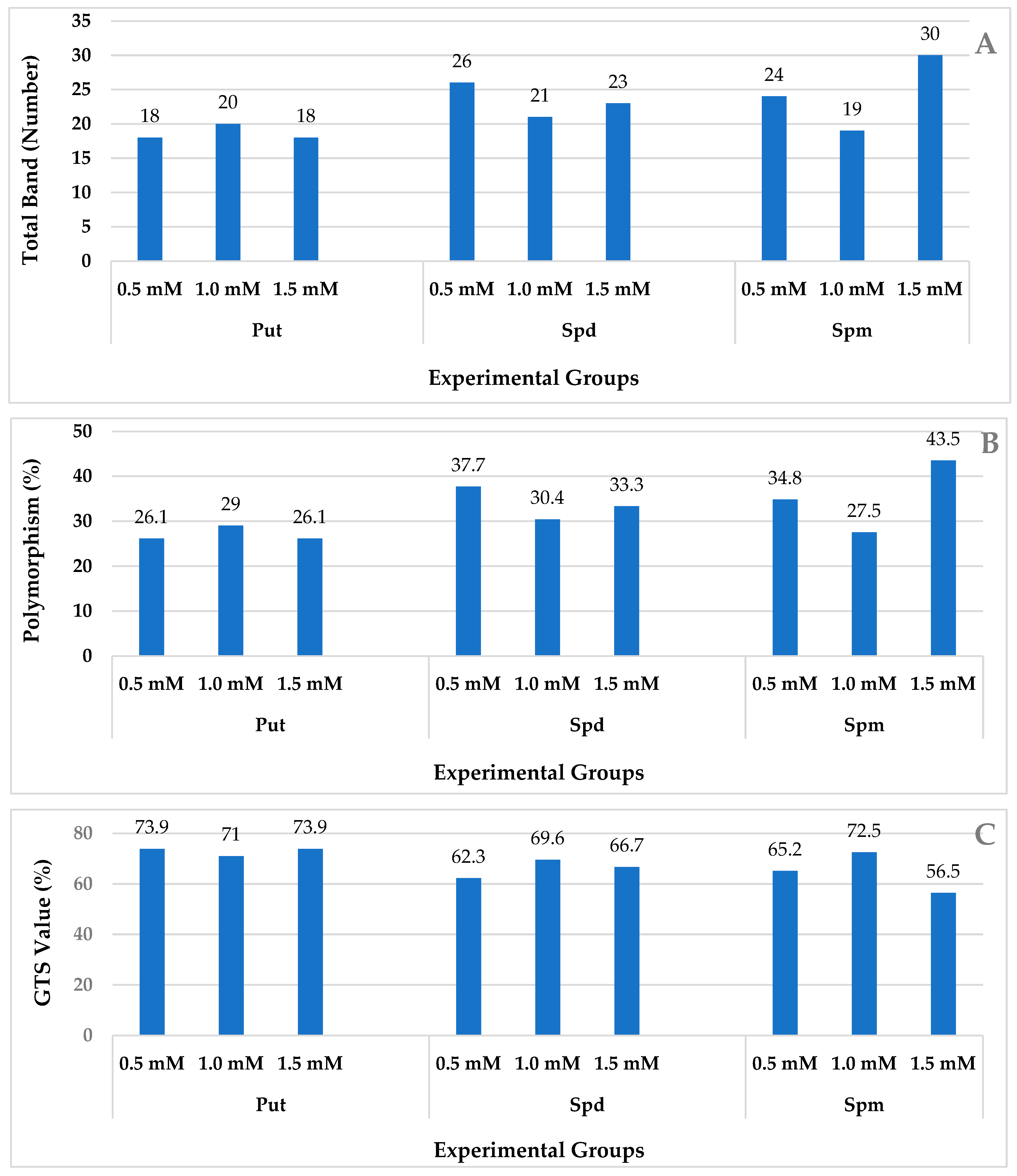
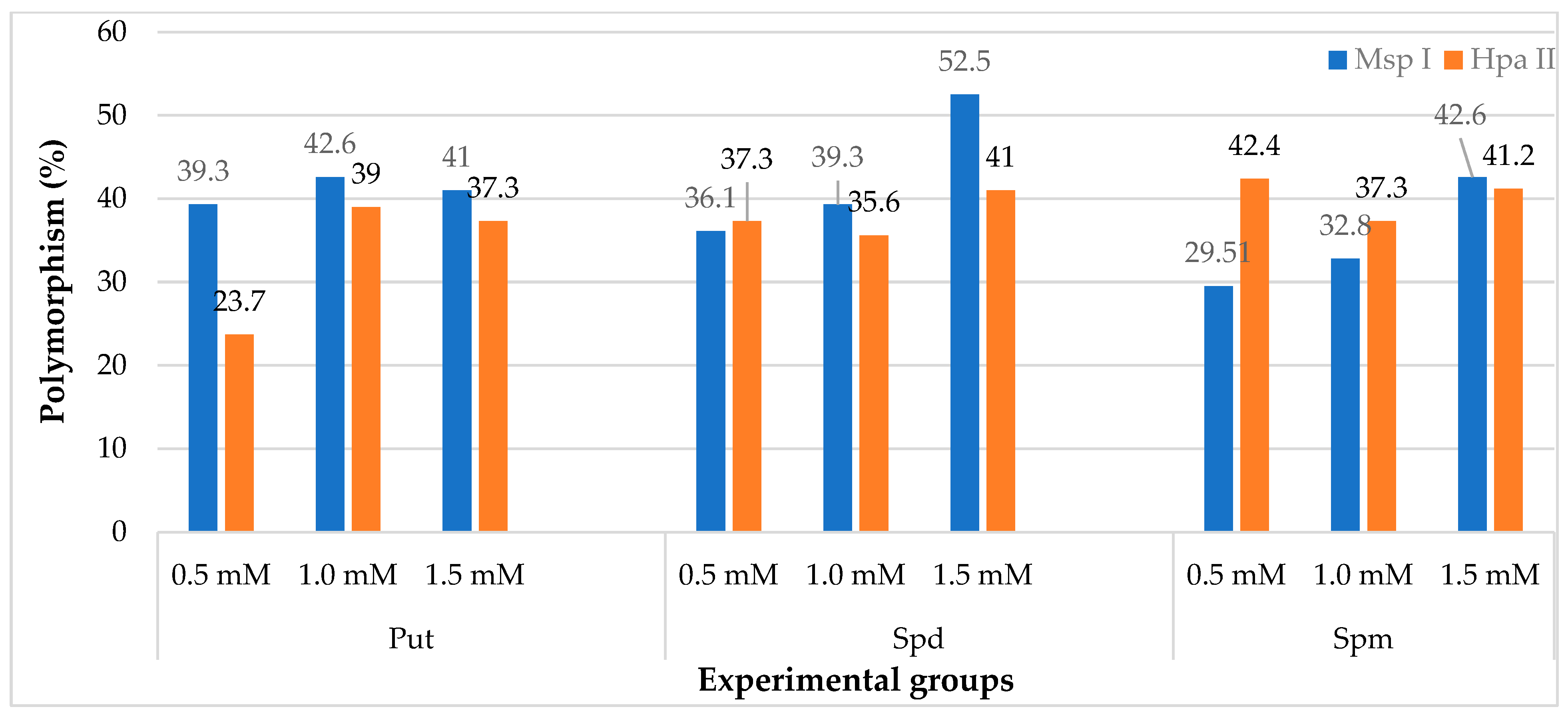
| Polyamine Types | Concentration (mM) | CI% 1 | ECI% 2 | REC% 3 | RE 4 | PN 5 |
|---|---|---|---|---|---|---|
| Putrescine (Put) | 0 | 92.50 abcd 6 | 77.50 bcd | 77.50 ab | 1.15 abc | 7.75 b |
| 0.5 | 95.00 ab | 79.30 bcd | 89.30 a | 1.14 abc | 8.50 b | |
| 1 | 97.50 a | 91.80 ab | 65.00 bc | 1.17 abc | 12.25 a | |
| 1.5 | 90.00 bcde | 85.00 abcd | 50.60 cde | 1.29 ab | 3.00 de | |
| Means | 93.75 A | 83.43 A | 70.62 A | 1.19 A | 7.87 A | |
| Spermidine (Spd) | 0 | 92.50 abcd | 77.50 bcd | 77.50 ab | 1.15 abc | 7.75 b |
| 0.5 | 93.75 abc | 77.50 bcd | 82.50 a | 0.95 c | 8.75 b | |
| 1 | 92.50 abcd | 87.50 abc | 92.50 a | 1.09 bc | 12.00 a | |
| 1.5 | 92.50 abcd | 100.00 a | 57.50 cd | 1.07 bc | 2.50 e | |
| Means | 92.812 A | 85.62 A | 77.50 A | 1.07 AB | 7.75 A | |
| Spermine (Spm) | 0 | 92.50 abcd | 77.50 bcd | 77.50 ab | 1.15 abc | 7.75 b |
| 0.5 | 87.50 cde | 67.50 d | 47.50 de | 1.01 bc | 5.25 c | |
| 1 | 83.75 e | 70.00 cd | 45.00 de | 1.40 a | 4.50 cd | |
| 1.5 | 86.25 de | 67.50 d | 35.00 e | 0.37 d | 2.00 e | |
| Means | 87.50 B | 70.625 B | 51.25 B | 0.98 B | 4.87 B | |
| Mean square of polyamine (P) | 181.77 ** | 1050.52 ** | 2964.58 ** | 0.17 ** | 46.08 ** | |
| The mean square of concentration (C) | 19.96 ns | 241.84 ns | 2076.04 ** | 0.22 ** | 110.5 ** | |
| The mean square of P × C | 38.71 ns | 235.59 ns | 619.79 ** | 0.29 ** | 15.91 ** | |
| iPBS Primer | ± 1 | Control 2 | Experimental Groups | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Putresince (Put) | Spermidine (Spd) | Spermine (Spm) | |||||||||
| 0.5 mM | 1.0 mM | 1.5 mM | 0.5 mM | 1.0 mM | 1.5 mM | 0.5 mM | 1.0 mM | 1.5 mM | |||
| 2075 | + | 6 | - | 590 | 689; 590 | 590 | 590 | - | 571 | - | 461 |
| - | - | - | - | - | - | - | - | - | - | ||
