New Insights into the Enhancement of Adventitious Root Formation Using N,N′-Bis(2,3-methylenedioxyphenyl)urea
Abstract
:1. Introduction
2. Results
2.1. Adventitious Rooting of Apple Slices
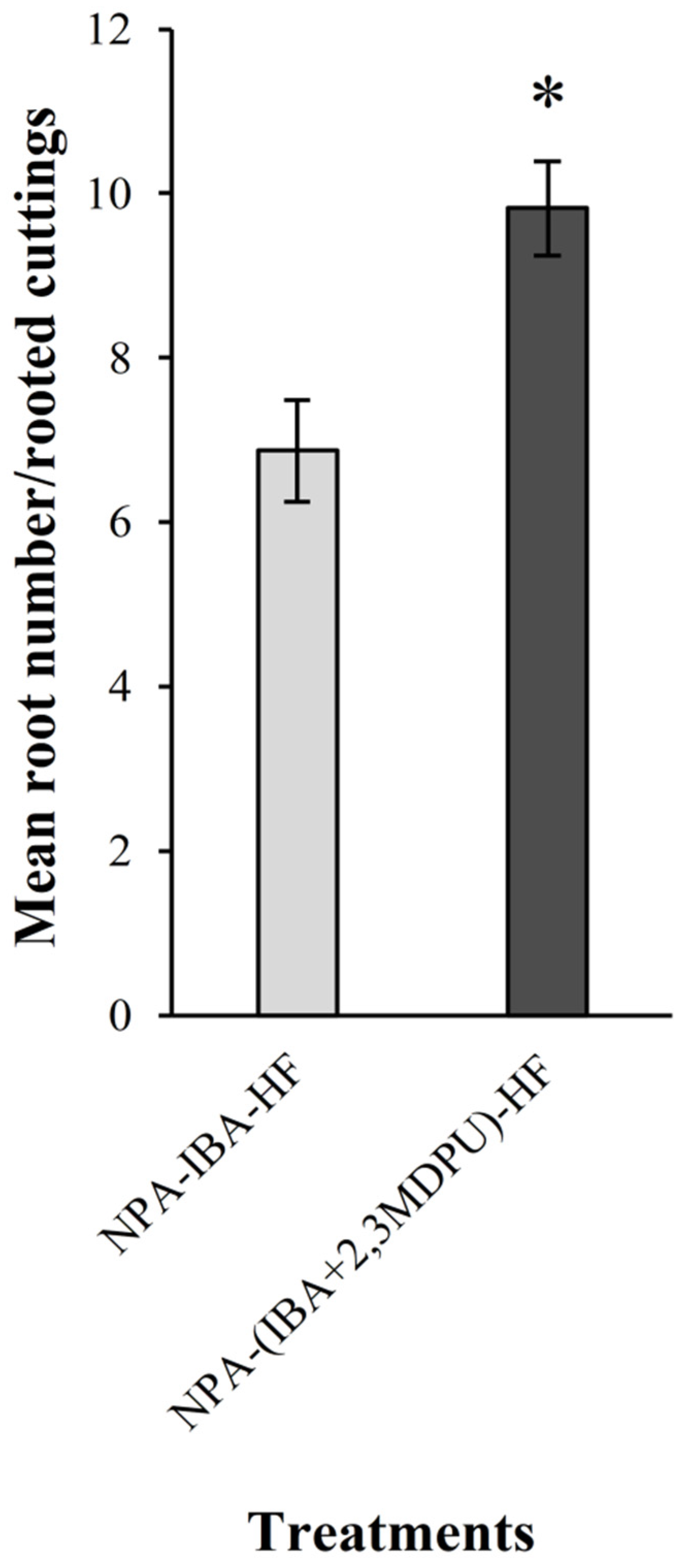
