Immune Responses and Transcriptomic Analysis of Nilaparvata lugens against Metarhizium anisopliae YTTR Mediated by Rice Ragged Stunt Virus
Abstract
1. Introduction
2. Results
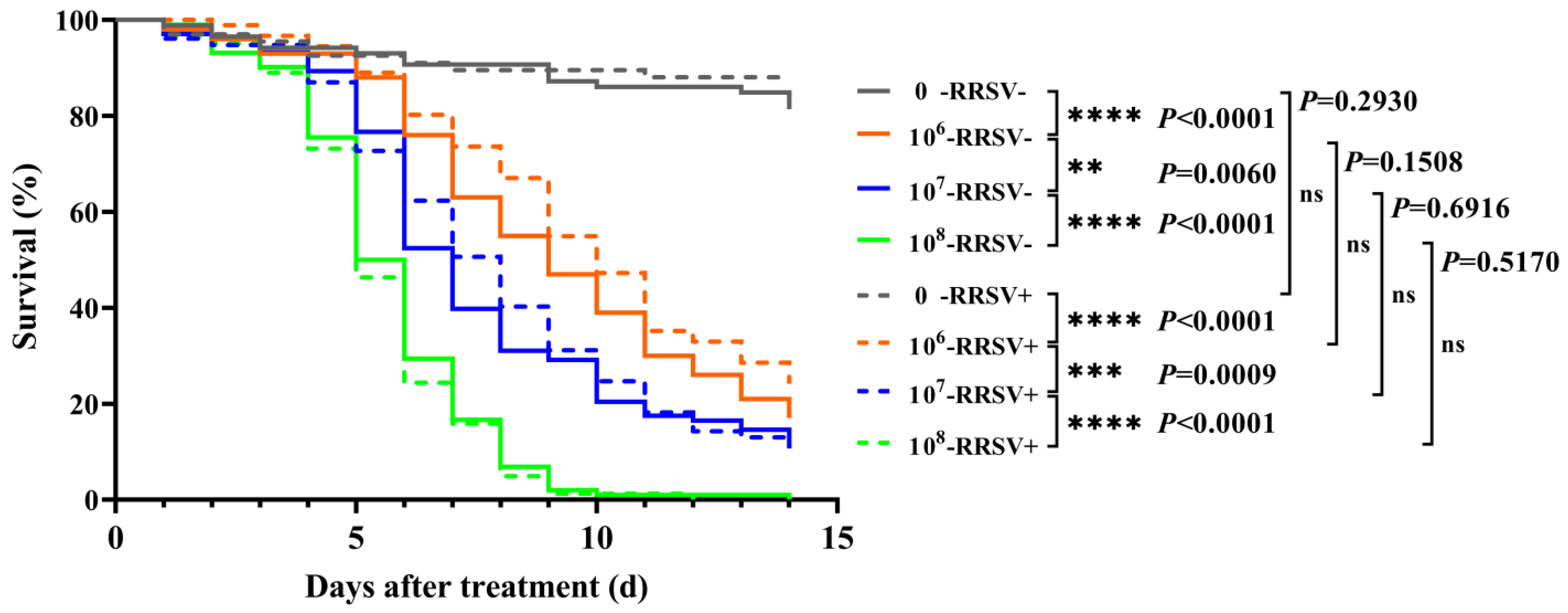
2.1. Effect of RRSV on the Lethality of Metarhizium anisopliae YTTR against BPH
2.2. Transcriptome Data Quality Control and Gene Annotation Analysis
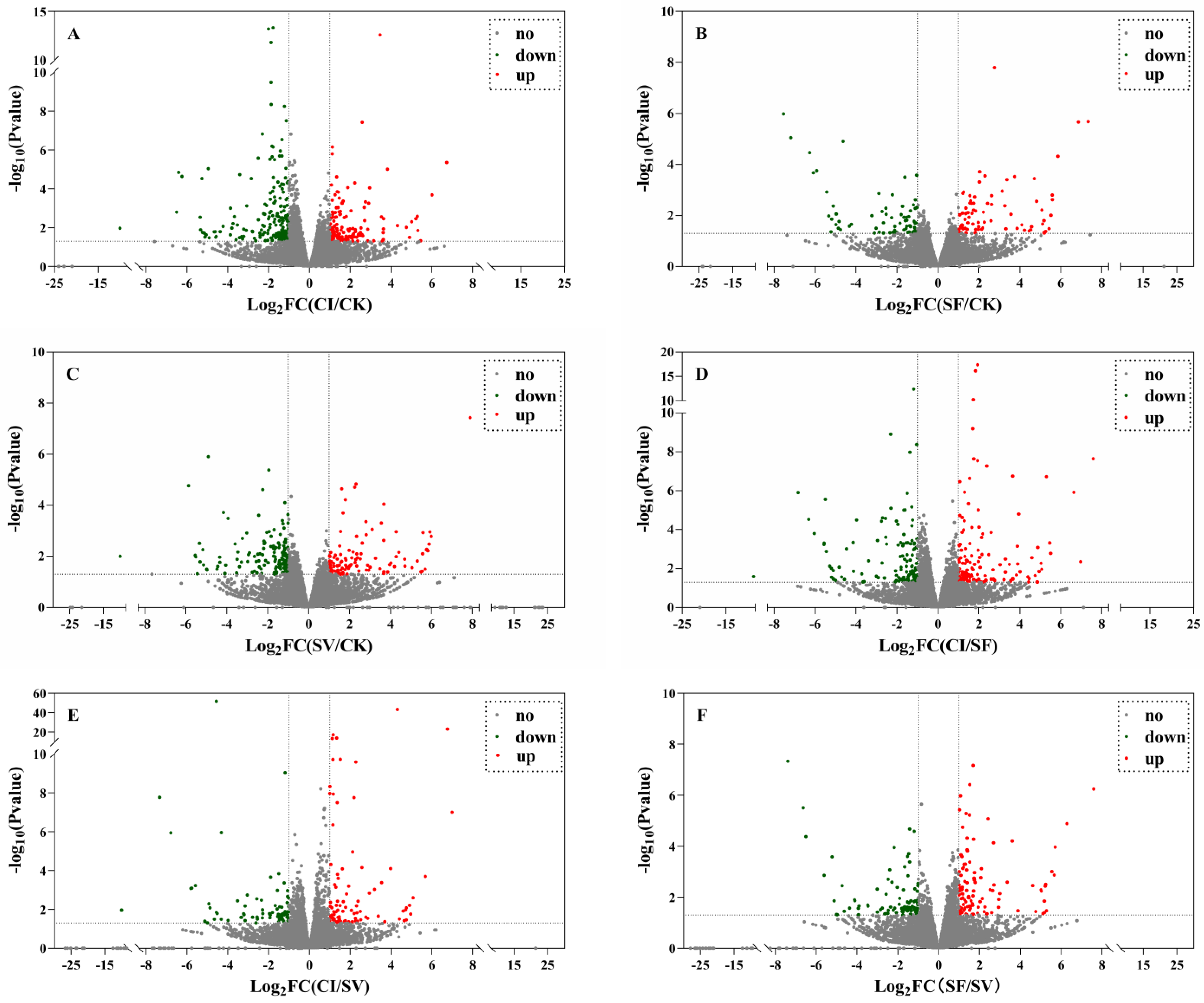
2.3. Differential Expressed Genes Analysis
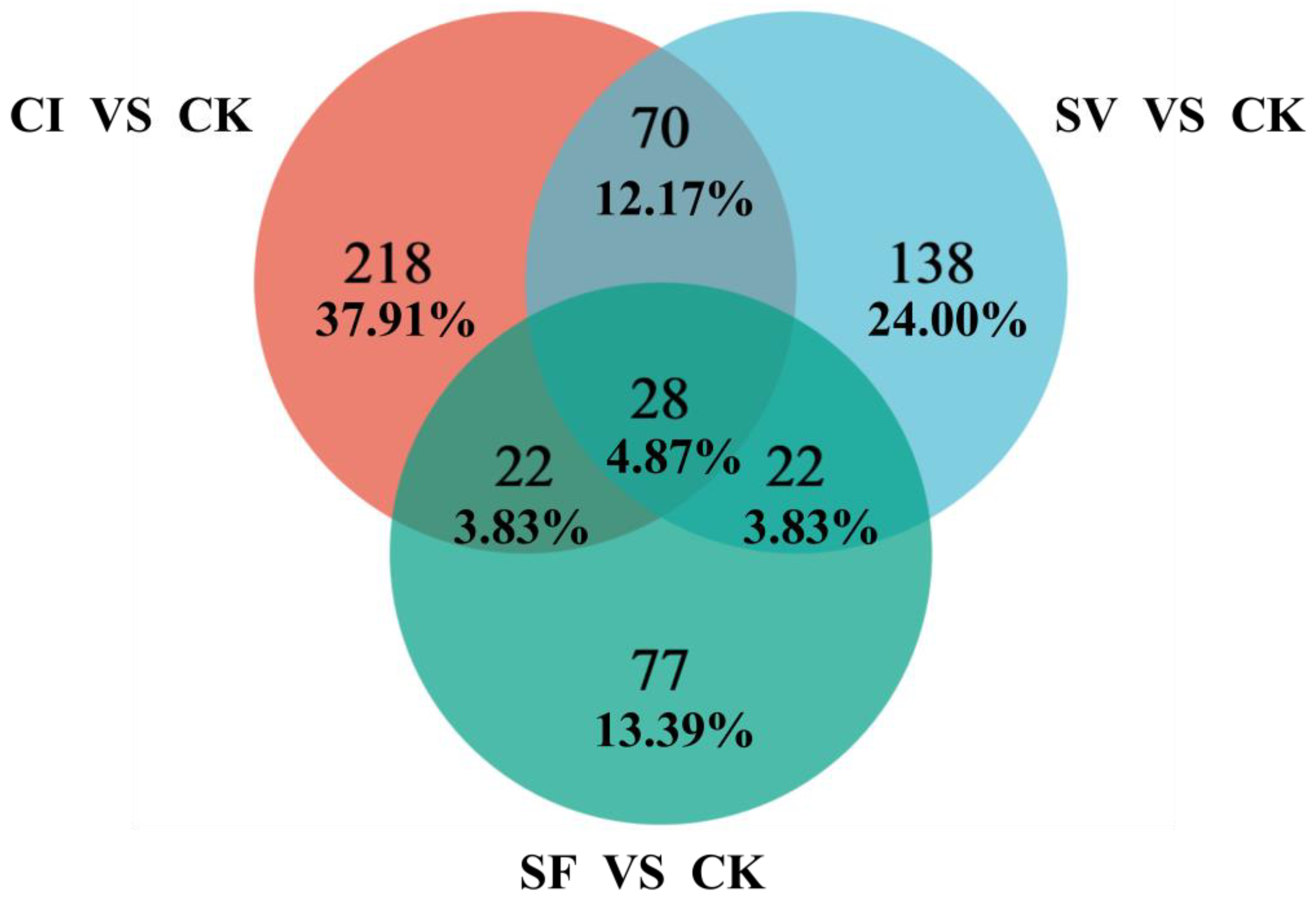
2.4. Venn’s Diagram Analysis
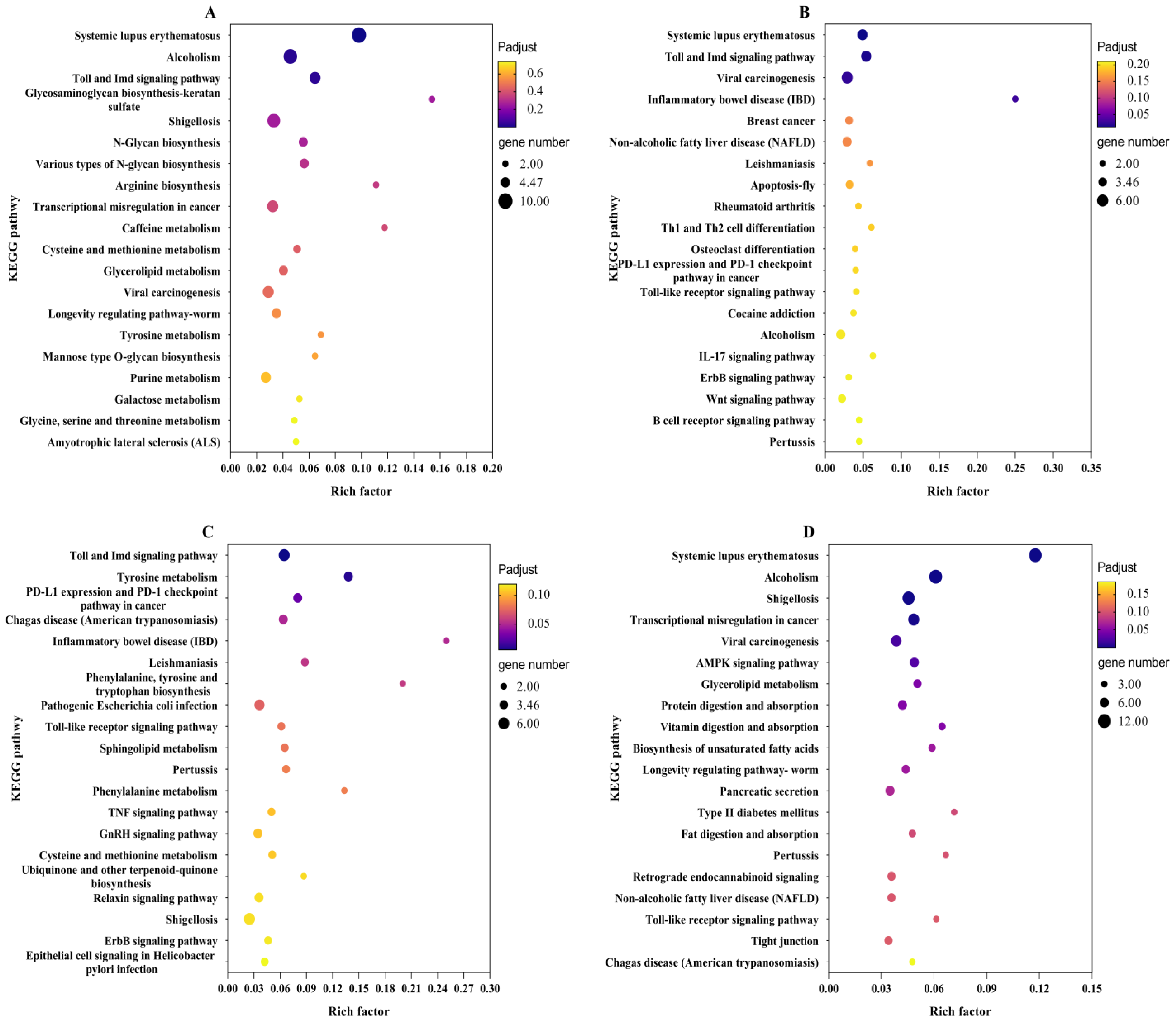
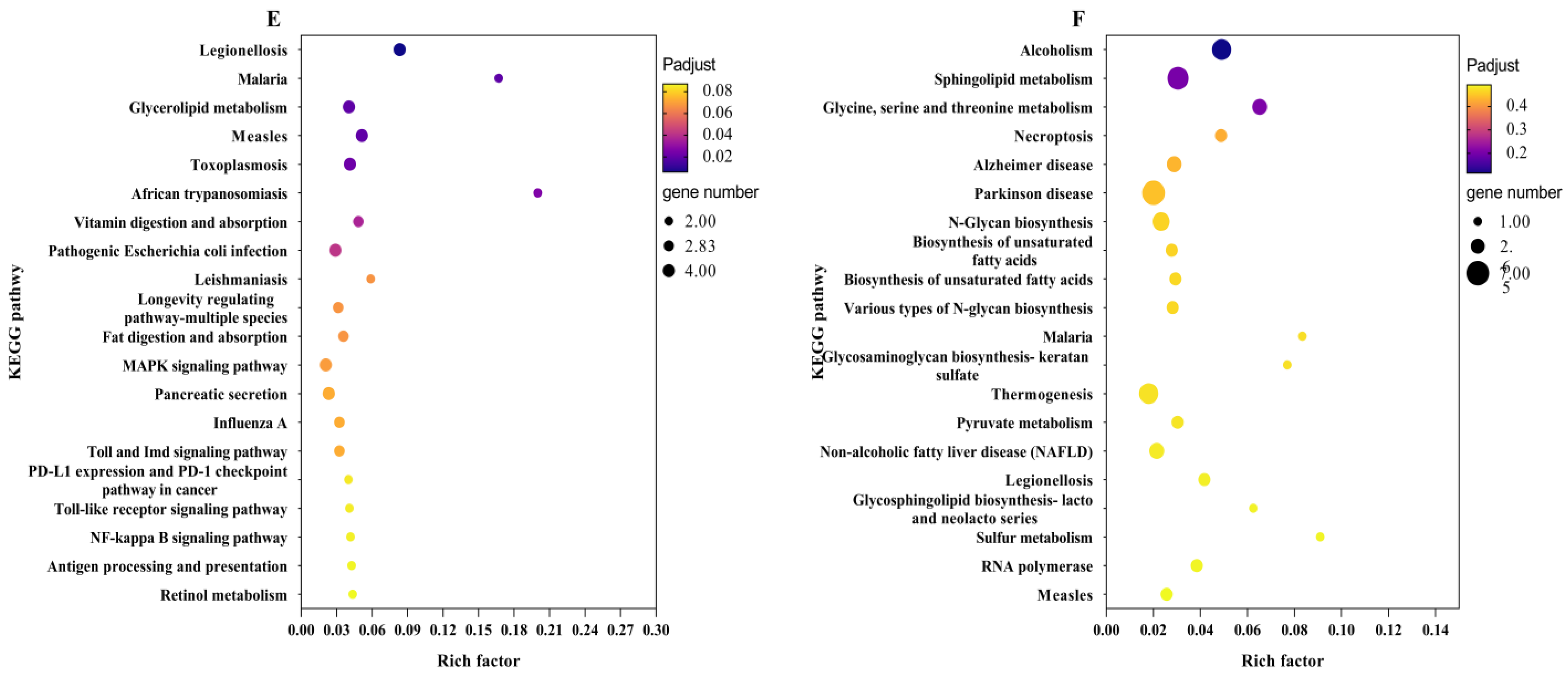
2.5. KEGG Enrichment Analysis
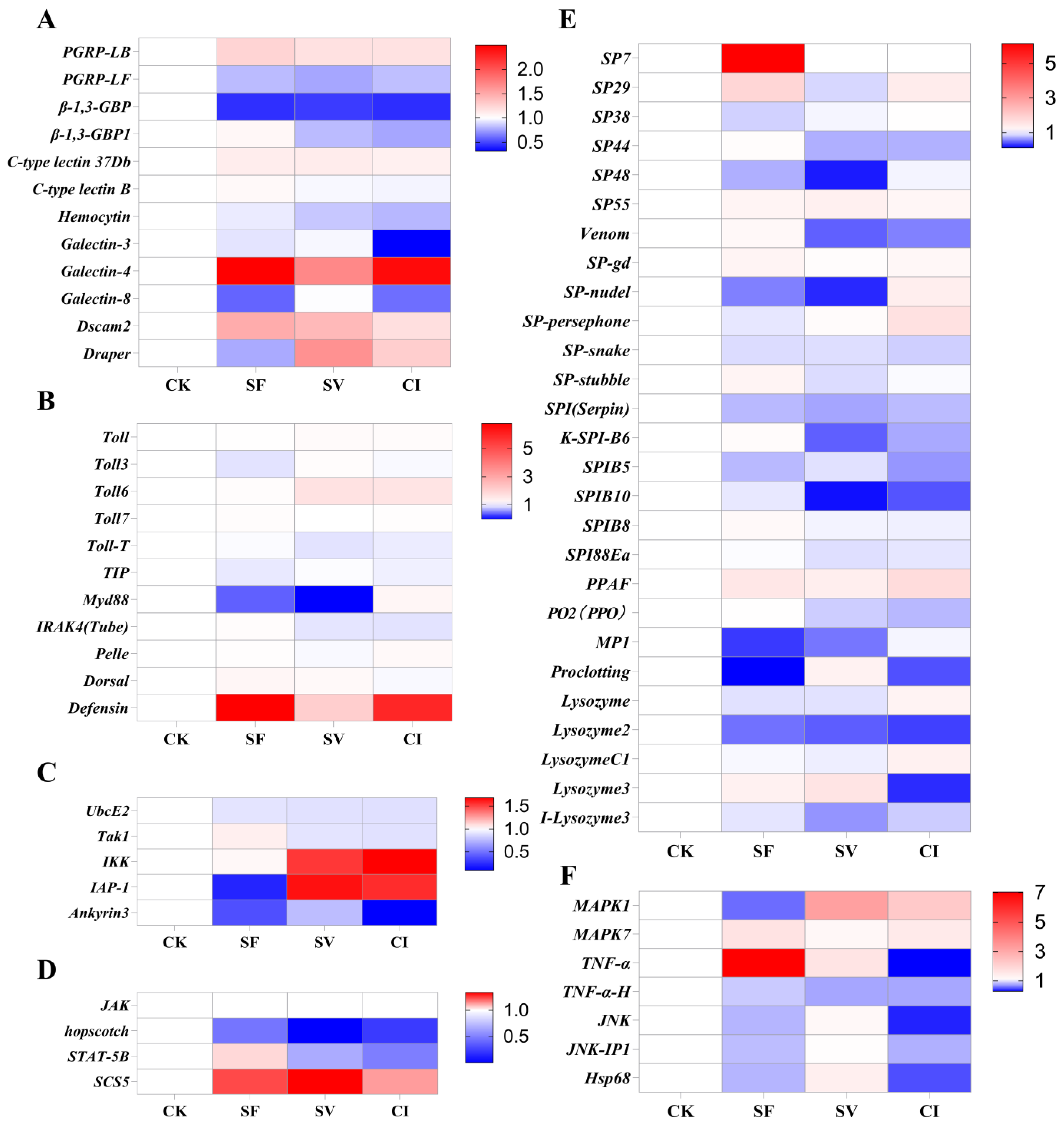
2.6. Immune-Related Genes’ Expression Level in Transcriptome Data
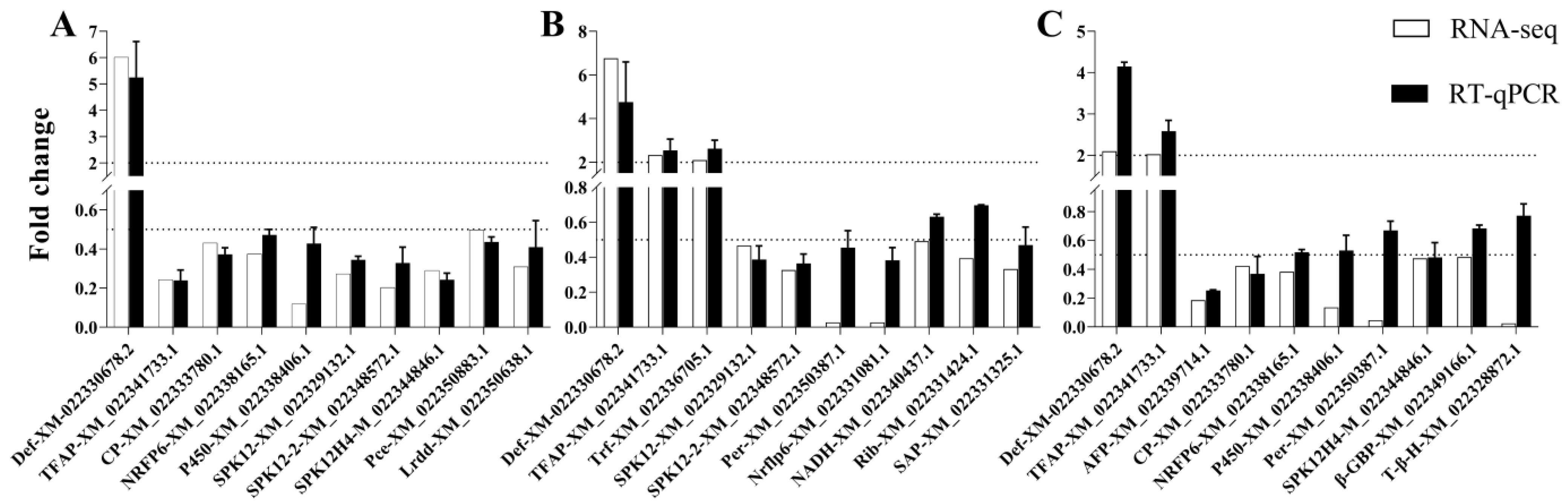
2.7. Transcriptomic Data Validation
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Materials
5.2. RRSV Meditated the Virulence of Metarhizium anisopliae YTTR against BPH
5.3. Transcriptomic Sequencing
5.4. Transcriptomic Data Analysis
5.5. Transcriptome Data Validation
5.6. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bottrell, D.G.; Schoenly, K.G. Resurrecting the ghost of green revolutions past: The brown planthopper as a recurring threat to high-yielding rice production in tropical Asia. J. Asia-Pac. Entomol. 2012, 15, 122–140. [Google Scholar] [CrossRef]
- Bao, Y.Y.; Zhang, C.X. Recent advances in molecular biology research of a rice pest, the brown planthopper. J. Integr. Agric. 2019, 18, 716–728. [Google Scholar] [CrossRef]
- Liao, X.; Xu, P.F.; Gong, P.P.; Wan, H.; Li, J.H. Current susceptibilities of brown planthopper Nilaparvata lugens to triflumezopyrim and other frequently used insecticides in China. Insect Sci. 2021, 28, 115–126. [Google Scholar] [CrossRef]