| 2077 | + | 6 | - | 1120; 918; 659; 467; 372 | 379 | 1600; 1100; 900; 475 | 1140; 843; 379 | 1600; 1060; 957; 475; 379 | 491; 379 | - | 1600; 1080 |
| - | 813; 700 | - | - | - | - | - | - | - | 311 | ||
| 2087 | + | 4 | - | 1600; 452; 416 | - | 1100; 480 | - | 425 | - | - | 1340 |
| - | - | - | - | 727; 648 | 831; 500 | - | 831; 727; 500 | 831; 500 | 727; 648; 500 | ||
| 2278 | + | 9 | - | 1200; 950 | 1250 | 1357 | 1357 | 1320; 1200; 950 | 1320 | 1320; 1200 | 1320; 1200 |
| - | 752; 520 | - | 520 | 520 | 752 | - | 520 | - | 752; 400; 310 | ||
| 2375 | + | 10 | - | 1371; 970; 852; 536 | 1371 | 1357 | 1357 | 1375; 1285; 1200; 1157; 985; 575 | 1371 | 1375; 1285; 1157 | 1375; 1285 |
| - | 779; 607 | - | 607 | 607 | 779 | - | 607 | - | 779; 718; 486; 308 | ||
| 2377 | + | 9 | - | - | - | - | - | - | - | - | 1214 |
| - | 1057; 970; 710; 582; 431 | 431 | 970; 582; 487; 431 | 582; 487; 431 | 900; 710; 637; 582; 520; 487; 431 | 487; 431 | 487; 431 | 710; 487; 431 | 487; 431 | ||
| 2380 | + | 5 | - | - | 548 | 1060; 837 | - | 1060; 920 | 1040; 858 | 858; 560 | - |
| - | - | - | - | - | - | - | - | - | - | ||
| 2381 | + | 9 | - | - | - | - | - | - | - | - | - |
| - | 1300; 1200; 833; 420; 376 | 472; 376 | 1300; 1200; 972; 833 | 1300; 1200; 972; 472; 420; 324 | 972; 376 | 472; 376 | 1300; 1200; 972; 833; 651; 472; 420; 376 | 1200; 833; 472; 376 | 1300; 1200; 833; 472; 376 | ||
| 2382 | + | 6 | 936 | 917 | - | 1000 | - | 1040; 413 | - | - | - |
| - | - | - | - | 521 | 521 | - | 589; 500 | 521 | 870; 589; 521 | ||
| 2384 | + | 5 | - | - | 525; 239 | - | - | - | - | - | - |
| - | 282 | 800 | - | - | 282 | - | - | 392; 282 | - | ||
| iPBS Primer | M/H 1 | ± 2 | Control 3 | Experimental Groups | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Putresince (Put) | Spermidine (Spd) | Spermine (Spm) | |||||||||||
