Abstract
Three carbon-chain extension genes associated with fatty acid synthesis in upland cotton (Gossypium hirsutum), namely GhKAR, GhHAD, and GhENR, play important roles in oil accumulation in cotton seeds. In the present study, these three genes were cloned and characterized. The expression patterns of GhKAR, GhHAD, and GhENR in the high seed oil content cultivars 10H1014 and 10H1041 differed somewhat compared with those of 10H1007 and 2074B with low seed oil content at different stages of seed development. GhKAR showed all three cultivars showed higher transcript levels than that of 2074B at 10-, 40-, and 45-days post anthesis (DPA). The expression pattern of GhHAD showed a lower transcript level than that of 2074B at both 10 and 30 DPA but a higher transcript level than that of 2074B at 40 DPA. GhENR showed a lower transcript level than that of 2074B at both 15 and 30 DPA. The highest transcript levels of GhKAR and GhENR were detected at 15 DPA in 10H1007, 10H1014, and 10H1041 compared with 2074B. From 5 to 45 DPA cotton seed, the oil content accumulated continuously in the developing seed. Oil accumulation reached a peak between 40 DPA and 45 DPA and slightly decreased in mature seed. In addition, GhKAR and GhENR showed different expression patterns in fiber and ovule development processes, in which they showed high expression levels at 20 DPA during the fiber elongation stage, but their expression level peaked at 15 DPA during ovule development processes. These two genes showed the lowest expression levels at the late seed maturation stage, while GhHAD showed a peak of 10 DPA in fiber development. Compared to 2074B, the oil contents of GhKAR and GhENR overexpression lines increased 1.05~1.08 folds. These results indicated that GhHAD, GhENR, and GhKAR were involved in both seed oil synthesis and fiber elongation with dual biological functions in cotton.
1. Introduction
Cotton (Gossypium hirsutum L.) is an important commercial crop that is grown worldwide as a source of fiber and edible oil [1,2,3,4]. Cotton fiber serves as an important raw material in the global textile industry and is a better alternative to synthetic fiber [5,6,7,8,9]. Cotton seed contains 28–40% oil, which could be used as edible oil and raw material for chemical production as well as a feedstock for biodiesel production [5,10]. Refined as edible oil, cottonseed oil is typically composed of about 26% palmitic acid (C16: 0), 15% oleic acid (C18: 1), and 58% linoleic acid (C18: 2) [11], with less total serum cholesterol but more effectively usage compared with corn (Zea mays) oil [12], and provides several kinds of benefits and essential fatty acids. Therefore, cottonseed oil is a valued raw material in the food industry because it contains a high quantity of saturated palmitic acid and lacks unstable linolenic acids, imparting good stability and flavor properties [13].
Annually, billions of barrels of fossil oil are consumed worldwide, so cottonseed oil could also serve as a potential raw material for the energy industry as environmentally friendly biodiesel [14,15]. In view of this, dissecting the genetic basis of cottonseed oil formation and increasing its content should be paid more attention. Thus, developing cotton cultivars with high oil content with desired yield and fiber quality traits reserved are the further direction of breeding in cotton [16]. Researchers have found that overexpression of the cotton gene GhDGAT1 with a seed-specific promoter in cotton seeds increased oil content to 13.9% in different transgenic lines [17]. Zhu et al. 2021 performed a comparative transcriptome analysis to identify the key genes for oil accumulation in cultivated tetraploid cotton [18]. They found three key transcription factors, WRI1, NF-YB6, and DPBF2, played a vital role in regulating seed oil in two tetraploid cotton. SAD6 and FATA genes were more important for oil biosynthesis in G. barbadense than that in G. hirsutum. In addition, GbSWEET and GbACBP6 could significantly increase the oil content in G. barbadense cotton [18]. Another study showed that the transcription factor GhHSL1 was involved in the regulation of cottonseed oil content based on the methods of quantitative trait loci (QTLs) and transcriptome analysis [19]. Bioinformatic and phylogenetic analyses also revealed that cotton stearoyl-acyl carrier protein desaturase (SAD) genes were involved in cottonseed oil biogenesis [20]. By comparative transcriptome analysis between two germplasms with the high-oil and low-oil contents of cottonseeds, a recent report found that GhCYSD1 was identified as an important key player in oil biosynthesis. The overexpression of GhCYSD1 in yeast resulted in increased oil content and altered fatty acid composition [21]. All these previously reported candidate genes might be used to increase oil content in cottonseed without affecting the yield and quality of cotton fiber.
However, in addition to the oil biosynthesis genes reported above, Type II fatty acid synthase (FAS) also catalyzes the synthesis of straight-chain fatty acids with 16 or 18 carbon atoms in cotton [22,23]. Cotton FASII consists of six kinds of enzymes: among them, β-ketone fatty acyl-ACP reductase (KAR), β-hydroxy fatty acyl-ACP dehydratase (HAD), and enoyl-ACP reductase (ENR) catalyzed the carbon-chain reduction, dehydration, and reduction reactions [24,25]. In Eustigmatos cf. polyphem (an oleaginous microalga), transcriptome data indicated that KAR, HAD, and ENR were key genes in fatty acid biosynthesis and metabolism [26]. Compared to date palm, the total transcript levels of KAR, HAD, and ENR at five stages of mesocarp development were significantly higher in the oil palm by 44, 34, and 17-fold, respectively, based on the transcriptome sequencing method, indicating that these three genes were involved in the regulation of oil content in the mesocarp tissue of palms [27]. Another report observed that KAR, HAD, and ENR showed consistent transcription level trends at four stages of seed development in four oilseed plants [28]. In peanuts, analysis of the transcript levels of AhKAR, AhHAD, and AhENR genes at different stages of ovule development showed that the expression pattern of AhHAD differed from those of AhKAR and AhENR [29].
In cotton, the full-length cDNAs of GhKAR, GhHAD, and GhENR have been cloned, and each encodes a 283, 221, and 394 amino acid protein, respectively. Bioinformatic analysis of GhKAR, GhHAD, and GhENR indicated that the three genes served important functions in oil accumulation and were involved in the response to physiological stress [30]. To deeply provide new insights into understanding the mechanism of GhKAR, GhHAD, and GhENR in regulating the fatty acid biosynthesis network, in this study, we examined the expression patterns of GhKAR, GhHAD, and GhENR in cotton during fiber and seed development. We found that the expression levels of GhKAR, GhHAD, and GhENR in high seed oil content cultivars differed from the low seed oil content cultivars at different stages of seed development. Thereinto, GhKAR and GhENR presented different expression patterns during fiber and ovule development processes. Overexpression of GhKAR and GhENR slightly increased the oil contents in cottonseeds. These results not only bring new perspectives for understanding the mechanism of seed oil accumulation and fiber quality in cotton but also provide three new genes for the potential increase in oil content in cottonseeds by genetic engineering methods.