- Wu, C.J.; Ge, L.Q.; Liu, F.; Song, Q.S.; Stanley, D. Pesticide-Induced planthopper population eesurgence in rice cropping systems. Ann. Rev. Entomol. 2020, 65, 409–429. [Google Scholar] [CrossRef]
- Ge, L.Q.; Zhou, Z.; Sun, K.D.; Huang, B.; Stanley, D.; Song, Q.S. The antibiotic jinggangmycin increases brown planthopper (BPH) fecundity by enhancing rice plant sugar concentrations and BPH insulin-like signaling. Chemosphere 2020, 249, 126463. [Google Scholar] [CrossRef]
- Visarto, P.; Zalucki, M.P.; Nesbitt, H.J.; Jahn, G.C. Effect of fertilizer, pesticide treatment, and plant variety on the realized fecundity and survival rates of brown planthopper, Nilaparvata lugens (Stål) (Homoptera: Delphacidae)—Generating outbreaks in Cambodia. J. Asia-Pac. Entomol. 2001, 4, 75–84. [Google Scholar] [CrossRef]
- Yang, Y.J.; Liu, K.; Han, H.L.; Xu, H.X.; Zhang, F.C.; Zheng, X.S.; Tian, J.C.; Wang, G.Y.; Chen, G.H.; Lu, Z.X. Impacts of nitrogen fertilizer on major insect pests and their predators in transgenic Bt rice lines T2A-1 and T1C-19. Entomol. Exp. Appl. 2016, 160, 281–291. [Google Scholar] [CrossRef]
- Wang, C.S.; Wang, S.B. Insect pathogenic fungi: Genomics, molecular interactions, and genetic improvements. Ann. Rev. Entomol. 2017, 62, 73–90. [Google Scholar] [CrossRef]
- Lacey, L.; Grzywacz, D.; Shapiro-Ilan, D.; Frutos, R.; Brownbridge, M.; Goettel, M. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef]
- Vega, F.E.; Meyling, N.V.; Luangsa-ard, J.J.; Blackwell, M. Fungal Entomopathogens. In Insect Pathology; Vega, F.E., Kaya, H.K., Eds.; Academic Press: Waltham, MA, USA, 2012; Volume 2, pp. 171–220. [Google Scholar]
- Shah, P.A.; Pell, J.K. Entomopathogenic fungi as biological control agents. Appl. Microbiol. Biotechnol. 2003, 61, 413–423. [Google Scholar] [CrossRef]
