Secondary Metabolites and Their Role in Strawberry Defense
Abstract
1. Introduction
2. Terpenoids
Terpene Synthases
3. Pathogenesis-Related Proteins
Strawberry PR-10 Allergens
4. Flavonoids
5. Strawberry Microbiome Composition and Its Role in Plant Growth and Defense
Influence in Plant Growth and Resistance to Biotic and Abiotic Stress
6. Potential Targets for Crop Improvement
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The Family of Terpene Synthases in Plants: A Mid-Size Family of Genes for Specialized Metabolism That Is Highly Diversified throughout the Kingdom. Plant J. Cell Mol. Biol. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.L.; Wai, C.M.; et al. Origin and Evolution of the Octoploid Strawberry Genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Uroz, S.; Courty, P.E.; Oger, P. Plant Symbionts Are Engineers of the Plant-Associated Microbiome. Trends Plant Sci. 2019, 24, 905–916. [Google Scholar] [CrossRef]
- Kessler, A.; Kalske, A. Plant Secondary Metabolite Diversity and Species Interactions. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 115–138. [Google Scholar] [CrossRef]
- Pang, Z.; Chen, J.; Wang, T.; Gao, C.; Li, Z.; Guo, L.; Xu, J.; Cheng, Y. Linking Plant Secondary Metabolites and Plant Microbiomes: A Review. Front. Plant Sci. 2021, 12, 621276. [Google Scholar] [CrossRef]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Guerrieri, A.; Dong, L.; Bouwmeester, H.J. Role and Exploitation of Underground Chemical Signaling in Plants. Pest Manag. Sci. 2019, 75, 2455–2463. [Google Scholar] [CrossRef]
- Mastan, A.; Bharadwaj, R.; Kushwaha, R.K.; Vivek Babu, C.S. Functional Fungal Endophytes in Coleus forskohlii Regulate Labdane Diterpene Biosynthesis for Elevated Forskolin Accumulation in Roots. Microb. Ecol. 2019, 78, 914–926. [Google Scholar] [CrossRef]
- Shulaev, V.; Sargent, D.J.; Crowhurst, R.N.; Mockler, T.C.; Folkerts, O.; Delcher, A.L.; Jaiswal, P.; Mockaitis, K.; Liston, A.; Mane, S.P.; et al. The Genome of Woodland Strawberry (Fragaria vesca). Nat. Genet. 2011, 43, 109–116. [Google Scholar] [CrossRef]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.I.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-Molecule Sequencing and Optical Mapping Yields an Improved Genome of Woodland Strawberry (Fragaria vesca) with Chromosome-Scale Contiguity. GigaScience 2018, 7, gix124. [Google Scholar] [CrossRef]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated Annotation of the Wild Strawberry Fragaria vesca V4 Genome. Hortic. Res. 2019, 6, 61. [Google Scholar] [CrossRef]
- Alger, E.I.; Platts, A.E.; Deb, S.K.; Luo, X.; Ou, S.; Cao, Y.; Hummer, K.E.; Xiong, Z.; Knapp, S.J.; Liu, Z.; et al. Chromosome-Scale Genome for a Red-Fruited, Perpetual Flowering and Runnerless Woodland Strawberry (Fragaria vesca). Front. Genet. 2021, 12, 671371. [Google Scholar] [CrossRef]