2. Results
2.1. Identification of Overexpressing GhKAR, GhHAD and GhENR Genes Plants
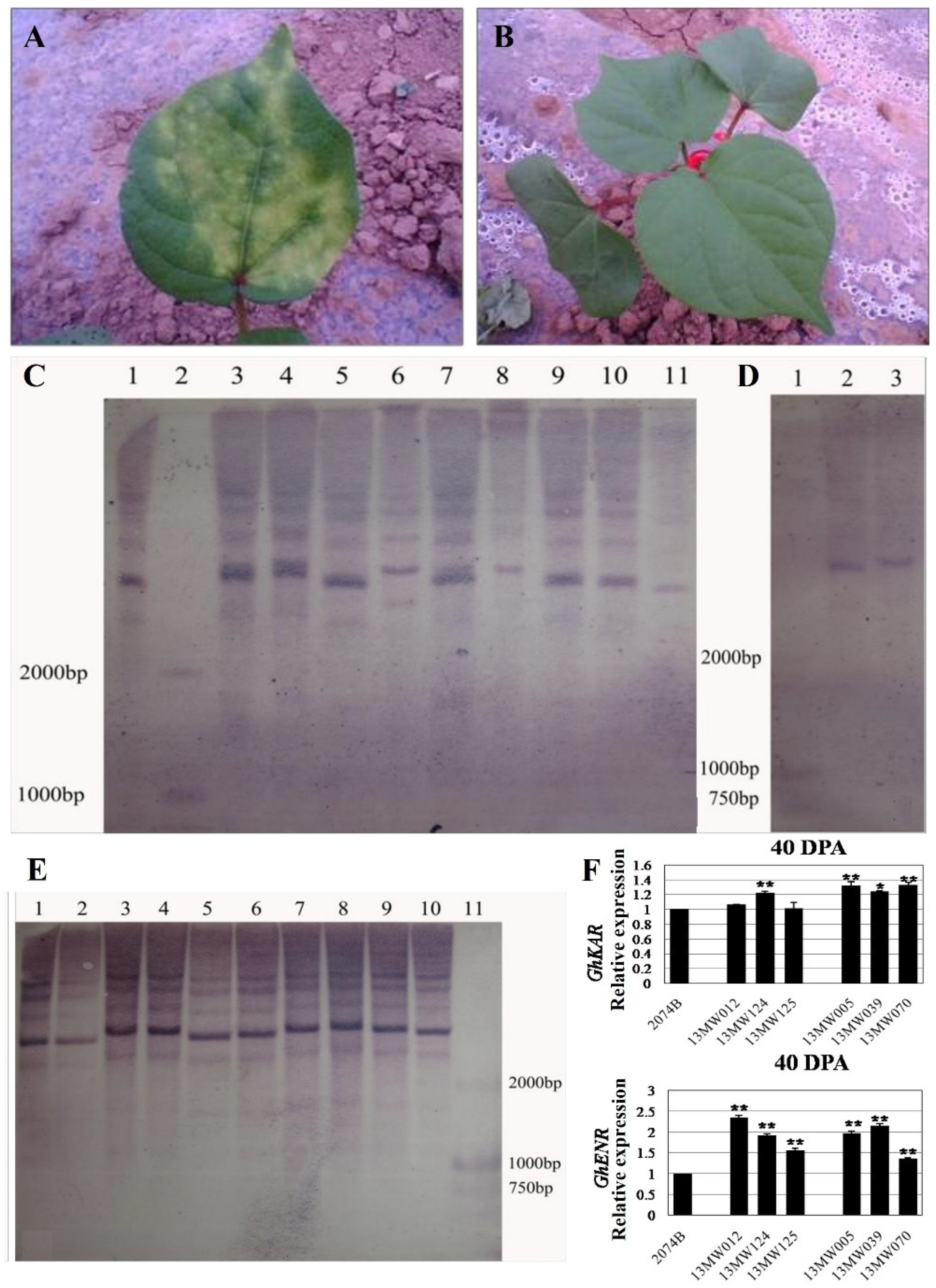
We primarily used a 5 g/L kanamycin solution to identify the transgenic cotton plants. After 3–5 days of application on leaves, those with yellow spots on the leaves were transgenic-negative plants (Figure 1A), while those without yellow spots were transgenic-positive candidate plants (Figure 1B). Then, the genomic DNA of transgenic positive plant leaves was extracted and digested by BamHI-HF endonuclease. The labeled probe was the fragment of kanamycin resistance encoding gene Kan. The copy number of transgenic positive plants was identified by the Roche Southern blot kit, and it was found that the copy number of the labeled probe in transgenic plants was between 1–4, as shown in Figure 1C–E. The results showed that GhKAR and GhENR overexpressed lines contained three and four copy transgene insertions (Figure 1C,E), while the copy number of overexpressed GhHAD plants was single (Figure 1D). The qRT-PCR analysis of 40DPA ovules revealed that the expression levels of six T3 transgenic lines that overexpression GhKAR (13MW012, 13MW124, and 13MW125) or GhENR (13MW005, 13MW039, and 13MW070) were all higher than the receptor cultivar 2074B (Figure 1F).
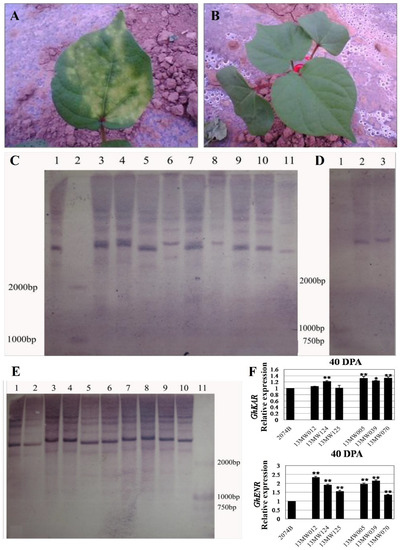
Figure 1.
Identification of overexpressing GhKAR, GhHAD and GhENR genes plants. (A) transgenic negative plants identified by kanamycin. (B) transgenic-positive plants identified by kanamycin. (C) Southern blotting detection of transgenic GhENR plants. One, 3–11 were the results of GhENR and 2 was Marker D2000; (D) Southern blotting detection of transgenic GhHAD plants. One: Marker D2000; 2 and 3: Southern blotting results of GhHAD plants. (E) Southern blotting detection of transgenic GhKAR plants. One–Ten: Southern blotting results; 11: Marker D2000. (F) Expression levels of GhKAR and GhENR at 40DPA ovules in six T3 transgenic lines that overexpression GhKAR (13MW012, 13MW124, and 13MW125) or GhENR (13MW005, 13MW039, and 13MW070) compared with the receptor cultivar 2074B (means of triplicates ± SD, * p < 0.05, ** p < 0.01, Student’s t-test).
2.2. Phenotypic Traits of the Plant Materials
Based on the seed oil content and gene copy number, four wild-type cotton cultivars (2074B, 10H1007, 10H1014, and 10H1041) and six T3 transgenic lines that overexpression GhKAR (13MW012, 13MW124, and 13MW125) or GhENR (13MW005, 13MW039, and 13MW070), each with single gene copy and high seed oil content, were selected to study the functions of GhKAR, and GhENR in cotton oil metabolism (Table 1). The results showed that two wild-type cotton cultivars (10H1014 and 10H1041) contained high oil contents up to 30.73% and 35.97%, respectively. However, another two wild types (10H1007 and 2074B) contained oil contents low to 26.09% and 27.32%, respectively. In this study, 2074B was the background material for the transgene receptor. Compared to 2074B, the oil contents of three GhKAR overexpression lines slightly increased by 1.05~1.07 folds, and GhENR overexpression lines slightly increased by 1.06~1.08 folds with no significant differences.

Table 1.
Oil content of the four wild-type and six T3 transgenic cotton lines used in the study.
The other important agronomic and economic traits of four wild-type cotton cultivars were also recorded. Among the four cultivars, 10H1041 was distinct, with a plant height of 86.8 ± 4.73 cm and the height of the first branch at 18.3 ± 2.21 cm, which were significantly lower than the others (Table 2). As for the six transgenic lines, the plant height, height of the first branch, branch number, and lint percentage were significantly higher, whereas the seed indices were significantly lower than those of the control line 2074B (Table 3). Among them, 13MW125 was a favorable line in which the plant height, seed oil content, and lint percentage were significantly higher compared with those of 2074B; however, its fiber quality was not significantly different (Table 3).

Table 2.
Phenotypic traits of the four non-transgenic cotton lines.

Table 3.
Phenotypic traits of six transgenic cotton lines and non-transgenic cotton 2074B.
2.3. Morphological Changes in Immature Ovules
Similar morphologically immature ovules at different stages were indistinguishable as seed maturity differed among bolls collected from different parts of the same plant. After the ovules were shelled and dried, the morphology of the ovules at different seed developmental stages differed notably. The sizes of dried ovules at different developmental stages between 10H1007 and 2074B were consistent (Figure S1A). Similarly, the sizes of dried ovules at different developmental stages also showed no significant difference between the GhKAR-overexpression line (13MW125), GhENR-overexpression line (13MW039), and 2074B (Figure S1B,C).
2.4. Moisture Content, Grain Weight, and Oil Content of Four Wild-Type Cotton Cultivars and Six T3 Transgenic Lines during Seed Development
Seed samples from the four wild-type cotton cultivars were collected at 20, 25, 30, 35, 40, and 45 DPA and mature stage. The seed moisture content at each developmental stage was determined. The changes in seed moisture content at the different seed developmental stages were similar among the four wild-type cotton cultivars (Table 4). The moisture content of ovules ranged from 81% to 87% at 20 and 25 DPA, 71–76% at 30 DPA, 61–65% at 35 DPA, about 55–59% at 40 DPA and 46–50% at 45 DPA. However, at maturity, the moisture content of ovules had decreased to 5–6%. When the cotton bolls developed after 45 DPA, the seeds were already mature. However, between 45 DPA and the maturity stage, the moisture content decreased by almost 50%, suggesting that seed ripening occurred.

Table 4.
Phenotypic traits for the ovule of four wild-type cotton cultivars and six T3 transgenic lines.
Moreover, the grain weight in four wild-type cotton cultivars increased by about 1 g every 5 d, but there existed no significant correlations between the grain weight and oil content in each line (Table 4). As also shown in Table 4, there were similar oil content accumulation trends among the four wild-type cultivars with different accumulation rates. Oil accumulation rapidly increased in ovules during the period of 20–30 DPA and peaked at 40 and 45 DPA, then decreased slightly at the mature stage. Among the four cultivars, 10H1041 contained the highest oil content, with oil content declining less at the maturity stage than that in the other three lines.
At the 20 DPA and 25 DPA, most grain weights of the six T3 transgenic lines were significantly higher than that of 2074B (Table 4). However, at the late developmental stages (30–45 DPA), most grain weights of the six T3 transgenic lines were significantly lower than that of 2074B, which were mainly caused by the decreased moisture content at the late stages (Table 4).
The oil contents of GhKAR, GhHAD, and GhENR overexpression transgenic cotton lines presented similar patterns (Table 4). The oil content of 13MW125 was the highest among the six transgenic lines at the mature stage and could be used as an important germplasm resource material. The net growth rate of oil content in most GhKAR and GhENR overexpression transgenic lines was higher than their receptor material (2074B) at 30–35 DPA (Table 5), indicating that GhKAR and GhENR may have important functions in oil accumulation at 30–35 DPA.