2.2. Adventitious Rooting of Apple Cuttings after Basal NPA Treatment
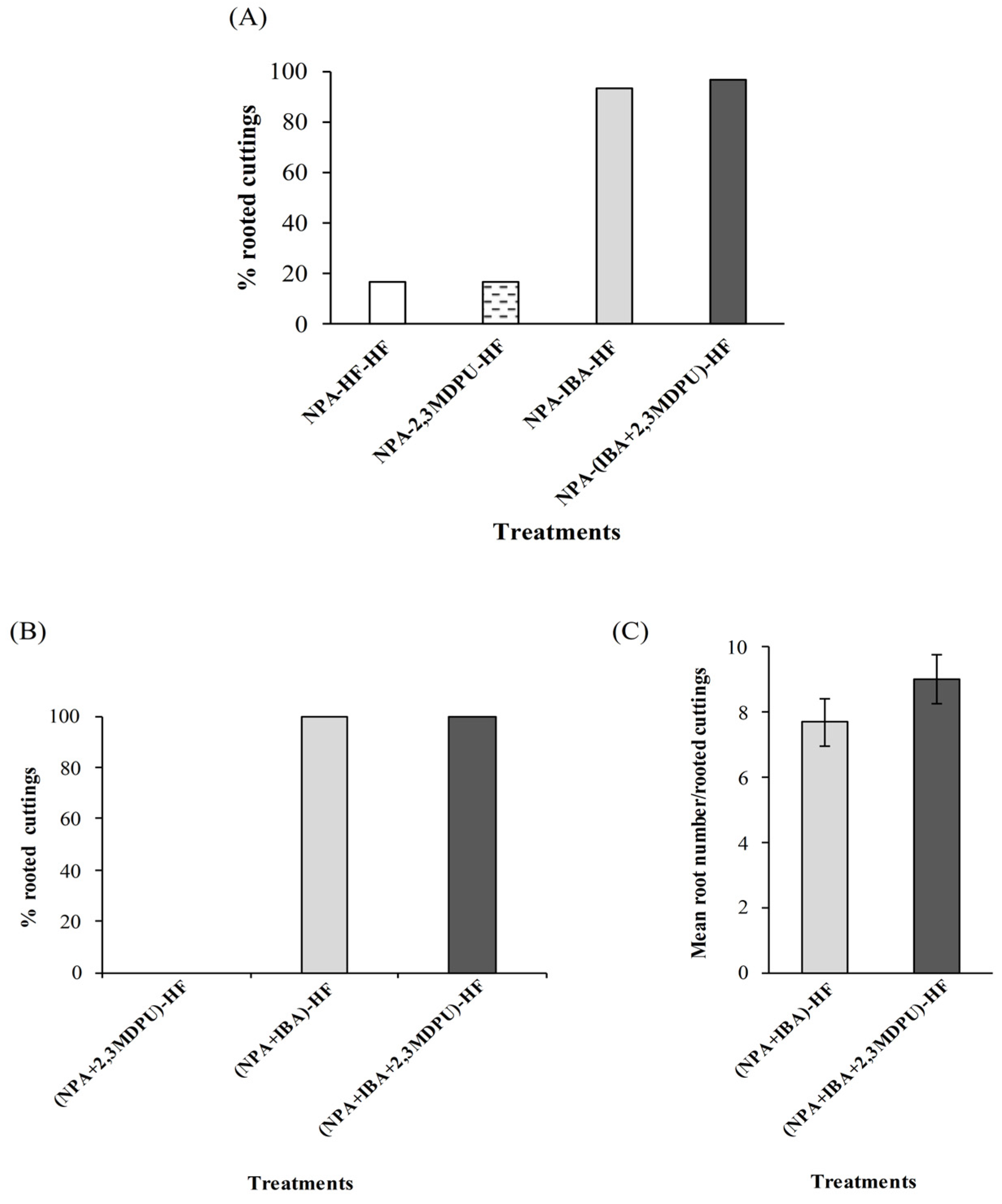
2.3. Adventitious Rooting of Apple Cuttings after Upside Down NPA Treatment
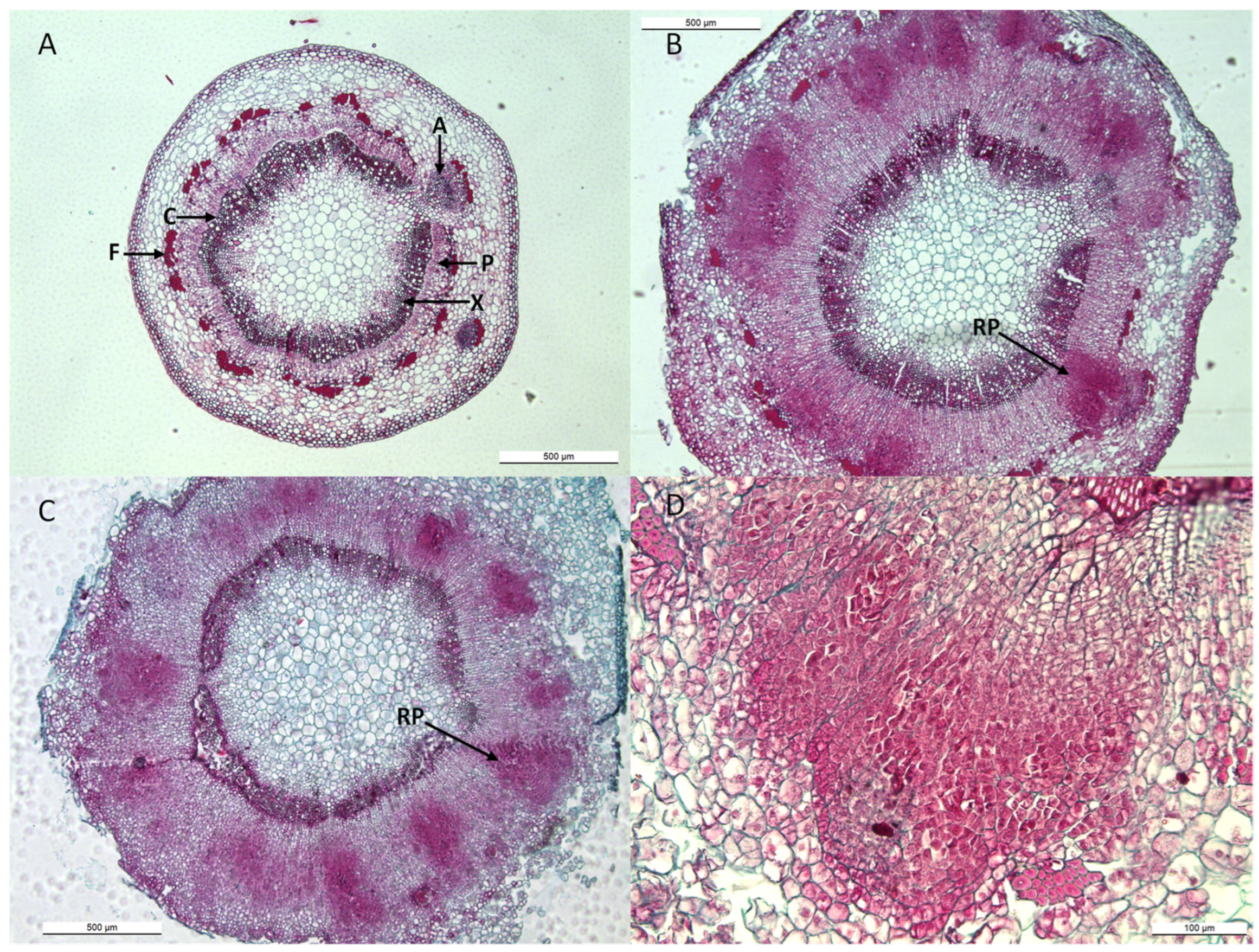