- Osorio, S.; Bombarely, A.; Giavalisco, P.; Usadel, B.; Stephens, C.; Aragüez, I.; Medina-Escobar, N.; Botella, M.A.; Fernie, A.R.; Valpuesta, V. Demethylation of Oligogalacturonides by FaPE1 in the Fruits of the Wild Strawberry Fragaria vesca Triggers Metabolic and Transcriptional Changes Associated with Defence and Development of the Fruit. J. Exp. Bot. 2011, 62, 2855–2873. [Google Scholar] [CrossRef]
- Toljamo, A.; Blande, D.; Kärenlampi, S.; Kokko, H. Reprogramming of Strawberry (Fragaria vesca) Root Transcriptome in Response to Phytophthora cactorum. PLoS ONE 2016, 11, e0161078. [Google Scholar] [CrossRef]
- Badmi, R.; Tengs, T.; Brurberg, M.B.; Elameen, A.; Zhang, Y.; Haugland, L.K.; Fossdal, C.G.; Hytönen, T.; Krokene, P.; Thorstensen, T. Transcriptional Profiling of Defense Responses to Botrytis cinerea Infection in Leaves of Fragaria vesca Plants Soil-Drenched with β-Aminobutyric Acid. Front. Plant Sci. 2022, 13, 1025422. [Google Scholar] [CrossRef]
- Gogoi, A.; Lysøe, E.; Eikemo, H.; Stensvand, A.; Davik, J.; Brurberg, M.B. Comparative Transcriptome Analysis Reveals Novel Candidate Resistance Genes Involved in Defence against Phytophthora cactorum in Strawberry. Int. J. Mol. Sci. 2023, 24, 10851. [Google Scholar] [CrossRef]
- Ormeño, E.; Viros, J.; Mévy, J.-P.; Tonetto, A.; Saunier, A.; Bousquet-Mélou, A.; Fernandez, C. Exogenous Isoprene Confers Physiological Benefits in a Negligible Isoprene Emitter (Acer monspessulanum L.) under Water Deficit. Plants 2020, 9, 159. [Google Scholar] [CrossRef]
- Toljamo, A.; Koistinen, V.; Hanhineva, K.; Kärenlampi, S.; Kokko, H. Terpenoid and Lipid Profiles Vary in Different Phytophthora cactorum–Strawberry Interactions. Phytochemistry 2021, 189, 112820. [Google Scholar] [CrossRef]
- Alvarez-Castellanos, P.P.; Bishop, C.D.; Pascual-Villalobos, M.J. Antifungal Activity of the Essential Oil of Flowerheads of Garland Chrysanthemum (Chrysanthemum coronarium) against Agricultural Pathogens. Phytochemistry 2001, 57, 99–102. [Google Scholar] [CrossRef]
- Himanen, S.; Vuorinen, T.; Tuovinen, T.; Holopainen, J.K. Effects of Cyclamen Mite (Phytonemus pallidus) and Leaf Beetle (Galerucella tenella) Damage on Volatile Emission from Strawberry (Fragaria × ananassa Duch.) Plants and Orientation of Predatory Mites (Neoseiulus cucumeris, N. californicus, and Euseius finlandicus). J. Agric. Food Chem. 2005, 53, 8624–8630. [Google Scholar] [CrossRef]
- Huang, A.C.; Jiang, T.; Liu, Y.-X.; Bai, Y.-C.; Reed, J.; Qu, B.; Goossens, A.; Nützmann, H.-W.; Bai, Y.; Osbourn, A. A Specialized Metabolic Network Selectively Modulates Arabidopsis Root Microbiota. Science 2019, 364, eaau6389. [Google Scholar] [CrossRef]
- Badmi, R.; Zhang, Y.; Tengs, T.; Brurberg, M.B.; Krokene, P.; Fossdal, C.G.; Hytönen, T.; Thorstensen, T. Induced and Primed Defence Responses of Fragaria vesca to Botrytis cinerea Infection. bioRxiv 2019, 692491. [Google Scholar] [CrossRef]
- Zhang, Z.; Lu, S.; Yu, W.; Ehsan, S.; Zhang, Y.; Jia, H.; Fang, J. Jasmonate Increases Terpene Synthase Expression, Leading to Strawberry Resistance to Botrytis cinerea Infection. Plant Cell Rep. 2022, 41, 1243–1260. [Google Scholar] [CrossRef]
