Proline, Cysteine and Branched-Chain Amino Acids in Abiotic Stress Response of Land Plants and Microalgae
Abstract
:1. Introduction
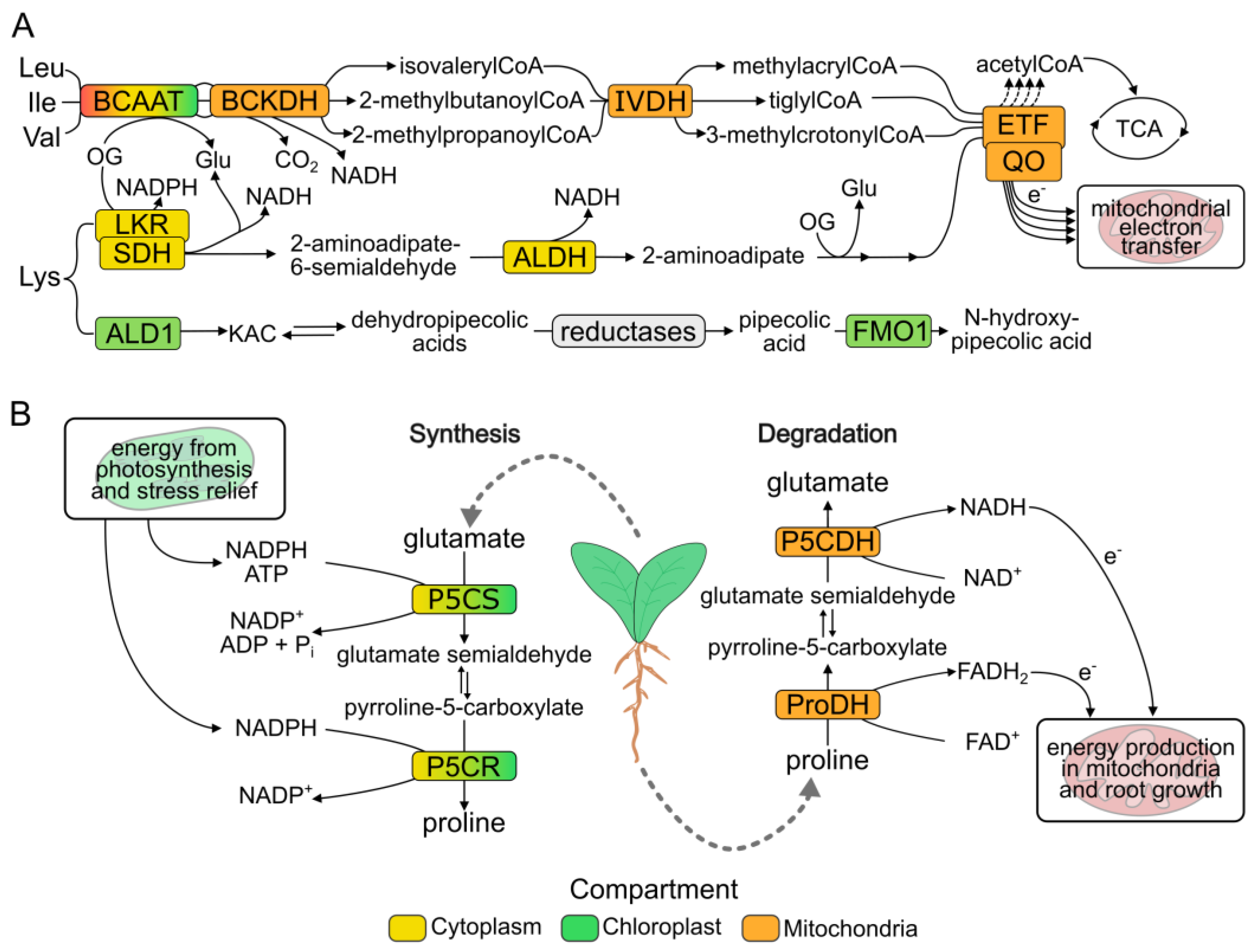
2. Amino Acid Degradation Provides Energy for Enduring Abiotic Stress in Land Plants
3. Amino Acids as Precursors for Secondary Metabolites
4. Proline Cycling for Osmotic Stress Response and Recovery
4.1. Proline Metabolic Pathway
4.2. Proline Provides Reducing Power to the Mitochondrial Electron Transport Chain
4.3. Proline Cycling Is Required for Drought Stress Tolerance in Arabidopsis thaliana
5. Cysteine and Cysteine Degradation in Drought Stress Response
5.1. Physico-Chemical Properties of Cysteine
5.2. Cysteine Biosynthesis
5.3. Cysteine Degradation Occurs through Different Pathways
5.4. The Role of Cysteine Metabolism in Drought Stress Response
6. Microalgae: Adaptation Strategies to Salinity Stress Include a Central Role for Amino Acids
7. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hildebrandt, T.M.; Nunes Nesi, A.; Araújo, W.L.; Braun, H.P. Amino Acid Catabolism in Plants. Mol. Plant. 2015, 8, 1563–1579. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Zeier, J. L-Lys metabolism to N-hydroxypipecolic acid: An integral immune-activating pathway in plants. Plant J. 2018, 96, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, B.; Hildebrandt, T.M. The role of amino acid metabolism in signaling and metabolic adaptation to stress-induced energy deficiency in plants. J. Exp. Bot. 2021, 72, 4634–4645. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.E.; Savouré, A.; Szabados, L. Proline metabolism as regulatory hub. Trends Plant Sci. 2022, 27, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Moormann, J.; Heinemann, B.; Hildebrandt, T.M. News about amino acid metabolism in plant–microbe interactions. Trends Biochem. Sci. 2022, 47, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Trovato, M.; Funck, D.; Forlani, G.; Okumoto, S.; Amir, R. Editorial: Amino Acids in Plants: Regulation and Functions in Development and Stress Defense. Front. Plant Sci. 2021, 12, 772810. [Google Scholar] [CrossRef]
- Miller, A.J.; Fan, X.; Shen, Q.; Smith, S.J. Amino acids and nitrate as signals for the regulation of nitrogen acquisition. J. Exp. Bot. 2008, 59, 111–119. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino Acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Ros, R.; Muñoz-Bertomeu, J.; Krueger, S. Serine in plants: Biosynthesis, metabolism, and functions. Trends Plant Sci. 2014, 19, 564–569. [Google Scholar] [CrossRef]
- Douce, R.; Bourguignon, J.; Neuburger, M.; Rébeillé, F. The glycine decarboxylase system: A fascinating complex. Trends Plant Sci. 2001, 6, 167–176. [Google Scholar] [CrossRef]
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184. [Google Scholar] [CrossRef] [PubMed]
- Viola, R.E. The central enzymes of the aspartate family of amino acid biosynthesis. Acc. Chem. Res. 2001, 34, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Soares da Costa, T.P.; Hall, C.J.; Panjikar, S.; Wyllie, J.A.; Christoff, R.M.; Bayat, S.; Hulett, M.D.; Abbott, B.M.; Gendall, A.R.; Perugini, M.A. Towards novel herbicide modes of action by inhibiting lysine biosynthesis in plants. Elife 2021, 27, e69444. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Ishida, H. An additional role for chloroplast proteins-an amino acid reservoir for energy production during sugar starvation. Plant Signal. Behav. 2019, 14, 1552057. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef]