Table 5.
Net increment in oil content of ovules of T3 transgenic cotton lines at three stages of ovular development.
2.5. Expression Patterns of GhKAR, GhHAD, and GhENR in 2074B
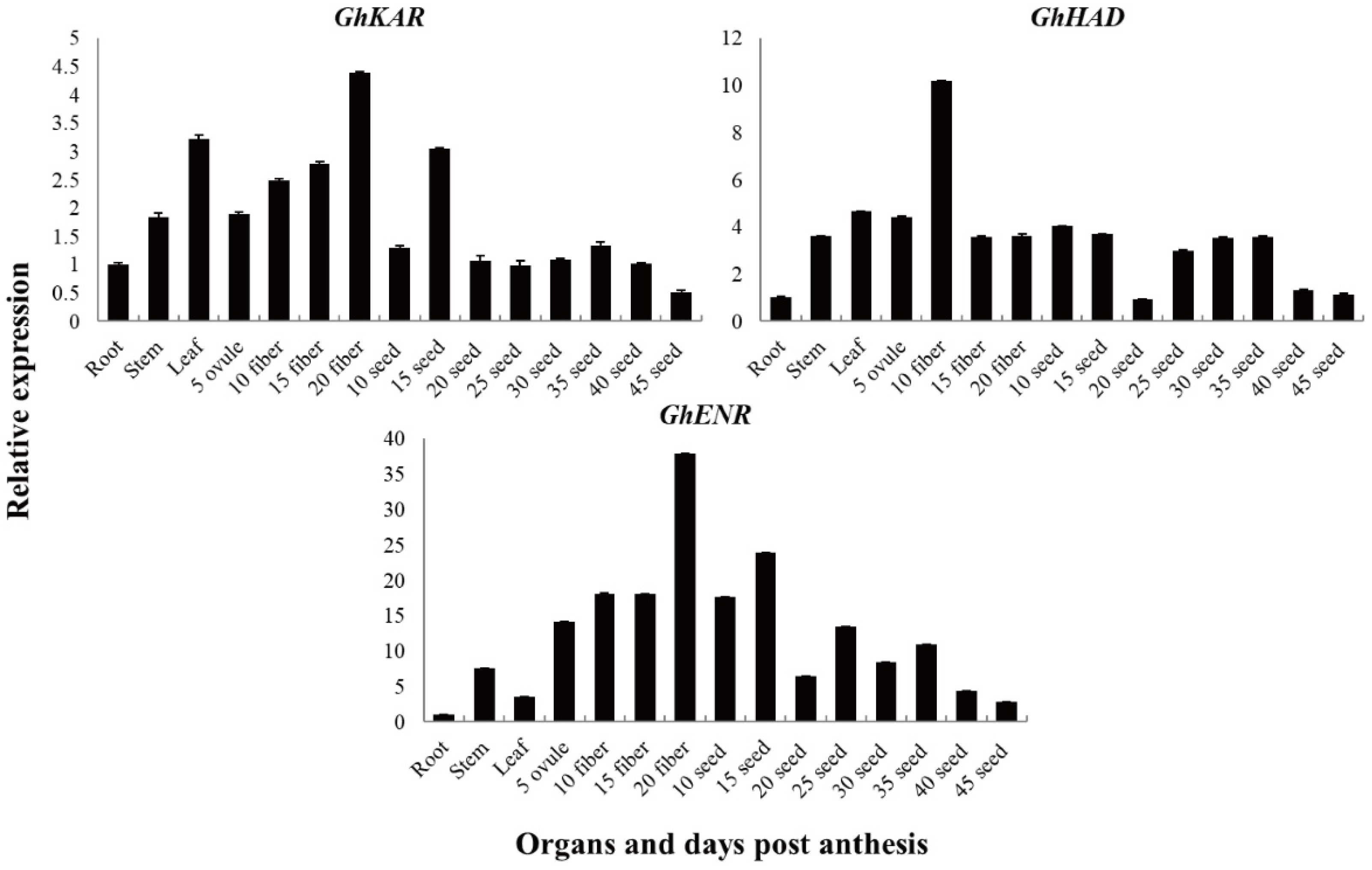
The expression patterns of GhKAR, GhHAD, and GhENR genes in different tissues were analyzed using quantitative real-time RT-PCR (qRT-PCR). To investigate the expression patterns of these three genes, which are associated with fatty acid synthesis and metabolism, in 2074B (Figure 2), total RNAs isolated from the roots, stems, leaves, fibers (5, 10, 15, and 20 DPA), and seeds (10, 15, 20, 25, 30, 35, 40, and 45 DPA), were used as templates, respectively. The housekeeping gene GhUBQ7 was used as an internal comparison gene.
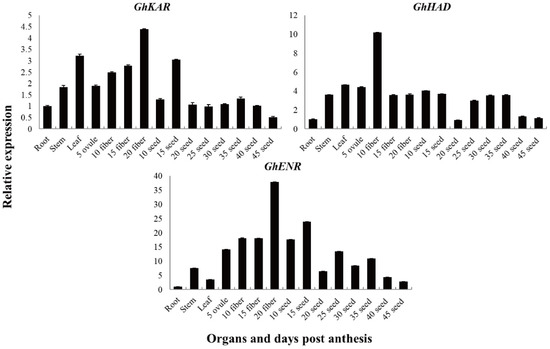
Figure 2.
Transcription patterns of GhKAR, GhHAD, and GhENR in vegetative organs, and during fiber and seed development in upland cotton 2074B. Numerals in fiber and ovule development stages indicate the number of days post-anthesis; the values and error bars the mean +/− SE of three biological replicates.
The genes GhKAR, GhHAD, and GhENR were constitutively expressed. The transcript levels of GhKAR and GhHAD were higher in the leaves than in the stems and roots, whereas the transcript levels of GhENR were significantly higher in the stems than in the leaves and roots (Figure 2). The transcript levels of GhKAR and GhENR were high in the fiber (especially in 20 DPA fiber tissue) and 15 DPA seeds. We, therefore, speculated that the expression of GhKAR and GhENR has important functions in seed oil synthesis and fiber elongation. The transcript level of GhHAD was high in fibers and seeds and especially high in 10 DPA fibers. GhHAD may have a similar function as GhKAR and GhENR but showed a different expression pattern.
2.6. Expression Patterns of GhKAR, GhHAD, and GhENR in Four Cotton Cultivars
2.6.1. Expression Patterns in Fibers
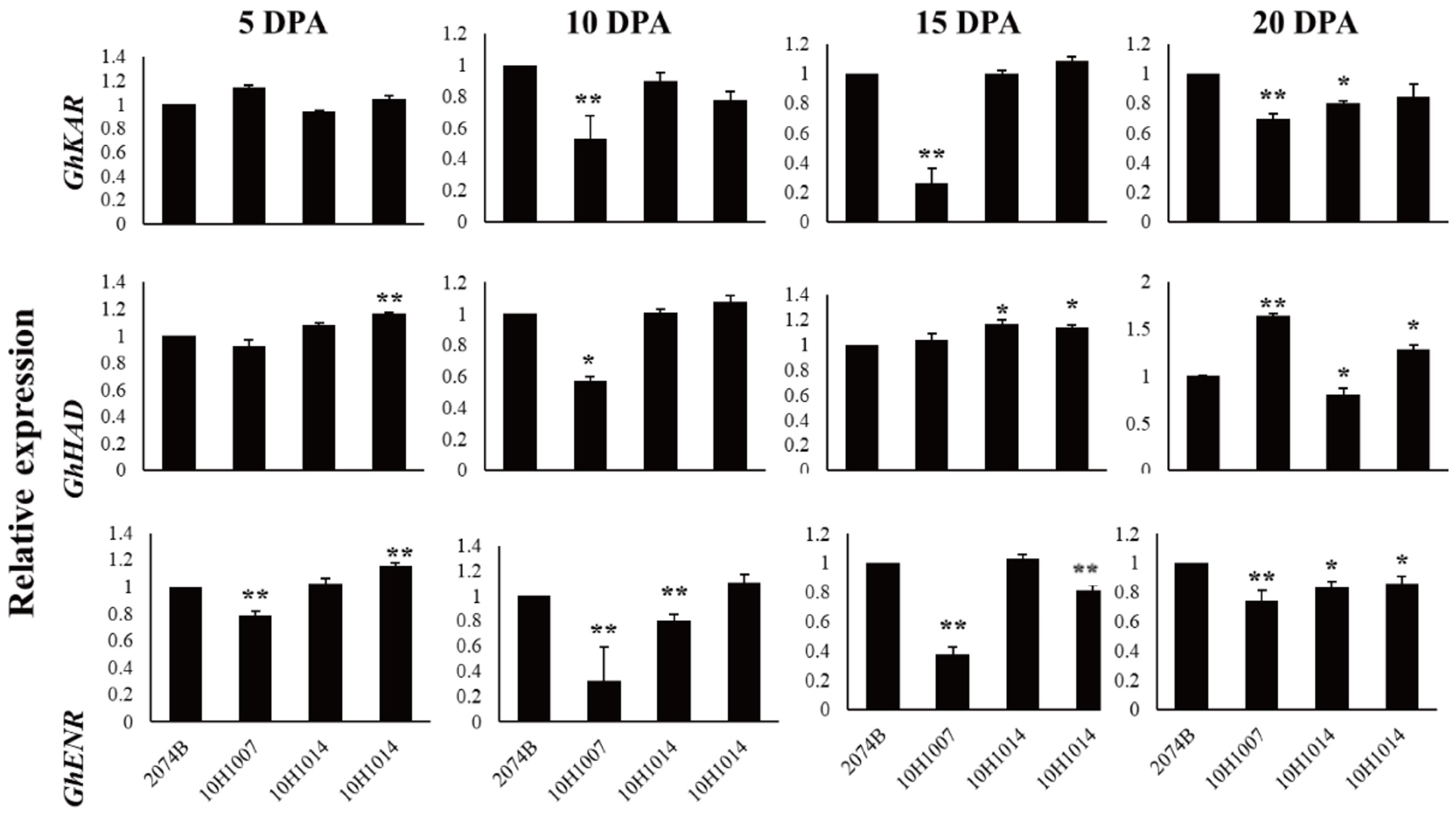
The expression patterns of GhKAR, GhHAD, and GhENR in fibers (5, 10, 15, and 20 DPA) were analyzed using qRT-PCR with the housekeeping gene GhUBQ7 as the internal comparison gene. Among the four materials, the expression patterns of the three genes varied during fiber development. The expression pattern of GhKAR and GhENR both showed an increasing trend that peaked at 20 DPA. However, the expression pattern of GhHAD differed with peaking at 10 DPA (Figure 3).