- Duan, W.; Peng, L.; Jiang, J.; Zhang, H.; Tong, G. Combined Transcriptome and Metabolome Analysis of Strawberry Fruits in Response to Powdery Mildew Infection. Agron. J. 2022, 114, 1027–1039. [Google Scholar] [CrossRef]
- Thimmappa, R.; Geisler, K.; Louveau, T.; O’Maille, P.; Osbourn, A. Triterpene Biosynthesis in Plants. Annu. Rev. Plant Biol. 2014, 65, 225–257. [Google Scholar] [CrossRef]
- Mehmood, N.; Yuan, Y.; Ali, M.; Ali, M.; Iftikhar, J.; Cheng, C.; Lyu, M.; Wu, B. Early Transcriptional Response of Terpenoid Metabolism to Colletotrichum gloeosporioides in a Resistant Wild Strawberry Fragaria nilgerrensis. Phytochemistry 2021, 181, 112590. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Z.; Wang, K.; Liu, Y.; Hong, Y.; Chen, C.; Guan, X.; Chen, Q. Co-Expression Network Analysis Uncovers Key Candidate Genes Related to the Regulation of Volatile Esters Accumulation in Woodland Strawberry. Planta 2020, 252, 55. [Google Scholar] [CrossRef]
- Falara, V.; Akhtar, T.A.; Nguyen, T.T.H.; Spyropoulou, E.A.; Bleeker, P.M.; Schauvinhold, I.; Matsuba, Y.; Bonini, M.E.; Schilmiller, A.L.; Last, R.L.; et al. The Tomato Terpene Synthase Gene Family. Plant Physiol. 2011, 157, 770–789. [Google Scholar] [CrossRef]
- Külheim, C.; Padovan, A.; Hefer, C.; Krause, S.T.; Köllner, T.G.; Myburg, A.A.; Degenhardt, J.; Foley, W.J. The Eucalyptus Terpene Synthase Gene Family. BMC Genom. 2015, 16, 450. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-MPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Bergman, M.E.; Davis, B.; Phillips, M.A. Medically Useful Plant Terpenoids: Biosynthesis, Occurrence, and Mechanism of Action. Molecules 2019, 24, 3961. [Google Scholar] [CrossRef]
- Oldfield, E.; Lin, F.-Y. Terpene Biosynthesis: Modularity Rules. Angew. Chem. Int. Ed. Engl. 2012, 51, 1124–1137. [Google Scholar] [CrossRef]
- Amil-Ruiz, F.; Blanco-Portales, R.; Muñoz-Blanco, J.; Caballero, J.L. The Strawberry Plant Defense Mechanism: A Molecular Review. Plant Cell Physiol. 2011, 52, 1873–1903. [Google Scholar] [CrossRef] [PubMed]
- Landi, L.; De Miccolis Angelini, R.M.; Pollastro, S.; Feliziani, E.; Faretra, F.; Romanazzi, G. Global Transcriptome Analysis and Identification of Differentially Expressed Genes in Strawberry after Preharvest Application of Benzothiadiazole and Chitosan. Front. Plant Sci. 2017, 8, 235. [Google Scholar] [CrossRef] [PubMed]
- Janotík, A.; Dadáková, K.; Lochman, J.; Zapletalová, M. L-Aspartate and L-Glutamine Inhibit Beta-Aminobutyric Acid-Induced Resistance in Tomatoes. Plants 2022, 11, 2908. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of Inducible Defense-Related Proteins in Infected Plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Besbes, F.; Franz-Oberdorf, K.; Schwab, W. Phosphorylation-Dependent Ribonuclease Activity of Fra a 1 Proteins. J. Plant Physiol. 2019, 233, 1–11. [Google Scholar] [CrossRef]
- Hoffmann-Sommergruber, K. Plant Allergens and Pathogenesis-Related Proteins. Int. Arch. Allergy Immunol. 2000, 122, 155–166. [Google Scholar] [CrossRef]