- Ingle, R.A. Histidine biosynthesis. Arab. Book 2011, 9, e0141. [Google Scholar] [CrossRef]
- Biswal, B.; Joshi, P.N.; Raval, M.K.; Biswal, U.C. Photosynthesis, a global sensor of environmental stress in green plants: Stress signalling and adaptation. Curr. Sci. 2011, 101, 47–56. [Google Scholar]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S. Regulation of Photosynthesis during Abiotic Stress-Induced Photoinhibition. Mol. Plant. 2015, 8, 1304–1320. [Google Scholar] [CrossRef]
- Hirota, T.; Izumi, M.; Wada, S.; Makino, A.; Ishida, H. Vacuolar Protein Degradation via Autophagy Provides Substrates to Amino Acid Catabolic Pathways as an Adaptive Response to Sugar Starvation in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1363–1376. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Jander, G. Abscisic acid-regulated protein degradation causes osmotic stress-induced accumulation of branched-chain amino acids in Arabidopsis thaliana. Planta 2017, 246, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, T.M. Synthesis versus degradation: Directions of amino acid metabolism during Arabidopsis abiotic stress response. Plant Mol. Biol. 2018, 98, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Hara, M.; Fukino, N.; Kakizaki, T.; Morimitsu, Y. Glucosinolate metabolism, functionality and breeding for the improvement of Brassicaceae vegetables. Breed Sci. 2014, 64, 48–59. [Google Scholar] [CrossRef]
- Evans, J.; Seemann, J.R. The allocation of protein nitrogen in the photosynthetic apparatus: Costs, consequences, and control. Photosynthesis 1989, 8, 183–205. [Google Scholar]
- Araújo, W.L.; Nunes-Nesi, A.; Nikoloski, Z.; Sweetlove, L.J.; Fernie, A.R. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 2011, 35, 1–21. [Google Scholar] [CrossRef]
- Engqvist, M.; Drincovich, M.F.; Flügge, U.I.; Maurino, V.G. Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J. Biol. Chem. 2009, 284, 25026–25037. [Google Scholar] [CrossRef]
- Araújo, W.L.; Ishizaki, K.; Nunes-Nesi, A.; Larson, T.R.; Tohge, T.; Krahnert, I.; Witt, S.; Obata, T.; Schauer, N.; Graham, I.A.; et al. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell. 2010, 22, 1549–1563. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Nam, H.G.; Lin, J.F.; Wu, S.H.; Swidzinski, J.; Ishizaki, K.; et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef]
- Lehmann, M.; Schwarzländer, M.; Obata, T.; Sirikantaramas, S.; Burow, M.; Olsen, C.E.; Tohge, T.; Fricker, M.D.; Møller, B.L.; Fernie, A.R.; et al. The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol. Plant. 2009, 2, 390–406. [Google Scholar] [CrossRef]
- Ishizaki, K.; Larson, T.R.; Schauer, N.; Fernie, A.R.; Graham, I.A.; Leaver, C.J. The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell. 2005, 17, 2587–2600. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, K.; Schauer, N.; Larson, T.R.; Graham, I.A.; Fernie, A.R.; Leaver, C.J. The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 2006, 45, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Angelovici, R.; Lipka, A.E.; Deason, N.; Gonzalez-Jorge, S.; Lin, H.; Cepela, J.; Buell, R.; Gore, M.A.; Dellapenna, D. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell. 2013, 25, 4827–4843. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Uygun, S.; Shiu, S.H.; Last, R.L. The Impact of the Branched-Chain Ketoacid Dehydrogenase Complex on Amino Acid Homeostasis in Arabidopsis. Plant Physiol. 2015, 169, 1807–1820. [Google Scholar] [CrossRef]