| 0.5 mM | 1.0 mM | 1.5 mM | 0.5 mM | 1.0 mM | 1.5 mM | 0.5 mM | 1.0 mM | 1.5 mM | |||||
| 2075 | M | + | 7 | - | - | - | - | - | - | 315 | - | - | |
| - | 393 | - | - | - | - | 393 | - | 393; 271 | 271 | ||||
| H | + | 7 | - | - | 376 | - | - | - | 626 | - | - | ||
| - | 400 | - | - | - | 400 | 400 | - | - | 266; 180 | ||||
| 2077 | M | + | 6 | 465 | 1416; 1216; 1033; 918; 465 | 1416; 1216 | 1433; 1050; 918 | 1416; 1016; 918 | 1416; 1016; 900; 613; 491; 377 | 1416; 1016; 880 | 1413; 1016 | 1433; 1033 | |
| - | - | - | - | 575; 415; 356; 312 | 415; 312 | 415; 312 | 415; 312 | 312 | |||||
| H | + | 6 | 818 | 1450; 1033; 936 | 1433; 1033; 918 | 1416; 1183; 1050 | 1433; 1050 | 1433; 1033; 984; 900; 800; 739 | 1433; 1300; 1033; | 1416; 1000; 918 | 1433; 1266; 1016 | ||
| - | - | - | 600 | 356; 318 | 318 | - | - | 318 | 415; 318 | ||||
| 2087 | M | + | 7 | 522; 400; 350 | - | - | - | - | - | - | - | - | |
| - | 1300; 1075 | 1300; 1075; 815 | 1075; 922; 815; 700; 600; 489 | 1075; 922; 815; 700; 489 | 1300; 1075; 922; 700; 600; | 1075; 922; 815; 700; 600; | 1300; 1075; 922; 700; 600 | 1075; 922; 815; 600; | 1300; 1075; 922; 815; 700; 600 | ||||
| H | + | 8 | 1100; 427 | 1250 | - | - | - | - | - | - | - | ||
| - | - | 700; 567; 418 | 1125; 629; 418 | 922; 815; 700; 567; 500 | 1125; 922; 815; 629; 567; 418 | 815; 567; 500; 418 | 1125; 922; 815;700; 629; 418 | 1125; 922; 815; 629; 567; 418 | 1125; 922; 815; 629; 567; 418 | ||||
| 2278 | M | + | 8 | 500 | - | - | - | - | - | - | - | - | |
| - | 1100; 900 | 1100; 900 | 900; 750; 650; 450 | 900; 815; 750; 450 | 1100; 900; 750; 450 | 900; 815; 750; 450 | 1100; 900; 750 | 900; 815; | 1100; 900; 815; 750 | ||||
| H | + | 5 | 1100; 427 | 1250 | - | - | - | - | - | - | - | ||
| - | - | 760; 465 | 1125; 465 | 1125; 760 | 760; 465 | 1125; 760; 465 | - | 1125;465 | 1125; 760 | ||||