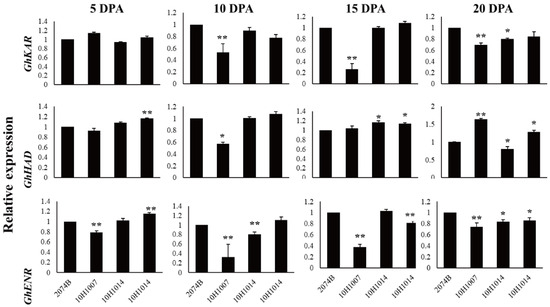
Figure 3.
Expression levels of GhKAR, GhHAD, and GhENR at different stages of fiber development in four non-transgenic cotton lines showing different seed oil contents. Values presented are the mean ± SE of three biological replicates. * and ** indicate significant differences in test of statistics (p < 0.05 and p < 0.01, respectively) compared with the 2074B value set at 1. Oil contents of the four lines were: 2074B, 26.41%; 10H1007, 28.09%; 10H1014, 30.88%; 10H1041, 35.29%. DPA, days post-anthesis.
2.6.2. Expression Patterns in Ovules
To investigate the gene expression patterns in relation to ovule development, the expression patterns of GhKAR, GhHAD, and GhENR genes in ovules (5, 10, 15, 20, 25, 30, 35, 40, and 45 DPA) were also analyzed using qRT-PCR with the housekeeping gene GhUBQ7 as an internal comparison gene.
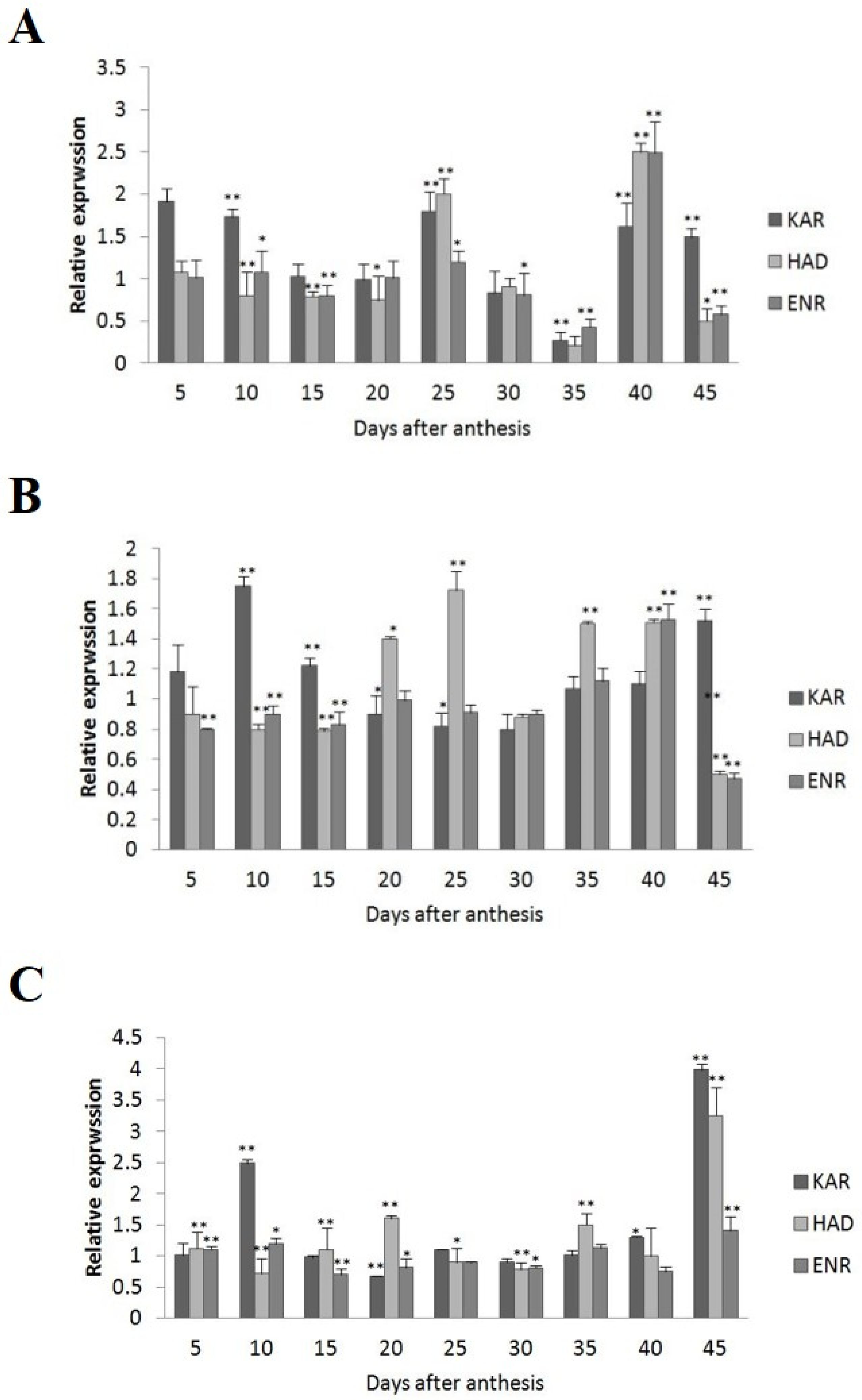
Among the four wild-type cultivars, seed oil contents were 26.41% in 2074B, 28.09% in 10H1007, 30.88% in 10H1014, and 35.29% in 10H1041. Meanwhile, the expression patterns of GhKAR, GhHAD, and GhENR genes were analyzed in developing ovules of the three wild-type cultivars compared to 2074B (Figure 4). Compared to the 2074B, GhKAR showed high expression levels at 5, 10, 25, 40, and 45 DPA, while the transcript levels of GhHAD and GhENR were higher at 25 and 40 DPA in 10H1014 (Figure 4A). As for 10H1007, the transcript level of GhKAR was almost two-fold 2074B at 10, 15, and 45 DPA; GhHAD showed high expression levels at 20, 25, 35, and 40 DPA, while GhENR only increased its transcript level at 40 DPA (Figure 4B). For 10H1041, GhKAR was expressed highly at 10, 40, and 45 DPA, GhHAD elevated its expressions at 20, 35, and 45 DPA, and GhENR increased only at 45 DPA compared with 2074B (Figure 4C). The above results showed that GhKAR, GhHAD, and GhENR genes almost presented higher expression levels at 10H1007, 10H1014, and 10H1041 cultivars with higher seed oil contents than 2074B. However, at this stage, the cultivars that showed high gene transcript levels were not necessarily consistent with the cultivars that showed low oil contents. Interestingly, at the late stage of oil accumulation (40–45 DPA), cultivars with high oil contents exhibited high gene transcript levels. We, therefore, hypothesized that the high transcript levels of GhKAR, GhHAD, and GhENR at the late oil accumulation stage may affect oil accumulation.