- González-Villagra, J.; Omena-Garcia, R.P.; Rodrigues-Salvador, A.; Nunes-Nesi, A.; Cohen, J.D.; Reyes-Díaz, M.M. Differential physiological and metabolic responses in young and fully expanded leaves of Aristotelia chilensis plants subjected to drought stress. Environ. Exp Bot. 2022, 196, 104814. [Google Scholar] [CrossRef]
- Shim, J.S.; Jeong, H.I.; Bang, S.W.; Jung, S.E.; Kim, G.; Kim, Y.S.; Redillas, M.C.F.R.; Oh, S.J.; Seo, J.S.; Kim, J.K. Drought-induced branched-chain amino acid aminotransferase enhances drought tolerance in rice. Plant Physiol. 2023, 191, 1435–1447. [Google Scholar] [CrossRef]
- Batista-Silva, W.; Heinemann, B.; Rugen, N.; Nunes-Nesi, A.; Araújo, W.L.; Braun, H.P.; Hildebrandt, T.M. The role of amino acid metabolism during abiotic stress release. Plant Cell. Environ. 2019, 42, 1630–1644. [Google Scholar] [CrossRef]
- Grubb, C.D.; Abel, S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006, 11, 89–100. [Google Scholar] [CrossRef]
- Ziegler, J.; Facchini, P.J. Alkaloid biosynthesis: Metabolism and trafficking. Annu. Rev. Plant. 2008, 59, 735–769. [Google Scholar] [CrossRef]
- Alcázar, R.; Bueno, M.; Tiburcio, A.F. Polyamines: Small Amines with Large Effects on Plant Abiotic Stress Tolerance. Cells 2020, 9, 2373. [Google Scholar] [CrossRef]
- Shao, J.; Huang, K.; Batool, M.; Idrees, F.; Afzal, R.; Haroon, M.; Noushahi, H.A.; Wu, W.; Hu, Q.; Lu, X.; et al. Versatile roles of polyamines in improving abiotic stress tolerance of plants. Front. Plant Sci. 2022, 13, 1003155. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, R.; Planas, J.; Saxena, T.; Zarza, X.; Bortolotti, C.; Cuevas, J.; Bitrián, M.; Tiburcio, A.F.; Altabella, T. Putrescine accumulation confers drought tolerance in transgenic Arabidopsis plants over-expressing the homologous Arginine decarboxylase 2 gene. Plant Physiol. Biochem. 2010, 48, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Rangel, D.; Chávez-Martínez, A.I.; Rodríguez-Hernández, A.A.; Maruri-López, I.; Urano, K.; Shinozaki, K.; Jiménez-Bremont, J.F. Simultaneous Silencing of Two Arginine Decarboxylase Genes Alters Development in Arabidopsis. Front. Plant Sci. 2016, 7, 300. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, R.; Cuevas, J.C.; Patron, M.; Altabella, T.; Tiburcio, A.F. Abscisic acid modulates polyamine metabolism under water stress in Arabidopsis thaliana. Physiol. Plant. 2006, 128, 448–455. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Kiyota, E.; Pena, I.A.; Arruda, P. The saccharopine pathway in seed development and stress response of maize. Plant Cell Environ. 2015, 38, 2450–2461. [Google Scholar] [CrossRef]
- Bernsdorff, F.; Döring, A.C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic Acid Orchestrates Plant Systemic Acquired Resistance and Defense Priming via Salicylic Acid-Dependent and -Independent Pathways. Plant Cell. 2016, 28, 102–129. [Google Scholar] [CrossRef]
- Arruda, P.; Barreto, P. Lysine Catabolism Through the Saccharopine Pathway: Enzymes and Intermediates Involved in Plant Responses to Abiotic and Biotic Stress. Front. Plant Sci. 2020, 11, 587. [Google Scholar] [CrossRef]
- Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef]
- Cai, J.; Aharoni, A. Amino acids and their derivatives mediating defense priming and growth tradeoff. Curr. Opin. Plant Biol. 2022, 69, 102288. [Google Scholar] [CrossRef]
- Moulin, M.; Deleu, C.; Larher, F.; Bouchereau, A. The lysine-ketoglutarate reductase-saccharopine dehydrogenase is involved in the osmo-induced synthesis of pipecolic acid in rapeseed leaf tissues. Plant Physiol. Biochem. 2006, 44, 474–482. [Google Scholar] [CrossRef]