- Ishibashi, M.; Nabe, T.; Nitta, Y.; Tsuruta, H.; Iduhara, M.; Uno, Y. Analysis of Major Paralogs Encoding the Fra a 1 Allergen Based on Their Organ-Specificity in Fragaria × ananassa. Plant Cell Rep. 2018, 37, 411–424. [Google Scholar] [CrossRef]
- Karlsson, A.L.; Alm, R.; Ekstrand, B.; Fjelkner-Modig, S.; Schiott, A.; Bengtsson, U.; Bjork, L.; Hjerno, K.; Roepstorff, P.; Emanuelsson, C.S. Bet v 1 Homologues in Strawberry Identified as IgE-Binding Proteins and Presumptive Allergens. Allergy 2004, 59, 1277–1284. [Google Scholar] [CrossRef]
- Muñoz, C.; Hoffmann, T.; Escobar, N.M.; Ludemann, F.; Botella, M.A.; Valpuesta, V.; Schwab, W. The Strawberry Fruit Fra a Allergen Functions in Flavonoid Biosynthesis. Mol. Plant 2010, 3, 113–124. [Google Scholar] [CrossRef]
- Hyun, T.K.; Kim, J.-S. Genomic Identification of Putative Allergen Genes in Woodland Strawberry (Fragaria vesca) and Mandarin Orange (Citrus clementina). Plant Omics J. 2011, 4, 428–434. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Treutter, D. Significance of Flavonoids in Plant Resistance: A Review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Tariq, H.; Asif, S.; Andleeb, A.; Hano, C.; Abbasi, B.H. Flavonoid Production: Current Trends in Plant Metabolic Engineering and De Novo Microbial Production. Metabolites 2023, 13, 124. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Smith, D.L. Flavonoids in Agriculture: Chemistry and Roles in, Biotic and Abiotic Stress Responses, and Microbial Associations. Agronomy 2020, 10, 1209. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.C.G.A.; Silva, A.M.S. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef]
- Wang, S.Y.; Lewers, K.S. Antioxidant Capacity and Flavonoid Content in Wild Strawberries. J. Am. Soc. Hortic. Sci. 2007, 132, 629–637. [Google Scholar] [CrossRef]
- Warner, R.; Wu, B.-S.; MacPherson, S.; Lefsrud, M. A Review of Strawberry Photobiology and Fruit Flavonoids in Controlled Environments. Front. Plant Sci. 2021, 12, 611893. [Google Scholar] [CrossRef]
- Gu, N.; Zhang, X.; Gu, X.; Zhao, L.; Godana, E.A.; Xu, M.; Zhang, H. Transcriptomic and Proteomic Analysis of the Mechanisms Involved in Enhanced Disease Resistance of Strawberries Induced by Rhodotorula mucilaginosa Cultured with Chitosan. Postharvest Biol. Technol. 2021, 172, 111355. [Google Scholar] [CrossRef]
- Häkkinen, S.H.; Kärenlampi, S.O.; Heinonen, I.M.; Mykkänen, H.M.; Törrönen, A.R. Content of the Flavonols Quercetin, Myricetin, and Kaempferol in 25 Edible Berries. J. Agric. Food Chem. 1999, 47, 2274–2279. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Reis, F.; Richards, C.L.; Bossdorf, O. Understanding Plant Microbiomes Requires a Genotype × Environment Framework. Am. J. Bot. 2021, 108, 1820–1823. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yue, H.; Ma, Z.; Feng, Z.; Feng, H.; Zhao, L.; Zhang, Y.; Deakin, G.; Xu, X.; Zhu, H.; et al. Influence of Plant Genotype and Soil on the Cotton Rhizosphere Microbiome. Front. Microbiol. 2022, 13, 1021064. [Google Scholar] [CrossRef] [PubMed]
- Olimi, E.; Kusstatscher, P.; Wicaksono, W.A.; Abdelfattah, A.; Cernava, T.; Berg, G. Insights into the Microbiome Assembly during Different Growth Stages and Storage of Strawberry Plants. Environ. Microbiome 2022, 17, 21. [Google Scholar] [CrossRef]