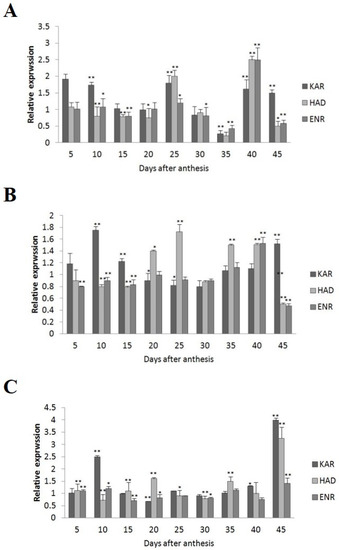
Figure 4.
Expression levels of GhKAR, GhHAD, and GhENR at different stages of ovular development in three cotton cultivars showing different seed oil contents. (A) 10H1014; (B) 10H1007; (C) 10H1041. Values presented are the mean ± SE of three biological replicates. * and ** indicate significant differences in test of statistics (p < 0.05 and p < 0.01, respectively) compared with the 2074B value set at 1. Seed oil contents of the four lines were: 2074B, 26.41%; 10H1007, 28.09%; 10H1014, 30.88%; and 10H1041, 35.29%. DPA, days post-anthesis.
3. Discussion
3.1. Genes Related to the Oil Biosynthesis and Accumulation in Cottonseed
Cotton is a leading natural resource for fiber and oil. A previous study investigated the genetic architecture of seed nutrients in upland cotton by a genome-wide association study (GWAS) method with a panel of 196 germplasm resources. They identified three candidate genes, Gh_D12G1161, Gh_D12G1162, and Gh_D12G1165, that were most likely involved in the formation of cottonseed protein and fatty acid compositions [31]. Another study used a genetic map with 388 molecular markers to identify QTL for oil content in cottonseed. Two candidate genes, Gh_A03G0701 and Gh_A03G0699, were screened at the overlapped qOil-3 region. These two genes encoded the 3-ketoacyl-CoA enzyme, which was closely related to the synthesis of oil in cottonseeds [32]. QTL-mapping and regulatory network analyses also suggested that Ghr-miR2949b, Ghr-miR2949c, and GhHSL1 were closely involved in the cottonseed oil content [19]. In addition, 83 representative upland cotton accessions grown in multiple environments were used to identify the quantitative trait loci (QTLs) underpinning cottonseed oil content and fatty acid components. Finally, three genes were screened as the candidate genes, and the ectopic expression of Gh_D01G2016 in yeast led to the sharp reduction in oil content, which suggested that the function of this gene could be verified by gene editing technology in oil biosynthesis of cottonseeds [33]. These results have widened our understanding of the genetic mechanism on cottonseed oil formation and provided important molecular tools to develop new cultivars with high fatty acid contents in cotton breeding by marker-assisted selection method.
With the advent of numerous high-quality genome sequences and omics technology platforms from Gossypium species, key genes involved in oil biosynthesis in cottonseeds have been identified. For instance, transcriptome data showed that WRI1, NF-YB6, and GhPDAT genes play important roles in the oil accumulation in fatty acid (FA) synthesis, FA desaturation, and triacylglycerol (TAG) biosynthesis [34]. Other early reports found that GhWRI1 was involved in oil metabolism. GhWRI1 showed high expressions in high oil content material, and overexpression of GhWRI1 increased seed oil content in transgenic A. thaliana, suggesting its crucial role in seed oil accumulation [35]. In addition, phosphoenolpyruvate carboxylase genes were also the potential targets for creating high cottonseed oil materials by RNAi strategy in cotton. For example, the cottonseed oil content in GhPEPC1 RNAi lines showed a significant increase without other phenotypic changes, and decreasing the GhPEPC1 expression led to the increased expression of triacylglycerol biosynthesis-related genes, which might contribute to the oil biosynthesis in cottonseeds [36]. Silencing the GhPEPC2 gene using RNAi could also increase oil accumulation in cotton seeds [37]. All these candidate genes could serve as the foundation for elucidating the molecular mechanisms of oil content formation and the genetic breeding for higher cottonseed oil content.
Genome-wide level analyses indicated that the omega-3 FAD gene family in cotton was characterized to be differentially expressed in seeds [38]. Meanwhile, the expression levels of GhCPS1 and GhCPS2 genes closely correlated with the total cyclopropenoid fatty acids (CFA) content in cottonseeds, and both of them can be considered potential targets for gene silencing to reduce undesirable seed CPE accumulation in cotton [39]. Another study showed that LPAAT genes were co-localized with quantitative trait loci (QTL) region for cottonseed oil. Overexpression of one LPAAT gene, Gh13LPAAT5, significantly increased the production of total TAG and other fatty acids [40]. Additionally, SNP variations from GhCIPKs genes were significantly associated with oil content in cotton, and overexpression of the GhCIPK6 gene reduced the oil content but increased C18:1 content in transgenic cotton [41]. These studies provided incentives for further underlying molecular mechanisms of oil accumulation in cottonseed oil but also brought new genes to increase cottonseed oil content through biotechnology.
3.2. The Application of Fatty Acid Synthase in Improving Seed Oil Content
Overexpression of Spinacia oleracea KASIII (SoKASIII) in tobacco leaves under the control of the 35S CaMV promoter and of Jatropha Curcas KASIII (ChKASIII) in A. thaliana and oilseed rape seeds under the control of the napin promoter, indicates that the 16:0 fatty acid contents of both transgenic plants were significantly increased [42]. In recent years, efforts have been made to manipulate key fatty acid synthetic genes in various species using transgenic technology [43,44,45,46,47]. Transformations using transgenes encoding key enzymes or enzyme subunits have resulted in the alteration in lipid levels to varying degrees [37,48], and in some cases, the oil content was reduced [49]. Four key enzymes were involved in the fatty acid carbon-chain extension process, namely KAS, KAR, HAD, and ENR. KAS can be divided into three categories (KASI, KASII, and KASIII) based on its catalytic substrates. KASIII serves an important function in the synthesis of C16:0 fatty acids [49]. KASII affects the C16:0 fatty acid content [50]. The mutation or deletion of KAS will affect not only the fatty acid synthesis but also the seed development [50,51]. Studies on KAR, HAD, and ENR were relatively limited. Transcriptional research works have shown that KAR, HAD, and ENR were involved in seed oil and fatty acid synthesis [27]. The ENR deletion mutant of A. thaliana suffered seed abortion and decreased fatty acid content. The deletion mutant of ENR in A. thaliana will cause seed abortion and fatty acid content decrement [30]. The mechanisms by which KAR, HAD, and ENR affect seed oil synthesis remain unclear; therefore, further studies are needed.
3.3. GhKAR, GhHAD, and GhENR Were Key Enzymes in the Synthesis of Fatty Acids
Oil accumulation is part of the seed maturation process, a highly controlled developmental program that sets in ovule tissues once morphogenesis has been achieved. The maturation process was characterized by the accumulation of storage compounds, acquisition of desiccation tolerance, and entry into a dormancy period of variable length [52]. Gene expression programs associated with these processes were activated during the maturation phase and were switched off during the vegetative phases of plant development. Studies of developing seeds and/or embryos have established that the biosynthetic pathways for fatty acids and TAGs were regulated at the transcription level [53,54,55]. In the present study, four cotton cultivars with different seed oil contents showed consistent oil accumulation trends during seed development. The overall trend of oil accumulation was that at 20–30 DPA, the oil content rapidly increased and thereafter showed slow accumulation. At 40 DPA, the oil content peaked and then slightly decreased. The decrease in oil content at maturity may be attributable to β-oxidation and several other physiological and biochemical processes, suggesting that the decline in oil accumulation during seed maturation might be overcome using molecular breeding techniques to achieve the goal of developing lines with high seed oil contents.
GhKAR, GhHAD, and GhENR were crucial genes involved in fatty acid carbon-chain extension that have been cloned from immature ovules of upland cotton [27]. The present analysis of transcript levels of these genes in different tissues and organs of 2074B indicated that the genes were highly expressed during fiber development and showed higher transcription levels than in most developing seeds. Therefore, these genes were not only involved in plant seed oil synthesis but also in the cotton fiber elongation process. The transcript levels of GhKAR and GhENR were particularly high at 20 DPA during fiber development and at 15 DPA during seed development. However, the transcript level of GhHAD was high only at 10 DPA during fiber development, which indicated that GhHAD, together with GhKAR and GhENR, showed different expression patterns during seed and fiber development. Analysis of the fatty acid compositions of the homozygous transgenic T3 and T4 generations of A. thaliana seed showed that overexpression of GhKAR, GhHAD, or GhENR could increase the total content of fatty acids in the seed. The total fatty acid content of transgenic GhKAR, GhHAD, and GhENR lines were 6.59%, 7.76%, and 3.86% higher than that of wild-type A. thaliana, respectively. Thus, overexpression of GhKAR, GhHAD, or GhENR may improve the total fatty acid content by increasing the concentration of every fatty acid because GhKAR, GhHAD, and GhENR are genes involved in fatty acid synthesis. Further studies were needed to verify this hypothesis and the molecular functions of the three genes.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Four cotton cultivars (2074B, 10H1004, 10H1007, and 10H1041), and T3 transgenic cotton lines that overexpression GhKAR (13MW012, 13MW124, and 13MW125) and GhENR (13MW005, 13MW039, and 13MW070), were used in our experiments.