- Deleu, C.; Coustaut, M.; Niogret, M.F.; Larher, F. Three new osmotic stress-regulated cDNAs identified by differential display polymerase chain reaction in rapeseed leaf discs. Plant Cell Environ. 1999, 22, 979–988. [Google Scholar] [CrossRef]
- Moulin, M.; Deleu, C.; Larher, F. L-Lysine catabolism is osmo-regulated at the level of lysine-ketoglutarate reductase and saccharopine dehydrogenase in rapeseed leaf discs. Plant Physiol. Biochem. 2000, 38, 577–585. [Google Scholar] [CrossRef]
- Yildiz, I.; Gross, M.; Moser, D.; Petzsch, P.; Köhrer, K.; Zeier, J. N-hydroxypipecolic acid induces systemic acquired resistance and transcriptional reprogramming via TGA transcription factors. Plant Cell Environ. 2023, 6, 1900–1920. [Google Scholar] [CrossRef]
- Návarová, H.; Bernsdorff, F.; Döring, A.C.; Zeier, J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell. 2012, 24, 5123–5141. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Holmes, E.C.; Rajniak, J.; Kim, J.G.; Tang, S.; Fischer, C.R.; Mudgett, M.B.; Sattely, E.S. N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4920–E4929. [Google Scholar] [CrossRef] [PubMed]
- Kishor, P.; Hong, Z.; Miao, G.H.; Hu, C.; Verma, D. Overexpression of [delta]-Pyrroline-5-Carboxylate Synthetase Increases Proline Production and Confers Osmotolerance in Transgenic Plants. Plant Physiol. 1995, 108, 1387–1394. [Google Scholar] [CrossRef]
- Hmida-Sayari, A.; Gargouri-Bouzid, R.; Bidani, A.; Jaoua, L.; Savoure, A.; Jaoua, S. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Sci. 2005, 169, 746–752. [Google Scholar] [CrossRef]
- Székely, G.; Abrahám, E.; Cséplo, A.; Rigó, G.; Zsigmond, L.; Csiszár, J.; Ayaydin, F.; Strizhov, N.; Jásik, J.; Schmelzer, E.; et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008, 53, 11–28. [Google Scholar] [CrossRef]
- Banu, M.N.; Hoque, M.A.; Watanabe-Sugimoto, M.; Islam, M.M.; Uraji, M.; Matsuoka, K.; Nakamura, Y.; Murata, Y. Proline and glycinebetaine ameliorated NaCl stress via scavenging of hydrogen peroxide and methylglyoxal but not superoxide or nitric oxide in tobacco cultured cells. Biosci. Biotechnol. Biochem. 2010, 74, 2043–2049. [Google Scholar] [CrossRef]
- Signorelli, S.; Coitiño, E.L.; Borsani, O.; Monza, J. Molecular mechanisms for the reaction between (˙)OH radicals and proline: Insights on the role as reactive oxygen species scavenger in plant stress. J. Phys. Chem. B 2014, 118, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.U.; Bashir, F.; Ayaydin, F.; Kóta, Z.; Páli, T.; Vass, I. Proline is a quencher of singlet oxygen and superoxide both in in vitro systems and isolated thylakoids. Physiol. Plant 2021, 172, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Sharma, S. Proline metabolism and its implications for plant-environment interaction. Arab. Book 2010, 8, e0140. [Google Scholar] [CrossRef]
- Cabassa-Hourton, C.; Schertl, P.; Bordenave-Jacquemin, M.; Saadallah, K.; Guivarc’h, A.; Lebreton, S.; Planchais, S.; Klodmann, J.; Eubel, H.; Crilat, E.; et al. Proteomic and functional analysis of proline dehydrogenase 1 link proline catabolism to mitochondrial electron transport in Arabidopsis thaliana. Biochem. J. 2016, 473, 2623–2634. [Google Scholar] [CrossRef]
- Kiyosue, T.; Yoshiba, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell. 1996, 8, 1323–1335. [Google Scholar] [CrossRef] [PubMed]
- Funck, D.; Eckard, S.; Müller, G. Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol. 2010, 10, 70. [Google Scholar] [CrossRef]
- Rizzi, Y.S.; Cecchini, N.M.; Fabro, G.; Alvarez, M.E. Differential control and function of Arabidopsis ProDH1 and ProDH2 genes on infection with biotrophic and necrotrophic pathogens. Mol. Plant Pathol. 2017, 18, 1164–1174. [Google Scholar] [CrossRef]