- Atta, B.; Rizwan, M.; Sabir, A.M.; Gogi, M.D.; Farooq, M.A.; Batta, Y.A. Efficacy of entomopathogenic fungi against brown planthopper Nilaparvata lugens (Stål) (Homoptera: Delphacidae) under controlled conditions. Gesunde Pflanz. 2020, 72, 101–112. [Google Scholar] [CrossRef]
- Peng, Y.F.; Tang, J.F.; Xie, J.Q. Transcriptomic analysis of the brown planthopper, Nilaparvata lugens, at different dtages after Metarhizium anisopliae challenge. Insects 2020, 11, 139. [Google Scholar] [CrossRef]
- San Aw, K.M.; Hue, S.M. Review mode of infection of Metarhizium spp. fungus and their potential as biological control agents. J. Fungi 2017, 3, 30. [Google Scholar]
- Roberts, D.W.; Leger, R.J.S. Metarhizium spp., cosmopolitan insect-pathogenic fungi: Mycological aspects. Adv. Appl. Microbiol. 2004, 54, 1–70. [Google Scholar]
- Tang, J.F.; Liu, X.Y.; Ding, Y.C.; Jiang, W.J.; Xie, J.Q. Evaluation of Metarhizium anisopliae for rice planthopper control and its synergy with selected insecticides. Crop Prot. 2019, 121, 132–138. [Google Scholar] [CrossRef]
- Jin, S.F.; Feng, M.G.; Chen, J.Q. Selection of global Metarhizium isolates for the control of the rice pest Nilaparvata lugens (Homoptera: Delphacidae). Pest Manag. Sci. 2008, 64, 1008–1014. [Google Scholar] [CrossRef]
- Jaber, L.R.; Ownley, B.H. Can we use entomopathogenic fungi as endophytes for dual biological control of insect pests and plant pathogens? Biol. Control. 2018, 116, 36–45. [Google Scholar] [CrossRef]
- Li, Y.D.; Duan, T.Y.; Li, Y.Z. Research progress in the interactions of fungal pathogens and insect pests during host plant colonization. J. Plant Dis. Prot. 2021, 128, 633–647. [Google Scholar] [CrossRef]
- Sheehan, G.; Farrell, G.; Kavanagh, K. Immune priming: The secret weapon of the insect world. Virulence 2020, 11, 238–246. [Google Scholar] [CrossRef]