2.4. Histology of Apple Cuttings
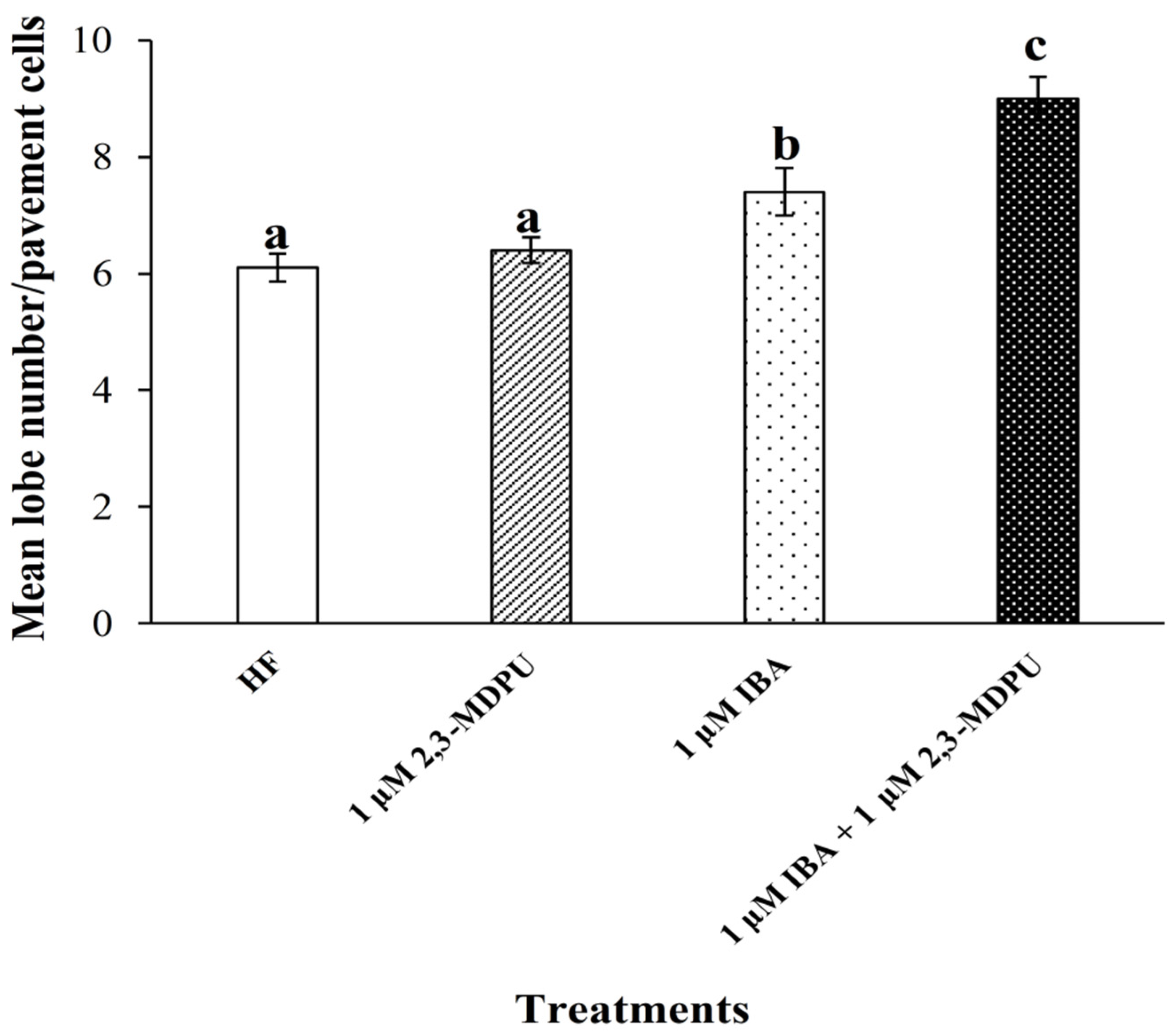
2.5. Analysis of Arabidopsis Pavement Cell Shape
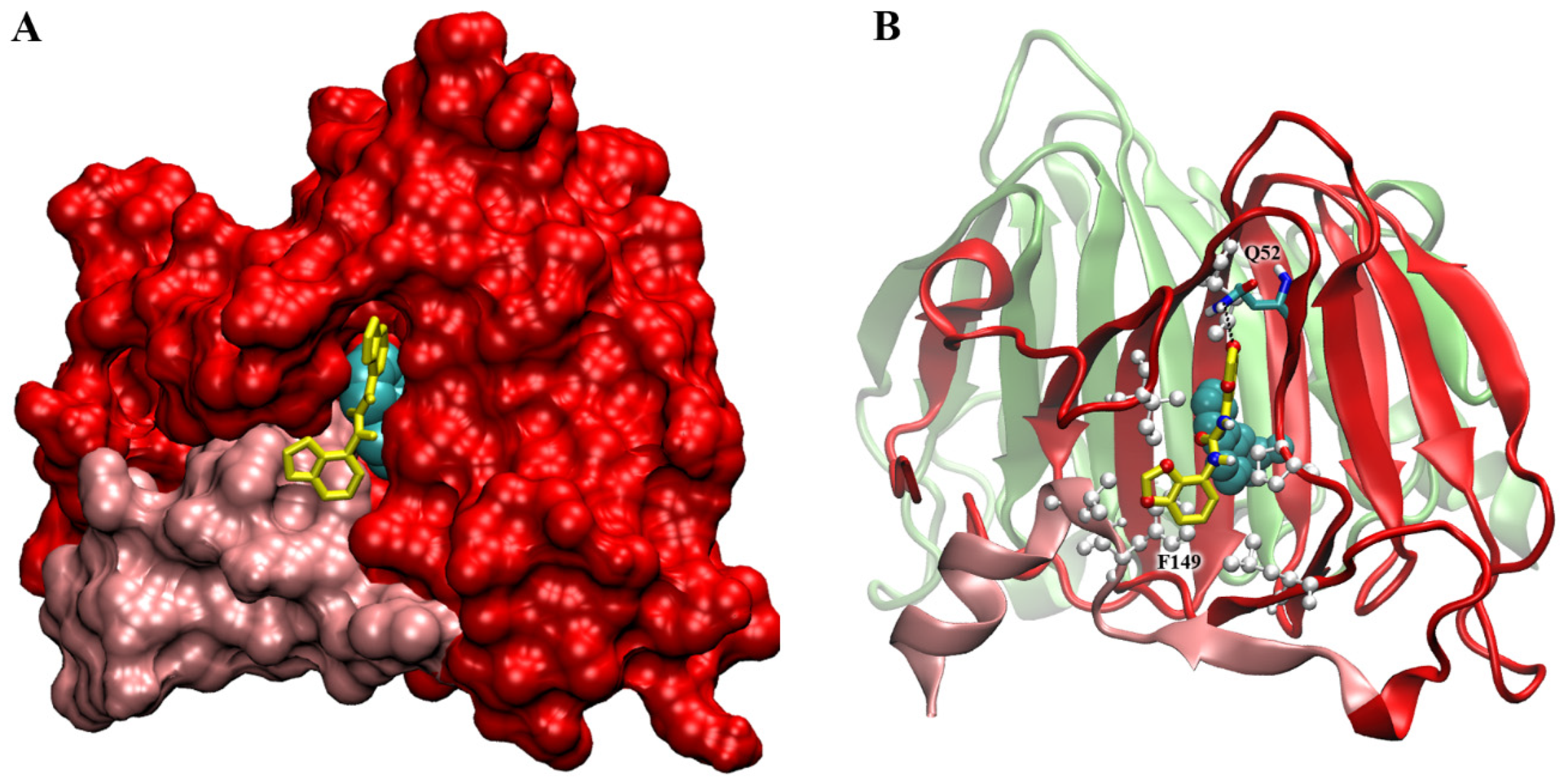
2.6. Docking Simulations on ABP1 Receptor
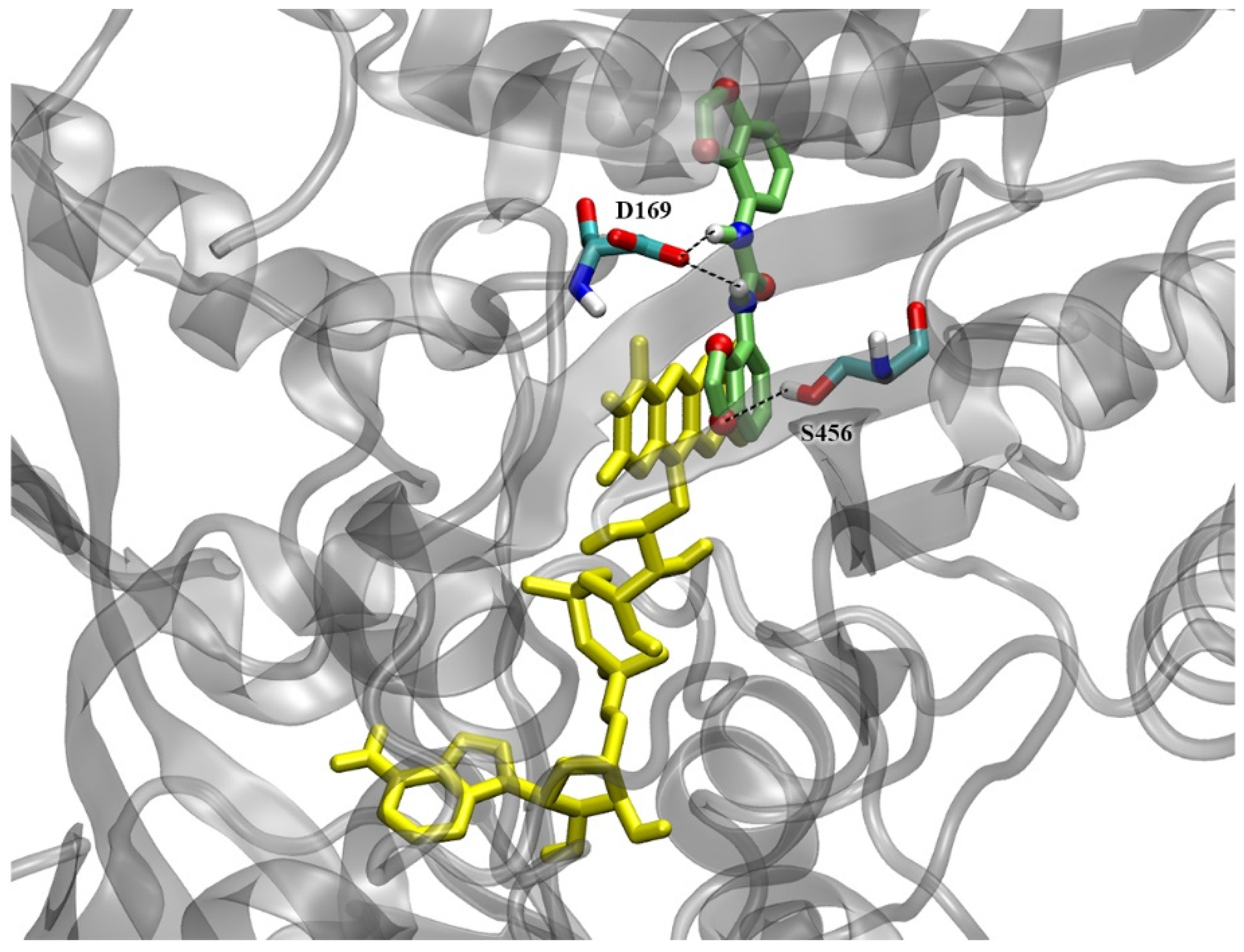