- Peng, Z.; Lu, Q.; Verma, D.P. Reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol. Gen. Genet. 1996, 253, 334–341. [Google Scholar] [CrossRef]
- Savouré, A.; Hua, X.J.; Bertauche, N.; Van Montagu, M.; Verbruggen, N. Abscisic acid-independent and abscisic acid-dependent regulation of proline biosynthesis following cold and osmotic stresses in Arabidopsis thaliana. Mol. Gen. Genet. 1997, 254, 104–109. [Google Scholar] [CrossRef]
- Strizhov, N.; Li, Y.; Rosso, M.G.; Viehoever, P.; Dekker, K.A.; Weisshaar, B. High-throughput generation of sequence indexes from T-DNA mutagenized Arabidopsis thaliana lines. BioTechniques 2003, 35, 1164–1168. [Google Scholar] [CrossRef]
- Launay, A.; Cabassa-Hourton, C.; Eubel, H.; Maldiney, R.; Guivarc’h, A.; Crilat, E.; Planchais, S.; Lacoste, J.; Bordenave-Jacquemin, M.; Clément, G.; et al. Proline oxidation fuels mitochondrial respiration during dark-induced leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 6203–6214. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, G.B.; Yang, T.H.; Verslues, P.E. Dynamic proline metabolism: Importance and regulation in water limited environments. Front. Plant Sci. 2015, 6, 484. [Google Scholar] [CrossRef] [PubMed]
- Shinde, S.; Villamor, J.G.; Lin, W.; Sharma, S.; Verslues, P.E. Proline Coordination with Fatty Acid Synthesis and Redox Metabolism of Chloroplast and Mitochondria. Plant Physiol. 2016, 172, 1074–1088. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Villamor, J.G.; Verslues, P.E. Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol. 2011, 157, 292–304. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Fermani, S.; Marchand, C.H.; Costa, A.; Sparla, F.; Rouhier, N.; Geigenberger, P.; Lemaire, S.D.; Trost, P. Redox Homeostasis in Photosynthetic Organisms: Novel and Established Thiol-Based Molecular Mechanisms. Antioxid. Redox Signal. 2019, 31, 155–210. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Carroll, K.S. Cysteine-mediated redox signaling chemistry, biology, and tools for discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef]
- García, I.; Arenas-Alfonseca, L.; Moreno, I.; Gotor, C.; Romero, L.C. HCN Regulates Cellular Processes through Posttranslational Modification of Proteins by S-cyanylation. Plant Physiol. 2019, 179, 107–123. [Google Scholar] [CrossRef]
- Wirtz, M.; Droux, M. Synthesis of the sulfur amino acids: Cysteine and methionine. Photosynth. Res. 2005, 86, 345–362. [Google Scholar] [CrossRef]
- Krueger, S.; Niehl, A.; Lopez Martin, M.C.; Steinhauser, D.; Donath, A.; Hildebrandt, T.; Romero, L.C.; Hoefgen, R.; Gotor, C.; Hesse, H. Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant Cell Environ. 2009, 32, 349–367. [Google Scholar] [CrossRef]
- Zechmann, B.; Liou, L.C.; Koffler, B.E.; Horvat, L.; Tomašić, A.; Fulgosi, H.; Zhang, Z. Subcellular distribution of glutathione and its dynamic changes under oxidative stress in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2011, 11, 631–642. [Google Scholar] [CrossRef]
- Park, S.; Imlay, J.A. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J. Bacteriol. 2003, 185, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Romero, L.C.; Aroca, M.Á.; Laureano-Marín, A.M.; Moreno, I.; García, I.; Gotor, C. Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Mol. Plant. 2014, 7, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Jez, J.M.; Dey, S. The cysteine regulatory complex from plants and microbes: What was old is new again. Curr. Opin. Struct. Biol. 2013, 23, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.Y.; Fujiwara, T.; Awazuhara, M.; Kimura, T.; Noji, M.; Saito, K. Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 2003, 33, 651–663. [Google Scholar] [CrossRef]