- Sheehan, G.; Garvey, A.; Croke, M.; Kavanagh, K. Innate humoral immune defences in mammals and insects: The same, with differences? Virulence 2018, 9, 1625–1639. [Google Scholar] [CrossRef]
- Qu, S.; Wang, S.B. Interaction of entomopathogenic fungi with the host immune system. Dev. Comp. Immunol. 2018, 83, 96–103. [Google Scholar] [CrossRef]
- Charles, H.M.; Killian, K.A. Response of the insect immune system to three different immune challenges. J. Insect Physiol. 2015, 81, 97–108. [Google Scholar] [CrossRef]
- Castillo, J.C.; Reynolds, S.E.; Eleftherianos, I. Insect immune responses to nematode parasites. Trends Parasitol. 2011, 27, 537–547. [Google Scholar] [CrossRef]
- Strand, M.R. The insect cellular immune response. Insect Sci. 2008, 15, 1–14. [Google Scholar] [CrossRef]
- Megan, B.K.; Kingsolver; Huang, Z.; Richard, W.H. Insect antiviral innate immunity: Pathways, effectors, and connections. J. Mol. Biol. 2013, 425, 4921–4936. [Google Scholar]
- Ding, J.L.; Hou, J.; Feng, M.G.; Ying, S.H. Transcriptomic analyses reveal comprehensive responses of insect hemocytes to mycopathogen Beauveria bassiana, and fungal virulence-related cell wall protein assists pathogen to evade host cellular defense. Virulence 2020, 11, 1352–1365. [Google Scholar] [CrossRef]
- Shelby, K.S.; Popham, H.J.R. RNA-Seq study of microbially induced hemocyte transcripts from larval Heliothis virescens (Lepidoptera: Noctuidae). Insects 2012, 3, 743–762. [Google Scholar] [CrossRef]
- Yi, Y.H.; Xu, H.; Li, M.; Wu, G.Q. RNA-seq profiles of putative genes involved in specific immune priming in Bombyx mori haemocytes. Infect. Genet. Evol. 2019, 74, 103921. [Google Scholar] [CrossRef]
- Jyoti, R.; Charu, C.; Tanwee, D.D.; Seena, K.; Punita, S.; Sanjay, T.; Karan, P.; Ashwani, K.; Mishra, K.C.; Pandey, N.S.; et al. Hemocyte RNA-Seq analysis of Indian malarial vectors Anopheles stephensi and Anopheles culicifacies: From similarities to differences. Gene 2021, 798, 145810. [Google Scholar]