2.7. Docking Simulations on CKX Enzymes
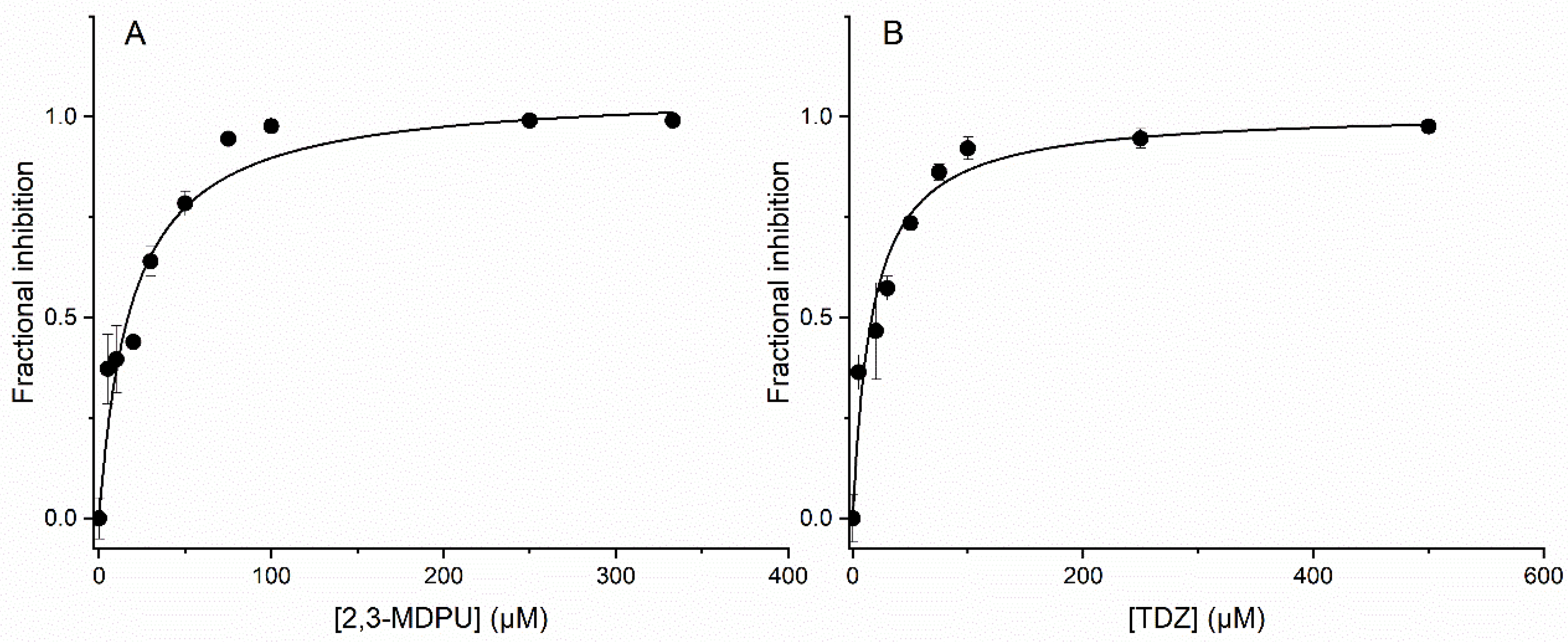
2.8. In Vitro Inhibition of ZmCKX1
3. Discussion
3.1. Interaction with ABP1
3.2. Interaction with CKX Enzymes
3.3. Adventitious Rooting of Apple Slices
3.4. Adventitious Rooting of Apple Cuttings
4. Materials and Methods
4.1. Chemicals
4.2. Plant Material and In Vitro Culture Conditions for Apple
4.3. Adventitious Rooting of Apple Slices
- -
- Groups of 25 slices were cultured in Petri dishes with the apical side on a nylon mesh put on an MP medium supplemented with 0.01, 0.1, 1 or 10 μM NPA and with 1 μM IBA as control.
- -
- Groups of 25 slices were cultured in the simultaneous presence of 0.01, 0.1, 1 or 10 μM NPA plus 1 μM IBA, while 1 μM IBA alone was used as control.
- -
- Groups of 25 slices were cultured in the simultaneous presence of 0.01, 0.1, 1 or 10 μM NPA plus 1 μM IBA plus 1 μM 2,3-MDPU, while 1 μM IBA plus 1 μM 2,3-MDPU was used as control.