- Olsen, L.R.; Huang, B.; Vetting, M.W.; Roderick, S.L. Structure of serine acetyltransferase in complexes with CoA and its cysteine feedback inhibitor. Biochemistry 2004, 43, 6013–6019. [Google Scholar] [CrossRef]
- García, I.; Castellano, J.M.; Vioque, B.; Solano, R.; Gotor, C.; Romero, L.C. Mitochondrial beta-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana. Plant Cell. 2010, 22, 3268–3279. [Google Scholar] [CrossRef]
- Álvarez, C.; García, I.; Romero, L.C.; Gotor, C. Mitochondrial sulfide detoxification requires a functional isoform O-acetylserine(thiol)lyase C in Arabidopsis thaliana. Mol Plant. 2012, 5, 1217–1226. [Google Scholar] [CrossRef]
- Machingura, M.; Salomon, E.; Jez, J.M.; Ebbs, S.D. The β-cyanoalanine synthase pathway: Beyond cyanide detoxification. Plant Cell Environ. 2016, 39, 2329–2341. [Google Scholar] [CrossRef]
- Krüßel, L.; Junemann, J.; Wirtz, M.; Birke, H.; Thornton, J.D.; Browning, L.W.; Poschet, G.; Hell, R.; Balk, J.; Braun, H.P.; et al. The mitochondrial sulfur dioxygenase ethylmalonic encephalopathy protein1 is required for amino acid catabolism during carbohydrate starvation and embryo development in Arabidopsis. Plant Physiol. 2014, 165, 92–104. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought Stress Responses and Resistance in Plants: From Cellular Responses to Long-Distance Intercellular Communication. Front. Plant Sci. 2020, 11, 556972. [Google Scholar] [CrossRef]
- Batool, S.; Uslu, V.V.; Rajab, H.; Ahmad, N.; Waadt, R.; Geiger, D.; Malagoli, M.; Xiang, C.B.; Hedrich, R.; Rennenberg, H.; et al. Sulfate is Incorporated into Cysteine to Trigger ABA Production and Stomatal Closure. Plant Cell. 2018, 30, 2973–2987. [Google Scholar] [CrossRef] [PubMed]
- Rajab, H.; Khan, M.S.; Malagoli, M.; Hell, R.; Wirtz, M. Sulfate-Induced Stomata Closure Requires the Canonical ABA Signal Transduction Machinery. Plants 2019, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.; Calo, L.; Romero, L.C.; García, I.; Gotor, C. An O-acetylserine(thiol)lyase homolog with L-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol. 2010, 152, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Scuffi, D.; Álvarez, C.; Laspina, N.; Gotor, C.; Lamattina, L.; García-Mata, C. Hydrogen sulfide generated by L-cysteine desulfhydrase acts upstream of nitric oxide to modulate abscisic acid-dependent stomatal closure. Plant Physiol. 2014, 166, 2065–2076. [Google Scholar] [CrossRef]
- Laureano-Marín, A.M.; García, I.; Romero, L.C.; Gotor, C. Assessing the transcriptional regulation of l-cysteine desulfhydrase 1 in Arabidopsis thaliana. Front. Plant Sci. 2014, 5, 683. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, H.; Zhou, M.; Ge, Z.; Zhang, F.; Foyer, C.H.; Yuan, X.; Xie, Y. The coordination of guard-cell autonomous ABA synthesis and DES1 function in situ regulates plant water deficit responses. J. Adv. Res. 2020, 27, 191–197. [Google Scholar] [CrossRef]
- Aroca, A.; Zhang, J.; Xie, Y.; Romero, L.C.; Gotor, C. Hydrogen sulfide signaling in plant adaptations to adverse conditions: Molecular mechanisms. J. Exp. Bot. 2021, 72, 5893–5904. [Google Scholar] [CrossRef]