- Bao, Y.Y.; Qu, L.Y.; Zhao, D.; Chen, L.B.; Jin, H.Y.; Xu, L.M.; Cheng, J.A.; Zhang, C.X. The genome- and transcriptome-wide analysis of innate immunity in the brown planthopper, Nilaparvata lugens. BMC Genom. 2013, 14, 1–23. [Google Scholar] [CrossRef]
- Wang, L.; Tang, N.; Gao, X.L.; Guo, D.Y.; Chang, Z.X.; Fu, Y.T.; Akinyemi, I.A.; Wu, Q.F. Understanding the immune system architecture and transcriptome responses to southern rice black-streaked dwarf virus in Sogatella furcifera. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.J.; Li, J.M.; Zhang, C.X.; Chen, J.P. Hemipteran-transmitted palnt viruses: Research progress and control strategies. Front. Agric. Sci. Eng. 2021, 9, 98–109. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Matsukawa, M.; Tanaka, T. Viral infection induces different detoxification enzyme activities in insecticide-resistant and-susceptible brown planthopper Nilaparvata lugens strains. J. Pestic. Sci. 2018, 43, 10–17. [Google Scholar] [CrossRef]
- Wei, J.; Jia, D.S.; Mao, Q.Z.; Zhang, X.F.; Chen, Q.; Wu, W.; Chen, H.Y.; Wei, T.Y. Complex interactions between insect-borne rice viruses and their vectors. Curr. Opin. Virol. 2018, 33, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.F.; Yu, J.T.; Wang, W.; Lu, H.; Kang, L.; Cui, F. A plant virus ensures viral stability in the hemolymph of vector insects through suppressing prophenoloxidase activation. MBio 2020, 11, e01453-20. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.J.; Bao, Y.Y.; Lao, S.H.; Huang, X.H.; Ye, Y.Z.; Wu, J.X.; Xu, H.J.; Zhou, X.P.; Zhang, C.X. Rice ragged stunt virus-induced apoptosis affects virus transmission from its insect vector, the brown planthopper to the rice plant. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, W.; Li, J.; Luo, L.; Kang, L.; Cui, F. The c-Jun N-terminal kinase pathway of a vector insect is activated by virus capsid protein and promotes viral replication. ELife 2017, 6, e26591. [Google Scholar] [CrossRef]