- Sangiorgio, D.; Cellini, A.; Donati, I.; Ferrari, E.; Tanunchai, B.; Fareed Mohamed Wahdan, S.; Sadubsarn, D.; Farneti, B.; Checcucci, A.; Buscot, F.; et al. Taxonomical and Functional Composition of Strawberry Microbiome Is Genotype-Dependent. J. Adv. Res. 2022, 42, 189–204. [Google Scholar] [CrossRef]
- Hassani, M.-A.; Gonzalez, O.; Hunter, S.S.; Holmes, G.; Hewavitharana, S.; Ivors, K.; Lazcano, C. Microbiome Network Connectivity and Composition Linked to Disease Resistance in Strawberry Plants. Phytobiomes J. 2023, PBIOMES-10-22-0069-R. [Google Scholar] [CrossRef]
- Gu, S.; Wei, Z.; Shao, Z.; Friman, V.-P.; Cao, K.; Yang, T.; Kramer, J.; Wang, X.; Li, M.; Mei, X.; et al. Competition for Iron Drives Phytopathogen Control by Natural Rhizosphere Microbiomes. Nat. Microbiol. 2020, 5, 1002–1010. [Google Scholar] [CrossRef]
- Prasad, P.; Kalam, S.; Sharma, N.K.; Podile, A.R.; Das, S.N. Phosphate Solubilization and Plant Growth Promotion by Two Pantoea Strains Isolated from the Flowers of Hedychium coronarium L. Front. Agron. 2022, 4, 990869. [Google Scholar] [CrossRef]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant Growth-Promoting Bacteria That Confer Resistance to Water Stress in Tomatoes and Peppers. Plant Sci. 2004, 166, 525–530. [Google Scholar] [CrossRef]
- Van Oosten, V.R.; Bodenhausen, N.; Reymond, P.; Van Pelt, J.A.; Van Loon, L.C.; Dicke, M.; Pieterse, C.M.J. Differential Effectiveness of Microbially Induced Resistance Against Herbivorous Insects in Arabidopsis. Mol. Plant-Microbe Interact. 2008, 21, 919–930. [Google Scholar] [CrossRef]
- Van der Ent, S.; Van Wees, S.C.M.; Pieterse, C.M.J. Jasmonate Signaling in Plant Interactions with Resistance-Inducing Beneficial Microbes. Phytochemistry 2009, 70, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kloepper, J.W.; Ryu, C.-M. Rhizosphere Bacteria Help Plants Tolerate Abiotic Stress. Trends Plant Sci. 2009, 14, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Lazcano, C.; Boyd, E.; Holmes, G.; Hewavitharana, S.; Pasulka, A.; Ivors, K. The Rhizosphere Microbiome Plays a Role in the Resistance to Soil-Borne Pathogens and Nutrient Uptake of Strawberry Cultivars under Field Conditions. Sci. Rep. 2021, 11, 3188. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Roskot, N.; Steidle, A.; Eberl, L.; Zock, A.; Smalla, K. Plant-Dependent Genotypic and Phenotypic Diversity of Antagonistic Rhizobacteria Isolated from Different Verticillium Host Plants. Appl. Environ. Microbiol. 2002, 68, 3328–3338. [Google Scholar] [CrossRef]
- De Tender, C.; Vandecasteele, B.; Verstraeten, B.; Ommeslag, S.; Kyndt, T.; Debode, J. Biochar-Enhanced Resistance to Botrytis cinerea in Strawberry Fruits (But Not Leaves) Is Associated With Changes in the Rhizosphere Microbiome. Front. Plant Sci. 2021, 12, 700479. [Google Scholar] [CrossRef]