| 2375 | M | + | 5 | 1040 | 1100; 851; 811 | 1100; 866 | - | 837 | 851 | - | 938; 811 | 824; 326 | |
| - | - | - | - | 405 | - | - | - | - | - | ||||
| H | + | 4 | - | 1280; 1060; 589 | 1040; 589; 468 | 1020; 589 | 1020; 454 | 1100; 1020; 589 | 1080; 1020; 651; 589 | 1020 | 1020; 680; 365 | ||
| - | 866;811; 709 | - | - | - | 709 | - | - | - | - | ||||
| 2377 | M | + | 6 | 640; 583; 492; 338; 264 | 964; 739; 682; 632 | 913 | - | 926 | 1075; 939; 762; 648; 616; 492 | 1100 | 1087 | 1112 | |
| - | - | - | - | - | - | - | - | 548 | 400 | ||||
| H | + | 6 | - | 1260; 1087; 762 | 1262 | 1262 | - | 1275; 1087 | - | 1287 | 1275 | ||
| - | - | - | 431; 394 | 431; 394 | 431 | - | - | 431; 394 | 729; 500 | ||||
| 2380 | M | + | 5 | - | 607 | 732; 607; 568; 450 | 875; 607; 527 | 607 | 944; 880; 613; 580 | - | 961 | 912 | |
| - | - | - | - | - | - | - | - | 400 | - | ||||
| H | + | 5 | - | 673 | 600; 500 | 607; 500 | - | 981 | 944; 527 | 1000; 555 | 591 | ||
| - | - | 846 | - | - | 400 | - | - | - | - | ||||
| 2381 | M | + | 5 | - | 1325; 1025; 750; 334 | - | - | - | - | - | 325; 269 | ||
| - | - | - | 833; 466 | 537 | 921; 633; 537 | 466 | 921; 633; 466; 409 | 537; 466; 409 | 921; 633 | ||||
| H | + | 4 | - | 1325; 1100; 679 | 900; 679 | - | - | - | - | - | - | ||
| - | - | - | - | 611 | 427 | - | 819; 427; 353 | 819; 353 | 819; 427; 353 | ||||
| 2382 | M | + | 4 | 753; 529 | 1016; 848 | 586; 369 | 408 | - | 545 | 529 | - | - | |
| - | - | - | - | - | 492 | - | - | - | - | ||||
| H | + | 8 | - | - | - | - | - | - | - | - | - | ||
| - | 940 | 940; | 753; 645 | 645 | 940; 537 | 940; 753 | 940; 753; 645; 537 | - | 940; 537 | ||||
| 2384 | M | + | 8 | - | - | - | - | - | - | - | - | - | |
| - | 958; 830; 540; 461; 391; 346 | 958; 346 | 958; 830 | - | 958; 461; 346 | 830; 461; 346 | - | - | 958; 346; 281 | ||||