- He, Y.J.; Lu, G.; Qi, Y.H.; Zhang, Y.; Zhang, X.D.; Huang, H.J.; Zhuo, J.C.; Sun, Z.T.; Yan, F.; Chen, J.P.; et al. Activation of Toll immune pathway in an insect vector induced by a plant virus. Front. Immunol. 2021, 11, 613957. [Google Scholar] [CrossRef]
- Zhou, X.; Hu, J.; Fu, M.L.; Jin, P.; Zhang, Y.Y.; Xiang, Y.; Li, Y.; Ma, F. Identification and characterization of a TLR13 gene homologue from Laodelphax striatellus involved in the immune response induced by rice stripe virus. J. Integr. Agric. 2020, 19, 183–192. [Google Scholar] [CrossRef]
- Garza-Hernández, J.A.; Rodríguez-Pérez, M.A.; Salazar, M.I.; Russell, T.L.; Adeleke, M.A.; de Luna-Santillana, E.J.; Reyes-Villanueva, F. Vectorial capacity of Aedes aegypti for dengue virus type 2 is reduced with co-infection of Metarhizium anisopliae. PLoS Negl. Trop. Dis. 2013, 7, e2013. [Google Scholar] [CrossRef]
- Dolezal, T.; Krejcova, G.; Bajgar, A.; Nedbalova, P.; Strasser, P. Molecular regulations of metabolism during immune response in insects. Insect Biochem. Mol. Biol. 2019, 109, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Van der Most, P.J.; de Jong, B.; Parmentier, H.K.; Verhulst, S. Trade-off between growth and immune function: A meta-analysis of selection experiments. Funct. Ecol. 2011, 25, 74–80. [Google Scholar] [CrossRef]
- Bird, L. Getting enough energy for immunity. Nat. Rev. Immunol. 2019, 19, 269. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Y.; He, H.J.; Zhao, X.F.; Wang, J.X. Immune responses of Helicoverpa armigera to different kinds of pathogens. BMC Immunol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.M.; Morton Jr, J.C.; Ramirez, J.L.; Souza-Neto, J.A.; Dimopoulos, G. The entomopathogenic fungus Beauveria bassiana activate toll and JAK-STAT pathway-controlled effector genes and anti-dengue activity in Aedes aegypti. Insect Biochem. Mol. Biol. 2012, 42, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Guo, K.Y.; Dong, Z.M.; Chen, Z.Y.; Zhu, H.T.; Zhang, Y.; Xia, Q.Y.; Zhao, P. Kunitz-type protease inhibitor BmSPI51 plays an antifungal role in the silkworm cocoon. Insect Biochem. Mol. Biol. 2019, 116, 103258. [Google Scholar] [CrossRef]
- Kanost, M.R. Serine proteinase inhibitors in arthropod immunity. Dev. Comp. Immunol. 1999, 23, 291–301. [Google Scholar] [CrossRef]
- De Gregorio, E.; Han, S.J.; Lee, W.J.; Baek, M.J.; Osaki, T.; Kawabata, S.I.; Lee, B.L.; Iwanaga, S.; Lemaitre, B.; Brey, P.T. An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev. Cell. 2002, 3, 581–592. [Google Scholar] [CrossRef]
- Jang, I.H.; Nam, H.J.; Lee, W.J. CLIP-domain serine proteases in Drosophila innate immunity. BMB Rep. 2008, 41, 102–107. [Google Scholar] [CrossRef]
- Wu, J.M.; Zheng, R.E.; Zhang, R.J.; Ji, J.L.; Yu, X.P.; Xu, Y.P. A clip domain serine protease involved in egg production in Nilaparvata lugens: Expression patterns and RNA interference. Insects 2019, 10, 378. [Google Scholar] [CrossRef]
- Wu, W.; Xu, H.L.; Wang, Z.L.; Yu, X.P. Cloning and Function Analysis of a Serine Protease Inhibitor Gene Nlserpin2 in Nilaparvata lugens. Sci. Agric. Sin. 2022, 55, 2338–2346. [Google Scholar]
- Zhang, C.; Shi, C.N.; Chen, D.; Wu, J.G. Rice ragged stunt virus propagation and infection on rice plants. Bio-Protocol 2018, 8, e3060. [Google Scholar] [CrossRef] [PubMed]
- Goettel, M.S.; Inglis, G.D. Fungi: Hyphomycetes. In Manual of Techniques in Insect Pathology; Academic Press: San Diego, CA, USA, 1997; pp. 213–249. [Google Scholar]