- Rodrigo, S.; Santamaria, O.; Halecker, S.; Lledó, S.; Stadler, M. Antagonism between Byssochlamys spectabilis (Anamorph Paecilomyces variotii) and Plant Pathogens: Involvement of the Bioactive Compounds Produced by the Endophyte: Antagonism between B. Spectabilis and Plant Pathogens. Ann. Appl. Biol. 2017, 171, 464–476. [Google Scholar] [CrossRef]
- Madhaiyan, M.; Poonguzhali, S.; Lee, J.-S.; Senthilkumar, M.; Lee, K.C.; Sundaram, S. Mucilaginibacter gossypii Sp. Nov. and Mucilaginibacter gossypiicola Sp. Nov., Plant-Growth-Promoting Bacteria Isolated from Cotton Rhizosphere Soils. Int. J. Syst. Evol. Microbiol. 2010, 60, 2451–2457. [Google Scholar] [CrossRef]
- Lombardi, N.; Salzano, A.M.; Troise, A.D.; Scaloni, A.; Vitaglione, P.; Vinale, F.; Marra, R.; Caira, S.; Lorito, M.; d’Errico, G.; et al. Effect of Trichoderma Bioactive Metabolite Treatments on the Production, Quality, and Protein Profile of Strawberry Fruits. J. Agric. Food Chem. 2020, 68, 7246–7258. [Google Scholar] [CrossRef]
- Hammerbacher, A.; Coutinho, T.A.; Gershenzon, J. Roles of Plant Volatiles in Defence against Microbial Pathogens and Microbial Exploitation of Volatiles. Plant Cell Environ. 2019, 42, 2827–2843. [Google Scholar] [CrossRef]
- Sharma, E.; Anand, G.; Kapoor, R. Terpenoids in Plant and Arbuscular Mycorrhiza-Reinforced Defence against Herbivorous Insects. Ann. Bot. 2017, mcw263. [Google Scholar] [CrossRef]
- Lin, J.; Wang, D.; Chen, X.; Köllner, T.G.; Mazarei, M.; Guo, H.; Pantalone, V.R.; Arelli, P.; Stewart, C.N.; Wang, N.; et al. An (E,E)-α-Farnesene Synthase Gene of Soybean Has a Role in Defence against Nematodes and Is Involved in Synthesizing Insect-Induced Volatiles. Plant Biotechnol. J. 2017, 15, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, H.; Yuan, J.S.; Köllner, T.G.; Chen, Y.; Guo, Y.; Zhuang, X.; Chen, X.; Zhang, Y.; Fu, J.; et al. The Rice Terpene Synthase Gene OsTPS19 Functions as an (S)-Limonene Synthase in Planta, and Its Overexpression Leads to Enhanced Resistance to the Blast Fungus Magnaporthe oryzae. Plant Biotechnol. J. 2018, 16, 1778–1787. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, Q.; Zhou, Z.; Yin, H.; Xie, Y.; Wei, Y. Two Terpene Synthases in Resistant Pinus massoniana Contribute to Defence against Bursaphelenchus xylophilus. Plant Cell Environ. 2021, 44, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, T.; Takai, T.; Hatanaka, H.; Mizuuchi, E.; Nagamune, T.; Okumura, K.; Ogawa, H. Multiple-Mutation at a Potential Ligand-Binding Region Decreased Allergenicity of a Mite Allergen Der f 2 without Disrupting Global Structure. FEBS Lett. 2005, 579, 1988–1994. [Google Scholar] [CrossRef]
- Zhang, Y.; De Stefano, R.; Robine, M.; Butelli, E.; Bulling, K.; Hill, L.; Rejzek, M.; Martin, C.; Schoonbeek, H. Different ROS-Scavenging Properties of Flavonoids Determine Their Abilities to Extend Shelf Life of Tomato. Plant Physiol. 2015, 86, 00346. [Google Scholar] [CrossRef][Green Version]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a Bacterial Speck Resistant Tomato by CRISPR/Cas9-Mediated Editing of SlJAZ2. Plant Biotechnol. J. 2019, 17, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.-Y.; Guo, Y.; Cheng, Y.; Hu, Y.; Xiao, S.; Wang, Y.; Wen, Y.-Q. CRISPR/Cas9-Mediated Mutagenesis of VvMLO3 Results in Enhanced Resistance to Powdery Mildew in Grapevine (Vitis vinifera). Hortic. Res. 2020, 7, 116. [Google Scholar] [CrossRef]