| H | + | 6 | - | - | - | - | 461 | - | 700; 480 | - | 1020; 452 | ||
| - | 958; 830; 551; 408 | 958 | - | 958 | - | 958; 830 | - | 958; 408 | - | ||||
| Traits 1 | ML Criteria 2 | SVM | RF | XGBoost | |||
|---|---|---|---|---|---|---|---|
| Test | Train | Test | Train | Test | Train | ||
| CI (%) | R2 | 0.254 | 0.407 | 0.379 | 0.486 | 0.383 | 0.529 |
| MSE | 4.364 | 4.121 | 3.981 | 3.838 | 3.889 | 3.673 | |
| MAPE | 3.816 | 3.401 | 3.662 | 3.204 | 3.289 | 3.057 | |
| MAD | 3.540 | 3.958 | 3.386 | 3.136 | 3.243 | 2.841 | |
| ECI (%) | R2 | 0.351 | 0.312 | 0.415 | 0.48 | 0.231 | 0.501 |
| MSE | 9.744 | 11.160 | 9.245 | 9.005 | 9.608 | 9.800 | |
| MAPE | 10.535 | 11.202 | 9.435 | 10.217 | 9.473 | 10.243 | |
| MAD | 7.624 | 7.528 | 7.063 | 6.087 | 7.064 | 6.977 | |
| REC (%) | R2 | 0.841 | 0.776 | 0.781 | 0.792 | 0.699 | 0.812 |
| MSE | 9.699 | 9.336 | 10.367 | 9.386 | 10.459 | 9.918 | |
| MAPE | 14.405 | 12.603 | 15.691 | 12.653 | 16.585 | 12.690 | |
| MAD | 7.197 | 6.950 | 7.783 | 7.184 | 8.282 | 7.490 | |
| RE | R2 | 0.482 | 0.356 | 0.646 | 0.548 | 0.738 | 0.613 |
| MSE | 0.271 | 0.198 | 0.224 | 0.166 | 0.193 | 0.154 | |
| MAPE | 42.415 | 15.433 | 34.323 | 14.739 | 23.748 | 11.776 | |
| MAD | 0.219 | 0.125 | 0.185 | 0.120 | 0.165 | 0.110 | |
| PN | R2 | 0.782 | 0.833 | 0.775 | 0.880 | 0.853 | 0.905 |
| MSE | 1.761 | 1.363 | 1.791 | 1.155 | 1.445 | 1.029 | |
| MAPE | 19.548 | 24.429 | 17.156 | 22.011 | 11.653 | 18.343 | |
| MAD | 1.137 | 0.899 | 1.192 | 0.815 | 0.926 | 0.770 | |
| Primer Name | Sequence (5′–3′) | Tm (°C) 1 | CG (%) 2 |
|---|---|---|---|
| iPBS-2075 | CTCATGATGCCA | 42.1 | 50.0 |
| iPBS-2077 | CTCACGATGCCA | 46.1 | 58.3 |
| iPBS-2087 | GCAATGGAACCA | 43.5 | 50.0 |
| iPBS-2278 | GCTCATGATACCA | 42.3 | 46.2 |
| iPBS-2375 | TCGCATCAACCA | 45.1 | 50.0 |