- Li, Y.; Cai, H.; Liu, P.; Wang, C.; Gao, H.; Wu, C.; Yan, K.; Zhang, S.; Huang, J.; Zheng, C. Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem. Biophys. Res. Commun. 2017, 484, 292–297. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, J.; Shen, J.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Fu, L.; Li, Z.; Yang, J.; et al. Hydrogen sulfide-linked persulfidation of ABI4 controls ABA responses through the transactivation of MAPKKK18 in Arabidopsis. Mol. Plant. 2012, 14, 921–936. [Google Scholar] [CrossRef]
- Geiger, D.; Scherzer, S.; Mumm, P.; Stange, A.; Marten, I.; Bauer, H.; Ache, P.; Matschi, S.; Liese, A.; Al-Rasheid, K.A.; et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 2009, 106, 21425–21430. [Google Scholar] [CrossRef]
- Sirichandra, C.; Gu, D.; Hu, H.C.; Davanture, M.; Lee, S.; Djaoui, M.; Valot, B.; Zivy, M.; Leung, J.; Merlot, S.; et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009, 583, 2982–2986. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jia, H.; Wang, X.; Shi, C.; Wang, X.; Ma, P.; Wang, J.; Ren, M.; Li, J. Hydrogen Sulfide Positively Regulates Abscisic Acid Signaling through Persulfidation of SnRK2.6 in Guard Cells. Mol. Plant. 2020, 13, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, J.; Zhou, M.; Zhou, H.; Cui, B.; Gotor, C.; Romero, L.C.; Fu, L.; Yang, J.; Foyer, C.H.; et al. Persulfidation-based Modification of Cysteine Desulfhydrase and the NADPH Oxidase RBOHD Controls Guard Cell Abscisic Acid Signaling. Plant Cell. 2020, 32, 1000–1017. [Google Scholar] [CrossRef] [PubMed]
- Papanatsiou, M.; Scuffi, D.; Blatt, M.R.; García-Mata, C. Hydrogen sulfide regulates inward-rectifying K+ channels in conjunction with stomatal closure. Plant Physiol. 2015, 168, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Jurado-Flores, A.; Aroca, A.; Romero, L.C.; Gotor, C. Sulfide promotes tolerance to drought through protein persulfidation in Arabidopsis. J. Exp. Bot. 2023, 74, 4654–4669. [Google Scholar] [CrossRef] [PubMed]
- Usher, P.K.; Ross, A.B.; Camargo-Valero, M.A.; Tomlin, A.S.; Gale, W.F. An overview of the potential environmental impacts of large-scale microalgae cultivation. Biofuels 2014, 5, 331–349. [Google Scholar] [CrossRef]
- Kholssi, R.; Lougraimzi, H.; Moreno-Garrido, I. Effects of global environmental change on microalgal photosynthesis, growth and their distribution. Mar. Environ. Res. 2023, 184, 105877. [Google Scholar] [CrossRef]
- Simas-Rodrigues, C.; Villela, H.D.; Martins, A.P.; Marques, L.G.; Colepicolo, P.; Tonon, A.P. Microalgae for economic applications: Advantages and perspectives for bioethanol. J. Exp. Bot. 2015, 66, 4097–4108. [Google Scholar] [CrossRef] [PubMed]
- Eltanahy, E.; Torky, A. Microalgae as Cell Factories: Food and Feed-grade High value Metabolites. In Microalgal Biotechnology; Shekh, A., Schenk, P., Sarada, R., Eds.; Royal Society of Chemistry: Cambridge, UK, 2021; pp. 1–35. [Google Scholar] [CrossRef]
- Kolackova, M.; Janova, A.; Dobesova, M.; Zvalova, M.; Chaloupsky, P.; Krystofova, O.; Adam, V.; Huska, D. Role of secondary metabolites in distressed microalgae. Environ. Res. 2023, 224, 115392. [Google Scholar] [CrossRef]
- Wang, N.; Qian, Z.; Luo, M.; Fan, S.; Zhang, X.; Zhang, L. Identification of Salt Stress Responding Genes Using Transcriptome Analysis in Green Alga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2018, 19, 3359. [Google Scholar] [CrossRef]
- Bazzani, E.; Lauritano, C.; Mangoni, O.; Bolinesi, F.; Saggiomo, M. Chlamydomonas Responses to Salinity Stress and Possible Biotechnological Exploitation. J. Mar. Sci. Eng. 2021, 9, 1242. [Google Scholar] [CrossRef]
- Qiao, T.; Zhao, Y.; Zhong, D.-B.; Yu, X. Hydrogen peroxide and salinity stress act synergistically to enhance lipids production in microalga by regulating reactive oxygen species and calcium. Algal Res. 2021, 53, 102017. [Google Scholar] [CrossRef]