- Lawrenson, T.; Hinchliffe, A.; Forner, M.; Harwood, W. Highly Efficient Genome Editing in Barley Using Novel Lb Cas12a Variants and Impact of SgRNA Architecture. bioRxiv 2022. [Google Scholar] [CrossRef]
- Lawrenson, T.; Chhetry, M.; Clarke, M.; Hundleby, P.; Harwood, W. Improved SpCas9 and LbCas12a Genome Editing Systems in Brassica oleracea and Brassica napus. bioRxiv 2022. [Google Scholar] [CrossRef]
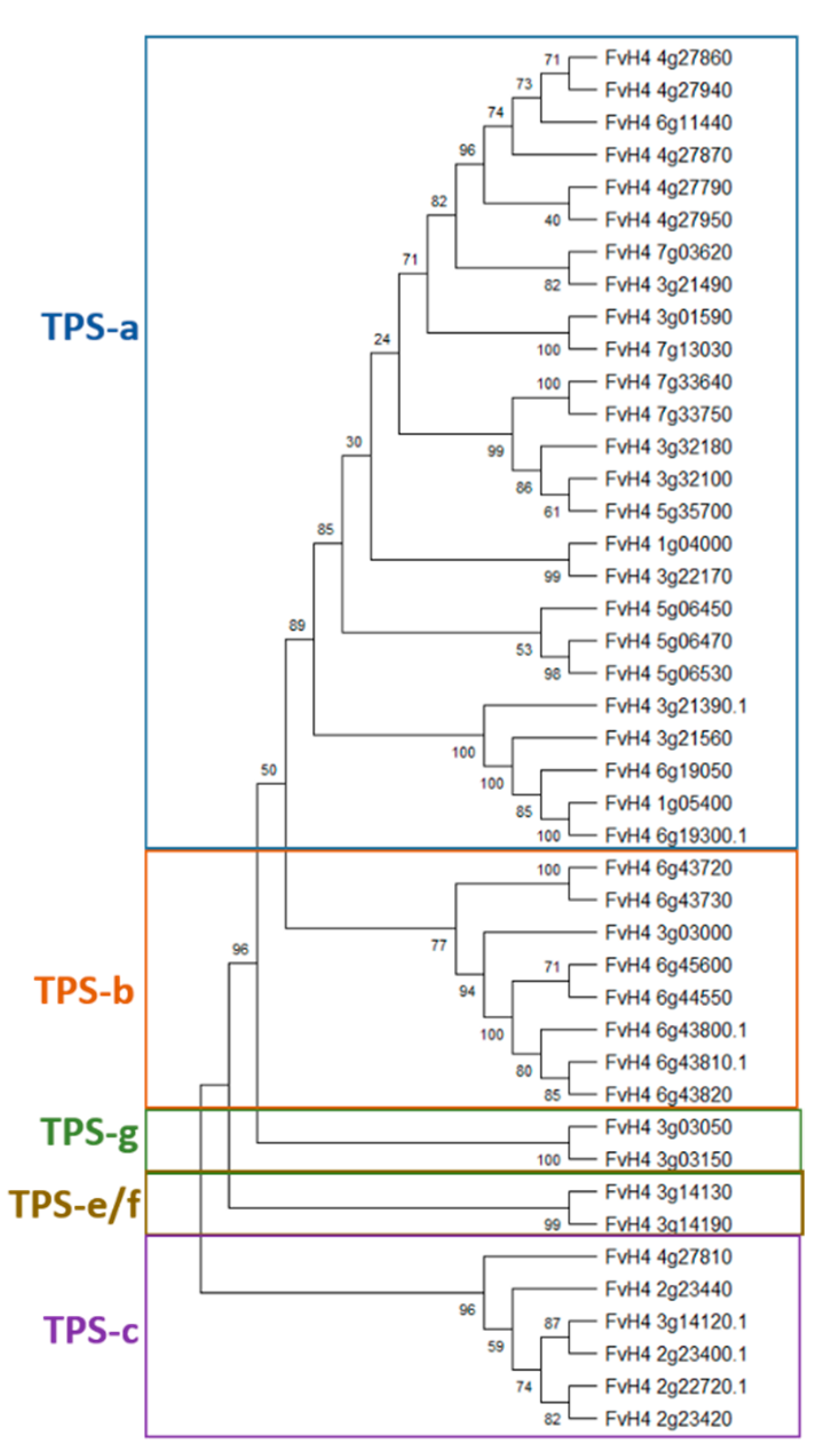


| TPS Clade | F. vesca IDs | Annotations | Plant mPLOC Prediction |
|---|---|---|---|
| TPSa | FvH4_1g04000 | (−)-germacrene D synthase-like | Chloroplast |
| FvH4_3g01590 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_3g21490 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_3g22170 | (−)-germacrene D synthase | Chloroplast | |
| FvH4_3g32100 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_3g32180 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_4g27790 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_4g27860 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_4g27870 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_4g27940 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_4g27950 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_5g06450 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_5g06470 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_5g06530 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_5g35700 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_6g11440 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_7g03620 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_7g13030 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_7g33640 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_7g33750 | (−)-germacrene D synthase-like | Chloroplast | |
| FvH4_1g05400 | (−)-alpha-pinene synthase-like | Chloroplast | |
| FvH4_3g21390 | (−)-alpha-pinene synthase-like | Chloroplast | |
| FvH4_3g21560 | (−)-alpha-pinene synthase-like | Chloroplast | |
| FvH4_6g19050 | (−)-alpha-pinene synthase-like | Chloroplast | |
| FvH4_6g19300 | (−)-alpha-pinene synthase-like | Chloroplast | |
| TPS-b | FvH4_6g43800 | tricyclene synthase EBOS and chloroplastic-like | Chloroplast |
| FvH4_6g43810 | tricyclene synthase EBOS and chloroplastic-like | Chloroplast | |
| FvH4_6g43820 | tricyclene synthase EBOS and chloroplastic-like | Chloroplast | |
| FvH4_6g44550 | tricyclene synthase EBOS and chloroplastic-like | Chloroplast | |
| FvH4_6g45600 | tricyclene synthase EBOS and chloroplastic-like | Chloroplast | |
| FvH4_3g03000 | alpha-farnesene synthase | Chloroplast | |
| FvH4_6g43720 | probable terpene synthase 9 | Chloroplast | |
| FvH4_6g43730 | probable terpene synthase 9 | Chloroplast | |
| TPS-c | FvH4_2g22720 | ent-copalyl diphosphate synthase and chloroplastic | Chloroplast |