Plants were grown on soil in a greenhouse (16 h light/8 h dark) at 28 °C. After three weeks, the roots, stems, and leaves of seedlings at the three-leaf stage were harvested, frozen in liquid nitrogen, and stored at −80 °C for DNA and total RNA extraction. Seeds were grown in the field in Hejian, from which samples of fibers (5, 10, 15, and 20 DPA), and ovules (5, 10, 15, 20, 25, 30, 35, 40, and 45 DPA) of the different materials were collected.
4.2. Field Experiments and Agronomic Trait Investigation
For field experiments, non-transgenic and transgenic plants were sown directly on April 27th at Hejian, Hebei Province (38°43′ N, 116°09′ E). The field planting followed a randomized complete block design with three replications. Two-row plots with 80 cm and 50 cm row spacing were used. The length of each plot was 4m. Field management followed conventional standard field practices. Data were collected from at least 10 plants in each line. Self-pollinated bells were harvested from each primary transgenic (T3) plant and analyzed for quality character of fibers and seed oil content. The cottonseed oil content was determined using the Soxhlet extraction method and near-infrared spectroscopy [30]. All results were statistically analyzed by three times repeats. LSD test and difference significance test of statistics method were adopted in final result analysis.
4.3. RNA Isolation and cDNA Synthesis
Total RNA was extracted using a modified CTAB-SDS method [56]. RNA samples were treated with DNase I (Ambion, Austin, TX, USA) in accordance with the manufacturer’s instructions to remove genomic DNA contaminants. Total RNA samples (1 µg per reaction) were reverse transcribed into cDNA by avian myeloblastosis virus (AMV) reverse transcriptase. The cDNAs were used as the template in subsequent qPCR reactions.
4.4. Quantitative Real-Time RT-PCR
Total RNA was extracted from young leaves, roots, stems, and developing fibers (5, 10, 15, and 20 DPA) and ovules (5, 10, 15, 20, 25, 30, 35, 40, and 45 DPA) as indicated above. Gene-specific primers were designed to amplify PCR products of ~200 bp in length (see Supplementary Table S1). The relative level of gene expression was estimated using the 2−△△CT method [57]. The analyses were performed with three biological replicates using samples from different plants. The SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa, Biotechnology (Dalian, China) Co., Ltd.) was used for RT-PCR.
The expression patterns of GhKAR, GhHAD, and GhENR in different tissues were studied using qRT-PCR. To investigate the expression patterns of the three genes in relation to fatty acid synthesis and metabolism in 2074B, total RNAs isolated from the root, stem, young leaves, and ovule, as well as from fibers (5, 10, 15, and 20 DPA), and ovules (5, 10, 15, 20, 25, 30, 35, 40, and 45 DPA), of 2074B were used as templates. In addition, G. hirsutum UBQ7 gene used to normalize served as an endogenous reference. Data are presented as the means (±SD) of three independent experiments.
4.5. Cotton Transformation
The coding sequences of GhKAR, GhHAD, and GhENR genes were amplified with the Long and Accurate polymerase (TaKaRa, Tokyo, Japan) from cDNA library and then inserted into the pCAMBIA 2301 vector. Cotton transformation was conducted using the pollen tube pathway method with cv. Sumian 20 as a receptor [58,59,60]. In brief, the flowers pollinated for 36–48 h were selected, and 0.1~0.2 μg plasmid DNA dissolved in double distilled water was injected into each ovule. The bolls injected with plasmid DNA were tagged. The T1 seeds of the transgenic plants were harvested and planted in the field, and then the transformants were primarily discriminated by checking the resistance to kanamycin (2 g/dm3) by spraying in the field during cotyledon and three leaves stages. The T1 plants showing kanamycin resistance were further selected for PCR and Southern blot analyses.
4.6. Identification of the Phenotypic Traits
Plant height was recorded by measuring the main stem height of individuals [61]. Fiber quality traits, including the fiber length (mm), fiber uniformity ratio (%), fiber strength (cN/tex), fiber elongation, and micronaire, were measured with an HVI 900 instrument (USTER HVISPECTRUM, SPINLAB, USA) at the Cotton Fiber Quality Inspection and Test Center of Ministry of Agriculture (Anyang, China) [62]. Boll samples were ginned for seed index, and one hundred cottonseeds were randomly selected from each line and weighed as seed index (SI, g) [14]. Cottonseeds delinted with concentrated sulphuric acid were used to measure oil content. The oil contents at the different stages of ovular development of the transgenic lines were measured using the Soxhlet extraction method [63]. The moisture content in cottonseed was analyzed by near-infrared reflectance spectroscopy (NIRS) technique [64]. Grain weight was recorded by weighing one hundred cotton ovules randomly.
4.7. Southern Blotting Analysis
Procedure for Southern blot was described briefly, as follows. Kan fragment was amplified by PCR (500 bp), and the concentration of recovered DNA was diluted to 60–70 ng/µL. Add 16 µL Kan fragment in a 200 µL centrifuge tube, bathe in boiling water for 10 min, quickly place on ice containing sodium chloride for 10 min, and centrifuge instantaneously. Adding 4 µL DIG-High-Prime, instantaneous centrifugation, and 37 °C water bath for 20 h; then, 2 µL 0.2 M EDTA (pH8.0) (or 10 min in 65 °C water bath) was added to terminate the reaction and stored at −20 °C. Probe denaturation: Add 5 µL labeled probe into 200 µL centrifuge tube, denaturate in water bath at 68 °C for 10 min after sealing with sealing film, and cool rapidly in ice water for 5 min (denaturation before use). The subsequent enzymatic digestion of cotton genomic DNA, treatment of digested products, electrophoresis, transmembrane and crosslinking, prehybridization and hybridization, rigor elution, detection reaction, and color reaction were finished according to the protocol of Roche Southern blot kit.
4.8. Statistical Analysis
The SPSS 13.0 statistical package (IBM Corporation, New York, NY, USA) was used for the analysis of variance and Student’s t-test. The normality test was checked by using the Shapiro-Wilk test to prove the data to satisfy the Gaussian distribution. The significance was tested using the least significant difference (LSD) at the 1% or 5% levels. Each sample included in the analysis was based on three biological replicates.
5. Conclusions
In this study, we observed that GhKAR and GhENR showed similar expression trends during fiber and seed development in three cultivars with different oil contents. The transcript levels of GhKAR and GhENR gradually increased and peaked at 20 DPA during fiber development. Considering that 10–20 DPA is a period of rapid fiber elongation, GhKAR and GhENR might have important functions at the late stage of rapid fiber elongation. However, the expression level of GhHAD peaked at 10 DPA, which suggested that GhHAD plays an important role in the early stage of rapid fiber elongation. Our results provided new insights into the fatty acid biosynthesis in cotton.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12193437/s1. Figure S1. Morphological changes in immature ovules of cotton at different developmental stages. (A) 10H1007 and 2074 (non-transgenic lines); (B) 13MW125 (GhKAR-overexpression line) and 2074B; (C) 13MW039 (GhENR-overexpression line) and 2074B. DPA, days post-anthesis. Figure S2. Phylogram of GhKAR protein sequences from different organisms; Figure S3. Phylogram of Gh HAD protein sequences from different organisms; Figure S4. Phylogram of GhENR protein sequences from different organisms; Table S1. Primers used in the experiment.
Author Contributions
Conceptualization, L.L., J.H., Y.Y. and Z.C.; Data curation, L.L., D.W., F.Z. and W.L.; Formal analysis, D.W., X.W., J.W., A.S. and Z.C.; Funding acquisition, J.H. and Y.Y.; Investigation, L.L., D.W., X.K., X.W., A.S. and F.Z.; Methodology, L.L., D.W., X.K., X.W., J.W., A.S. and F.Z.; Project administration, J.H. and Y.Y.; Resources, L.L., D.W. and W.L.; Software, L.L., D.W., X.K., X.W., A.S. and Z.C.; Supervision, J.H., Y.Y. and Z.C.; Validation, L.L., D.W., X.K., X.W., J.W., F.Z., W.L. and Z.C.; Visualization, J.H. and Z.C.; Writing—original draft, L.L. and Z.C.; Writing—review and editing, J.H., Y.Y. and Z.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the National Natural Science Foundation of China (NO. 31360349), and the National Key Research and Development Program of China (No. 2022YFD1200300). The funding bodies did not participate in the design of the study; the collection, analysis, or interpretation of the data; or in the writing of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are deeply indebted to Lida Zhang for helpful suggestions and comments on bioinformatic analyses, and we thank Yi Huang for valuable comments on previous versions of the manuscript. We are also grateful to two anonymous reviewers for their helpful suggestions and comments.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
TAG: Triacylglycerol; FAS, Fatty acid synthase; β-ketone fatty acyl-ACP reductase (KAR); β-hydroxy fatty acyl-ACP dehydratase (HAD), and enoyl-ACP reductase (ENR); β-ketone fatty acyl-ACP synthase (KAS); Acetyl-CoA carboxylase (ACCase); acetyl-coenzyme A (CoA)-acyl carrier protein (ACP) transferase (ACAT); malonic acid single acyl-CoA-ACP transferase (MCAT); DPA, days post-anthesis; G. hirsutum, Gossypium hirsutum; A. thaliana, Arabidopsis thaliana; qRT-PCR, quantitative real-time polymerase chain reaction.