- Ren, Y.; Yu, G.; Shi, C.P.; Liu, L.M.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.X.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Liu, S.H.; Yang, B.J.; Gu, J.H.; Yao, X.M.; Zhang, Y.X.; Song, F.; Liu, Z.W. Molecular cloning and characterization of a juvenile hormone esterase gene from brown planthopper, Nilaparvata lugens. J. Insect Physiol. 2008, 54, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.S. Calculating the LC50 of insecticides with software SPSS. Chin. Bull. Entomol. 2006, 43, 414–417. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhang, B.; Zou, J.; Li, Q.; Liu, J.; Cai, S.; Akutse, K.S.; You, M.; Lin, S. Immune Responses and Transcriptomic Analysis of Nilaparvata lugens against Metarhizium anisopliae YTTR Mediated by Rice Ragged Stunt Virus. Plants 2023, 12, 345. https://doi.org/10.3390/plants12020345
Li X, Zhang B, Zou J, Li Q, Liu J, Cai S, Akutse KS, You M, Lin S. Immune Responses and Transcriptomic Analysis of Nilaparvata lugens against Metarhizium anisopliae YTTR Mediated by Rice Ragged Stunt Virus. Plants. 2023; 12(2):345. https://doi.org/10.3390/plants12020345
Chicago/Turabian StyleLi, Xuewen, Bang Zhang, Jiaxing Zou, Qianqian Li, Jianli Liu, Shouping Cai, Komivi Senyo Akutse, Minsheng You, and Sheng Lin. 2023. "Immune Responses and Transcriptomic Analysis of Nilaparvata lugens against Metarhizium anisopliae YTTR Mediated by Rice Ragged Stunt Virus" Plants 12, no. 2: 345. https://doi.org/10.3390/plants12020345
APA StyleLi, X., Zhang, B., Zou, J., Li, Q., Liu, J., Cai, S., Akutse, K. S., You, M., & Lin, S. (2023). Immune Responses and Transcriptomic Analysis of Nilaparvata lugens against Metarhizium anisopliae YTTR Mediated by Rice Ragged Stunt Virus. Plants, 12(2), 345. https://doi.org/10.3390/plants12020345