| FvH4_2g23400 | ent-copalyl diphosphate synthase and chloroplastic-like | Chloroplast | |
| FvH4_2g23420 | ent-copalyl diphosphate synthase and chloroplastic-like | Chloroplast | |
| FvH4_2g23440 | ent-copalyl diphosphate synthase and chloroplastic-like | Chloroplast | |
| FvH4_3g14120 | ent-copalyl diphosphate synthase and chloroplastic-like | Chloroplast | |
| FvH4_4g27810 | (−)-germacrene D synthase-like | Nucleus | |
| TPS-e/f | FvH4_3g14130 | ent-kaur-16-ene synthase and chloroplastic | Chloroplast |
| FvH4_3g14190 | ent-kaur-16-ene synthase and chloroplastic-like | Chloroplast | |
| TPS-g | FvH4_3g03050 | (3S,6E)-nerolidol synthase 1-like | Chloroplast |
| FvH4_3g03150 | (3S,6E)-nerolidol synthase 1-like | Chloroplast |
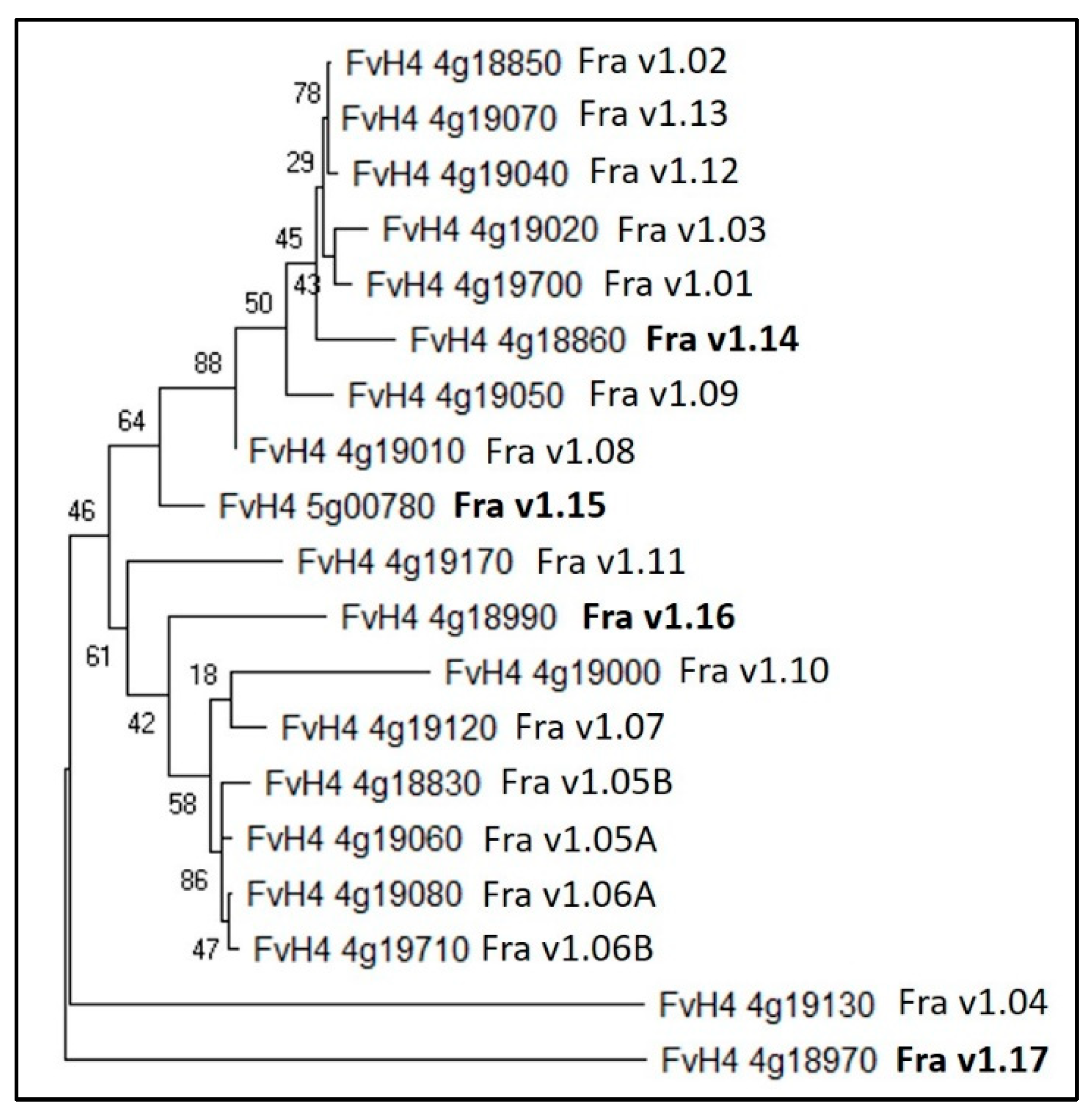
| This Review | Hyun and Kim [42] | F_vesca_v4.0 | Annotations | Plant mPLOC Prediction |
|---|---|---|---|---|
| Fra v1.05B | FvH4_4g18830 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.02 | FvH4_4g18850 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.14 | FvH4_4g18860 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.17 | FvH4_4g18970 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.16 | FvH4_4g18990 | Pathogenesis-related protein PR10 | Cytoplasm | |
| Fra v1.10 | FvH4_4g19000 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.08 | FvH4_4g19010 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.03 | FvH4_4g19020 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.12 | FvH4_4g19040 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.09 | FvH4_4g19050 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.05A | FvH4_4g19060 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.13 | FvH4_4g19070 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.06A | FvH4_4g19080 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.07 | FvH4_4g19120 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.04 | FvH4_4g19130 | Major allergen Pru ar 1-like | Cytoplasm and nucleus | |
| Fra v1.11 | FvH4_4g19170 | Major allergen Pru av 1-like | Cytoplasm | |
| Fra v1.01 | FvH4_4g19700 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.06B | FvH4_4g19710 | Major allergen Pru ar 1-like | Cytoplasm | |
| Fra v1.15 | FvH4_5g00780 | Major allergen Pru ar 1-like | Cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badmi, R.; Gogoi, A.; Doyle Prestwich, B. Secondary Metabolites and Their Role in Strawberry Defense. Plants 2023, 12, 3240. https://doi.org/10.3390/plants12183240
Badmi R, Gogoi A, Doyle Prestwich B. Secondary Metabolites and Their Role in Strawberry Defense. Plants. 2023; 12(18):3240. https://doi.org/10.3390/plants12183240
Chicago/Turabian StyleBadmi, Raghuram, Anupam Gogoi, and Barbara Doyle Prestwich. 2023. "Secondary Metabolites and Their Role in Strawberry Defense" Plants 12, no. 18: 3240. https://doi.org/10.3390/plants12183240
APA StyleBadmi, R., Gogoi, A., & Doyle Prestwich, B. (2023). Secondary Metabolites and Their Role in Strawberry Defense. Plants, 12(18), 3240. https://doi.org/10.3390/plants12183240