| iPBS-2377 | ACGAAGGGACCA | 47.2 | 58.3 |
| iPBS-2380 | CAACCTGATCCA | 41.4 | 50.0 |
| iPBS-2381 | GTCCATCTTCCA | 49.9 | 50.0 |
| iPBS-2382 | TGTTGGCTTCCA | 44.9 | 50.0 |
| iPBS-2384 | GTAATGGGTCCA | 40.9 | 50.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eren, B.; Türkoğlu, A.; Haliloğlu, K.; Demirel, F.; Nowosad, K.; Özkan, G.; Niedbała, G.; Pour-Aboughadareh, A.; Bujak, H.; Bocianowski, J. Investigation of the Influence of Polyamines on Mature Embryo Culture and DNA Methylation of Wheat (Triticum aestivum L.) Using the Machine Learning Algorithm Method. Plants 2023, 12, 3261. https://doi.org/10.3390/plants12183261
Eren B, Türkoğlu A, Haliloğlu K, Demirel F, Nowosad K, Özkan G, Niedbała G, Pour-Aboughadareh A, Bujak H, Bocianowski J. Investigation of the Influence of Polyamines on Mature Embryo Culture and DNA Methylation of Wheat (Triticum aestivum L.) Using the Machine Learning Algorithm Method. Plants. 2023; 12(18):3261. https://doi.org/10.3390/plants12183261
Chicago/Turabian StyleEren, Barış, Aras Türkoğlu, Kamil Haliloğlu, Fatih Demirel, Kamila Nowosad, Güller Özkan, Gniewko Niedbała, Alireza Pour-Aboughadareh, Henryk Bujak, and Jan Bocianowski. 2023. "Investigation of the Influence of Polyamines on Mature Embryo Culture and DNA Methylation of Wheat (Triticum aestivum L.) Using the Machine Learning Algorithm Method" Plants 12, no. 18: 3261. https://doi.org/10.3390/plants12183261
APA StyleEren, B., Türkoğlu, A., Haliloğlu, K., Demirel, F., Nowosad, K., Özkan, G., Niedbała, G., Pour-Aboughadareh, A., Bujak, H., & Bocianowski, J. (2023). Investigation of the Influence of Polyamines on Mature Embryo Culture and DNA Methylation of Wheat (Triticum aestivum L.) Using the Machine Learning Algorithm Method. Plants, 12(18), 3261. https://doi.org/10.3390/plants12183261