- Chen, T.H.; Murata, N. Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr. Opin. Plant Biol. 2002, 5, 250–257. [Google Scholar] [CrossRef]
- Nikitashina, V.; Stettin, D.; Pohnert, G. Metabolic adaptation of diatoms to hypersalinity. Phytochemistry 2022, 201, 113267. [Google Scholar] [CrossRef] [PubMed]
- Khona, D.K.; Shirolikar, S.M.; Gawde, K.K.; Hom, E.; Deodhar, M.M.; D’Souza, J.S. Characterization of salt stress-induced palmelloids in the green alga, Chlamydomonas reinhardtii. Algal Res. 2016, 16, 434–448. [Google Scholar] [CrossRef]
- Kaçka, A.; Dönmez, G. Isolation of Dunaliella spp. from a hypersaline lake and their ability to accumulate glycerol. Bioresour Technol. 2008, 99, 8348–8352. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Dell’Aversana, E.; Carillo, P. Spatial and Temporal Profile of Glycine Betaine Accumulation in Plants Under Abiotic Stresses. Front Plant Sci. 2019, 10, 230. [Google Scholar] [CrossRef]
- Gulyás, Z.; Simon-Sarkadi, L.; Badics, E.; Novák, A.; Mednyánszky, Z.; Szalai, G.; Galiba, G.; Kocsy, G. Redox regulation of free amino acid levels in Arabidopsis thaliana. Physiol Plantarum. 2017, 159, 264–276. [Google Scholar] [CrossRef]
- Chokshi, K.; Pancha, I.; Ghosh, A.; Mishra, S. Salinity induced oxidative stress alters the physiological responses and improves the biofuel potential of green microalgae Acutodesmus dimorphus. Bioresour Technol. 2017, 244, 1376–1383. [Google Scholar] [CrossRef]
- Rezayian, M.; Niknam, V.; Faramarzi, M.A. Antioxidative responses of Nostoc ellipsosporum and Nostoc piscinale to salt stress. J. Appl. Phycol. 2019, 31, 157–169. [Google Scholar] [CrossRef]
- Fal, S.; Aasfar, A.; Rabie, R.; Smouni, A.; El Arroussi, H. Salt induced oxidative stress alters physiological, biochemical and metabolomic responses of green microalga Chlamydomonas reinhardtii. Heliyon. 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Pancha, I.; Chokshi, K.; Maurya, R.; Trivedi, K.; Patidar, S.K.; Ghosh, A.; Mishra, S. Salinity induced oxidative stress enhanced biofuel production potential of microalgae Scenedesmus sp. CCNM 1077. Biores. Technol. 2015, 189, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yuan, Y.; Cheng, D.; Gao, J.; Kong, L.; Zhao, Q.; Wei, W.; Sun, Y. Exploring stress tolerance mechanism of evolved freshwater strain Chlorella sp. S30 under 30 g/L salt. Bioresour Technol. 2018, 250, 495–504. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ingrisano, R.; Tosato, E.; Trost, P.; Gurrieri, L.; Sparla, F. Proline, Cysteine and Branched-Chain Amino Acids in Abiotic Stress Response of Land Plants and Microalgae. Plants 2023, 12, 3410. https://doi.org/10.3390/plants12193410
Ingrisano R, Tosato E, Trost P, Gurrieri L, Sparla F. Proline, Cysteine and Branched-Chain Amino Acids in Abiotic Stress Response of Land Plants and Microalgae. Plants. 2023; 12(19):3410. https://doi.org/10.3390/plants12193410
Chicago/Turabian StyleIngrisano, Rachele, Edoardo Tosato, Paolo Trost, Libero Gurrieri, and Francesca Sparla. 2023. "Proline, Cysteine and Branched-Chain Amino Acids in Abiotic Stress Response of Land Plants and Microalgae" Plants 12, no. 19: 3410. https://doi.org/10.3390/plants12193410
APA StyleIngrisano, R., Tosato, E., Trost, P., Gurrieri, L., & Sparla, F. (2023). Proline, Cysteine and Branched-Chain Amino Acids in Abiotic Stress Response of Land Plants and Microalgae. Plants, 12(19), 3410. https://doi.org/10.3390/plants12193410