References
- Zhang, F.; Li, S.; Yang, S.; Wang, L.; Guo, W. Overexpression of a cotton annexin gene, GhAnn1, enhances drought and salt stress tolerance in transgenic cotton. Plant Mol. Biol. 2015, 87, 47–67. [Google Scholar] [CrossRef]
- Gao, L.; Chen, W.; Xu, X.; Zhang, J.; Singh, T.K.; Liu, S.; Zhang, D.; Tian, L.; White, A.; Shrestha, P.; et al. Engineering trienoic fatty acids into cottonseed oil improves low-temperature seed germination, plant photosynthesis and cotton fiber quality. Plant Cell Physiol. 2020, 61, 1335–1347. [Google Scholar] [CrossRef]
- Khan, A.; Tan, D.K.Y.; Munsif, F.; Afridi, M.Z.; Shah, F.; Wei, F.; Fahad, S.; Zhou, R. Nitrogen nutrition in cotton and control strategies for greenhouse gas emissions: A review. Environ. Sci. Pollut. Res. Int. 2017, 24, 23471–23487. [Google Scholar] [CrossRef]
- Mubeen, M.; Ahmad, A.; Hammad, H.M.; Awais, M.; Farid, H.U.; Saleem, M.; ul Din, M.S.; Amin, A.; Ali, A.; Fahad, S.; et al. Evaluating the climate change impact on water use efficiency of cotton-wheat in semi-arid conditions using DSSAT model. J. Water Clim. Change 2019, 11, 1661–1675. [Google Scholar] [CrossRef]
- Lee, M.K.; Zhang, Y.; Zhang, M.; Goebel, M.; Kim, H.J.; Triplett, B.A.; Stelly, D.M.; Zhang, H.B. Construction of a plant-transformation-competent BIBAC library and genome sequence analysis of polyploid Upland cotton (Gossypium hirsutum L.). BMC Genom. 2013, 14, 208. [Google Scholar] [CrossRef]
- Wendel, J.F.; Grover, C.E. Taxonomy and evolution of the cotton genus. In Cotton Agronomy; Monograph 24; Fang, D., Percy, R., Eds.; ASA-CSSA-SSSA: Madison, WI, USA, 2015. [Google Scholar]
- Wang, K.; Wendel, J.F.; Hua, J. Designations for individual genomes and chromosomes in Gossypium. J. Cotton Res. 2018, 1, 3. [Google Scholar] [CrossRef]
- Chen, Z.; Grover, C.E.; Li, P.; Wang, Y.; Nie, H.; Zhao, Y.; Wang, M.; Liu, F.; Zhou, Z.; Wang, X.; et al. Molecular evolution of the plastid genome during diversification of the cotton genus. Mol. Phylogenet. Evol. 2017, 112, 268–276. [Google Scholar] [CrossRef]
- Huang, G.; Huang, J.Q.; Chen, X.Y.; Zhu, Y.X. Recent advances and future perspectives in cotton research. Annu. Rev. Plant Biol. 2021, 72, 437–462. [Google Scholar] [CrossRef]
- Khan, A.; Tan, D.K.Y.; Afridi, M.Z.; Luo, H.; Tung, S.A.; Ajab, M.; Fahad, S. Nitrogen fertility and abiotic stresses management in cotton crop: A review. Environ. Sci. Pollut. Res. Int. 2017, 24, 14551–14566. [Google Scholar] [CrossRef]
- Liu, Q.; Singh, S.P.; Green, A.G. High-stearic and High-oleic cottonseed oils produced by hairpin RNA-mediated post-transcriptional gene silencing. Plant Physiol. 2002, 129, 1732–1743. [Google Scholar] [CrossRef]
- Radcliffe, J.D.; King, C.C.; Czajka-Narins, D.M.; Imrhan, V. Serum and liver lipids in rats fed diets containing corn oil, cottonseed oil, or a mixture of corn and cottonseed oils. Plant Foods Hum. Nutr. 2001, 56, 51–60. [Google Scholar] [CrossRef]
- Royon, D.; Daz, M.; Ellenrieder, G.; Locatelli, S. Enzymatic production of biodiesel from cotton seed oil using t-butanol as a solvent. Bioresour. Technol. 2007, 98, 648–653. [Google Scholar] [CrossRef]
- Shang, L.; Abduweli, A.; Wang, Y.; Hua, J. Genetic analysis and QTL mapping of oil content and seed index using two recombinant inbred lines and two backcross populations in Upland cotton. Plant Breed. 2016, 135, 224–231. [Google Scholar] [CrossRef]
- Nabi, M.N.; Rahman, M.M.; Akhter, M.S. Biodiesel from cotton seed oil and its effect on engine performance and exhaust emissions. Appl. Therm. Eng. 2009, 29, 2265–2270. [Google Scholar] [CrossRef]
- Wu, M.; Pei, W.; Wedegaertner, T.; Zhang, J.; Yu, J. Genetics, breeding and genetic engineering to improve cottonseed oil and protein: A review. Front. Plant Sci. 2022, 13, 864850. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Xu, X.; Li, J.; Zhang, J.; Chang, S.; Yang, X.; Guo, X. Seed-specific overexpression of cotton GhDGAT1 gene leads to increased oil accumulation in cottonseed. Crop J. 2021, 9, 487–490. [Google Scholar] [CrossRef]
- Zhu, D.; Le, Y.; Zhang, R.; Li, X.; Lin, Z. A global survey of the gene network and key genes for oil accumulation in cultivated tetraploid cottons. Plant Biotechnol. J. 2021, 19, 1170–1182. [Google Scholar] [CrossRef]
- Zhang, Z.; Gong, J.; Zhang, Z.; Gong, W.; Li, J.; Shi, Y.; Liu, A.; Ge, Q.; Pan, J.; Fan, S.; et al. Identification and analysis of oil candidate genes reveals the molecular basis of cottonseed oil accumulation in Gossypium hirsutum L. Theor. Appl. Genet. 2022, 135, 449–460. [Google Scholar] [CrossRef]
- Shang, X.; Cheng, C.; Ding, J.; Guo, W. Identification of candidate genes from the SAD gene family in cotton for determination of cottonseed oil composition. Mol. Genet. Genom. 2017, 292, 173–186. [Google Scholar] [CrossRef]
- Song, J.; Pei, W.; Wang, N.; Ma, J.; Xin, Y.; Yang, S.; Wang, W.; Chen, Q.; Zhang, J.; Yu, J.; et al. Transcriptome analysis and identification of genes associated with oil accumulation in upland cotton. Physiol. Plant 2022, 174, e13701. [Google Scholar] [CrossRef]
- Brown, A.P.; Slabas, A.R.; Rafferty, J.B. Fatty acid biosynthesis in plants—Metabolic pathways, structure and organization. In Lipids in Photosynthesis: Essential and Regulatory Functions; Wada, H., Murata, N., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 11–34. [Google Scholar]
- Ryall, K.; Harper, J.T.; Keeling, P.J. Plastid-derived Type II fatty acid biosynthetic enzymes in chromists. Gene 2003, 313, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Rogalski, M.; Carrer, H. Engineering plastid fatty acid biosynthesis to improve food quality and biofuel production in higher plants. Plant Biotechnol. J. 2011, 9, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhao, Y.; Wang, Y.; Liu, Z.; Ijaz, B.; Huang, Y.; Hua, J. Genome-wide identification and expression analysis of the biotin carboxyl carrier subunits of heteromeric acetyl-CoA carboxylase in Gossypium. Front. Plant Sci. 2017, 8, 624. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Han, J.; Sang, M.; Li, A.; Wu, H.; Yin, S.; Zhang, C. De novo transcriptomic analysis of an oleaginous microalga: Pathway description and gene discovery for production of next-generation biofuels. PLoS ONE 2012, 7, e35142. [Google Scholar] [CrossRef] [PubMed]
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.F.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef]
- Troncoso-Ponce, M.A.; Kilaru, A.; Cao, X.; Durrett, T.P.; Fan, J.; Jensen, J.K.; Thrower, N.A.; Pauly, M.; Wilkerson, C.; Ohlrogge, J.B. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011, 68, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ji, Y.; Wang, Y.; Zhang, X.; Yu, Y. Cloning and characterization of a novel β-ketoacyl-ACP reductase from Comamonas testosteroni. Chem. Biol. Interact. 2015, 234, 213–220. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, P.; Wang, D.; Liu, Z.-J.; Wang, Y.-M.; Hua, J. Molecular cloning and expression analysis of three chain extension genes related to fatty acid synthesis in Upland cotton (Gossypium hirsutum L.). Sci. Agric. Sin. 2013, 46, 3523–3533. [Google Scholar]
- Yuan, Y.; Wang, X.; Wang, L.; Xing, H.; Wang, Q.; Saeed, M.; Tao, J.; Feng, W.; Zhang, G.; Song, X.-L.; et al. Genome-wide association study identifies candidate genes related to seed oil composition and protein content in Gossypium hirsutum L. Front. Plant Sci. 2018, 9, 1359. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, L.; Mei, L.; Quampah, A.; He, Q.; Zhang, B.; Sun, W.; Zhang, X.; Shi, C.; Zhu, S. qOil-3, a major QTL identification for oil content in cottonseed across genomes and its candidate gene analysis. Ind. Crops Prod. 2020, 145, 112070. [Google Scholar] [CrossRef]
- Xin, Y.; Ma, J.; Song, J.; Jia, B.; Yang, S.; Wu, L.; Huang, L.; Pei, W.; Wang, L.; Yu, J.; et al. Genome wide association study identifies candidate genes related to fatty acid components in upland cotton (Gossypium hirsutum L.). Ind. Crops Prod. 2022, 183, 114999. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Y.; Huang, Y.; Cui, Y.; Hua, J. Gene network of oil accumulation reveals expression profiles in developing embryos and fatty acid composition in Upland cotton. J. Plant Physiol. 2018, 228, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, Z.; Wang, X.; Wang, Y.; Hua, J. Molecular characterization and expression analysis of GhWRI1 in Upland cotton. J. Plant Biol. 2018, 61, 186–197. [Google Scholar] [CrossRef]
- Xu, Z.; Li, J.; Guo, X.; Jin, S.; Zhang, X. Metabolic engineering of cottonseed oil biosynthesis pathway via RNA interference. Sci. Rep. 2016, 6, 33342. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Huang, Y.; Wang, Y.; Cui, Y.; Liu, Z.; Hua, J. RNA interference of GhPEPC2 enhanced seed oil accumulation and salt tolerance in Upland cotton. Plant Sci. 2018, 271, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, O.P.; Park, S.; Ilut, D.C.; Inmon, J.J.; Millhollon, J.C.; Liechty, Z.; Page, J.T.; Jenks, M.A.; Chapman, K.D.; Udall, J.A.; et al. Genome-wide analysis of the omega-3 fatty acid desaturase gene family in Gossypium. BMC Plant Biol. 2014, 14, 312. [Google Scholar] [CrossRef]
- Yu, X.-H.; Rawat, R.; Shanklin, J. Characterization and analysis of the cotton cyclopropane fatty acid synthase family and their contribution to cyclopropane fatty acid synthesis. BMC Plant Biol. 2011, 11, 97. [Google Scholar] [CrossRef]
- Wang, N.; Ma, J.; Pei, W.; Wu, M.; Li, H.; Li, X.; Yu, S.; Zhang, J.; Yu, J. A genome-wide analysis of the lysophosphatidate acyltransferase (LPAAT) gene family in cotton: Organization, expression, sequence variation, and association with seed oil content and fiber quality. BMC Genom. 2017, 18, 218. [Google Scholar] [CrossRef]
- Cui, Y.; Su, Y.; Wang, J.; Jia, B.; Wu, M.; Pei, W.; Zhang, J.; Yu, J. Genome-wide characterization and analysis of CIPK gene family in two cultivated allopolyploid cotton species: Sequence variation, association with seed oil content, and the role of GhCIPK6. Int. J. Mol. Sci. 2020, 21, 863. [Google Scholar] [CrossRef]
- Yu, N.; Xiao, W.F.; Zhu, J.; Chen, X.Y.; Peng, C.C. The Jatropha curcas KASIII gene alters fatty acid composition of seeds in Arabidopsis thaliana. Biol. Plant 2015, 59, 773–782. [Google Scholar] [CrossRef]
- Zou, J.; Katavic, V.; Giblin, E.M.; Barton, D.L.; MacKenzie, S.L.; Keller, W.A.; Hu, X.; Taylor, D.C. Modification of seed oil content and acyl composition in the brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell 1997, 9, 909–923. [Google Scholar] [CrossRef] [PubMed]
- Arias-Gaguancela, O.; Aziz, M.; Chapman, K.D. Fatty acid amide hydrolase and 9-lipoxygenase modulate cotton seedling growth by ethanolamide oxylipin levels. Plant Physiol. 2022, 191, 1234–1253. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Chen, X.; Xue, H.; Jia, T.; Meng, F.; Liu, Y.; Luo, X.; Xiao, G.; Zhu, S. GhBZR3 suppresses cotton fiber elongation by inhibiting very-long-chain fatty acid biosynthesis. Plant J. 2022, 111, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Shorrosh, B.S.; Roesler, K.R.; Shintani, D.; van de Loo, F.J.; Ohlrogge, J.B. Structural analysis, plastid localization, and expression of the biotin carboxylase subunit of acetyl-coenzyme A carboxylase from tobacco. Plant Physiol. 1995, 108, 805–812. [Google Scholar] [CrossRef]
- Jako, C.; Kumar, A.; Wei, Y.; Zou, J.; Barton, D.L.; Giblin, E.M.; Covello, P.S.; Taylor, D.C. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001, 126, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hu, X.; Wang, P.; Gao, L.; Pei, Y.; Ge, Z.; Ge, X.; Li, F.; Hou, Y. GhPLP2 positively regulates cotton resistance to verticillium wilt by modulating fatty acid accumulation and jasmonic acid signaling pathway. Front. Plant Sci. 2021, 12, 749630. [Google Scholar] [CrossRef]
- Dehesh, K.; Tai, H.; Edwards, P.; Byrne, J.; Jaworski, J.G. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001, 125, 1103–1114. [Google Scholar] [CrossRef]
- Pidkowich, M.S.; Nguyen, H.T.; Heilmann, I.; Ischebeck, T.; Shanklin, J. Modulating seed β-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc. Natl. Acad. Sci. USA 2007, 104, 4742–4747. [Google Scholar] [CrossRef]
- Wu, G.Z.; Xue, H.W. Arabidopsis β-ketoacyl-[acyl carrier protein] synthase i is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell 2010, 22, 3726–3744. [Google Scholar] [CrossRef]
- Baud, S.; Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 2010, 49, 235–249. [Google Scholar] [CrossRef]
- Baud, S.; Graham, I.A. A spatiotemporal analysis of enzymatic activities associated with carbon metabolism in wild-type and mutant embryos of Arabidopsis using in situ histochemistry. Plant J. 2006, 46, 155–169. [Google Scholar] [CrossRef]
- Fawcett, T.; Simon, W.J.; Swinhoe, R.; Shanklin, J.; Nishida, I.; Christie, W.W.; Slabas, A.R. Expression of mRNA and steady-state levels of protein isoforms of enoyl-ACP reductase from Brassica napus. Plant Mol. Biol. 1994, 26, 155–163. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, P.; Slabas, A.R.; Fawcett, T. Fatty acid and lipid biosynthetic genes are expressed at constant molar ratios but different absolute levels during embryogenesis. Plant Physiol. 2002, 129, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Vennapusa, A.R.; Somayanda, I.M.; Doherty, C.J.; Jagadish, S.V.K. A universal method for high-quality RNA extraction from plant tissues rich in starch, proteins and fiber. Sci. Rep. 2020, 10, 16887. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, B.; Wang, Q. Cotton transformation via pollen tube pathway. In Transgenic Cotton: Methods and Protocols; Zhang, B., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 71–77. [Google Scholar]
- Wang, M.; Sun, R.; Zhang, B.; Wang, Q. Pollen tube pathway-mediated cotton transformation. Methods Mol. Biol. 2019, 1902, 67–73. [Google Scholar]
- Liu, Z.J.; Zhao, Y.P.; Liang, W.; Cui, Y.P.; Wang, Y.M.; Hua, J.P. Over-expression of transcription factor GhWRI1 in upland cotton. Biol. Plant 2018, 62, 335–342. [Google Scholar] [CrossRef]
- Shang, L.; Liu, F.; Wang, Y.; Abduweli, A.; Cai, S.; Wang, K.; Hua, J. Dynamic QTL mapping for plant height in Upland cotton (Gossypium hirsutum). Plant Breed. 2015, 134, 703–712. [Google Scholar] [CrossRef]
- Shang, L.; Liang, Q.; Wang, Y.; Wang, X.; Wang, K.; Abduweli, A.; Ma, L.; Cai, S.; Hua, J. Identification of stable QTLs controlling fiber traits properties in multi-environment using recombinant inbred lines in Upland cotton (Gossypium hirsutum L.). Euphytica 2015, 205, 877–888. [Google Scholar] [CrossRef]
- García-Ayuso, L.E.; Velasco, J.; Dobarganes, M.C.; Luque de Castro, M.D. Determination of the oil content of seeds by focused microwave-assisted soxhlet extraction. Chromatographia 2000, 52, 103–108. [Google Scholar] [CrossRef]
- Zhang, X.F.; Yu, X.; Yan, J.X.; Ni, Y. Measurement of moisture and oil content in gross cottonseed based on near-infrared reflectance technique by open detecting mode. Guang Pu Xue Yu Guang Pu Fen Xi 2007, 27, 473–476. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).