4.4. Adventitious Rooting of Apple Cuttings after Basal NPA Treatment
4.5. Adventitious Rooting of Apple Cuttings after Upside Down NPA Treatment
4.6. Histology of Apple Cuttings
4.7. Analysis of Arabidopsis Pavement Cell Shape
4.8. Inhibition Assays of ZmCKX1
4.9. Molecular Docking Simulations
4.9.1. ABP1 Receptor
4.9.2. CKX Enzymes
4.9.3. MDPU Ligand
4.10. Statistical Analyses
4.10.1. Adventitious Rooting of Apple Slices
4.10.2. Adventitious Rooting of Apple Cuttings after Upside Down NPA Treatment
4.10.3. Analysis of Arabidopsis Pavement Cell Shape
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geiss, G.; Gutierrez, L.; Bellini, C. Adventitious root formation: New insights and perspectives. In Annual Plant Reviews: Root Development; Beeckman, T., Ed.; Wiley-Blackwell: Oxford, UK, 2009; Volume 37, pp. 127–156. [Google Scholar]
- Li, S.W.; Xue, L.; Xu, S.; Feng, H.; An, L. Mediators, genes and signaling in adventitious rooting. Bot. Rev. 2009, 75, 230–247. [Google Scholar] [CrossRef]
- da Costa, C.T.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.S.; Fett-Neto, A.G. When stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133–151. [Google Scholar] [CrossRef]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious Roots and Lateral Roots: Similarities and Differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, I.; Schotte, S.; Geelen, D. Hypocotyl adventitious root organogenesis differs from lateral root development. Front. Plant Sci. 2014, 5, 495–507. [Google Scholar] [CrossRef]
- Druege, U.; Franken, P.; Hajirezaei, M.R. Plant hormone homeostasis, signaling, and function during adventitious root formation in cuttings. Front. Plant Sci. 2016, 7, 381–394. [Google Scholar] [CrossRef]
- Druege, U.; Hilo, A.; Perez-Perez, J.M.; Klopotek, Y.; Acosta, M.; Shahinnia, F.; Zerche, S.; Franken, P.; Hajirezaei, M.R. Molecular and physiological control of adventitious rooting in cuttings: Phytohormone action meets resource allocation. Ann. Bot. 2019, 123, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Gonin, M.; Bergougnoux, V.; Nguyen, T.D.; Gantet, P.; Champion, A. What makes adventitious roots? Plants 2019, 8, 240–264. [Google Scholar] [CrossRef] [PubMed]
- Kevers, C.; Hausman, J.F.; Faivre-Rampant, O.; Dommes, J.; Gaspar, T. What we have learned about the physiology of in vitro adventitious rooting of woody plants and how it is related to improvements in the practice. In Adventitious Root Formation of Forest Trees and Horticultural Plants—From Genes to Applications; Niemi, K., Ed.; Research Signpost: Trivandrum, India, 2009; pp. 209–225. [Google Scholar]
- Oinam, G.; Yeung, E.; Kurepin, L.; Haslam, T.; Lopez-Villalobos, A. Adventitious root formation in ornamental plants: I. General overview and recent successes. Propag. Ornam. Plants 2011, 11, 78–90. [Google Scholar]
- Hackett, W.P. Donor plant maturation and adventitious root formation. In Adventitious Root Formation in Cuttings; Advances in plant sciences series; Davies, T.M., Haissig, B.E., Sankhla, N., Eds.; Dioscorides: Portland, OR, USA, 1988; Volume 2, pp. 11–28. [Google Scholar]
- Altamura, M.M. Root histogenesis in herbaceous and woody explants cultured in vitro. A critical review. Agron. EDP Sci. 1996, 16, 589–602. [Google Scholar] [CrossRef]
- De Klerk, G.J.; Van Der Krieken, W.; De Jong, J.C. The formation of adventitious roots: New concepts, new possibilities. In Vitro Cell. Dev. Biol. Plant 1999, 35, 189–199. [Google Scholar] [CrossRef]
- De Klerk, G.J.; Hanecakova, J.; Jasik, J. The role of cytokinins in rooting of stem slices cut from apple microcuttings. Plant Biosyst. 2001, 135, 79–84. [Google Scholar] [CrossRef]
- Kevers, C.; Hausman, J.F.; Faivre-Rampant, O.; Evers, D.; Gaspar, T. Hormonal control of adventitious rooting: Progress and questions. Angew. Bot. 1997, 71, 71–79. [Google Scholar]
- Pacurar, D.I.; Perrone, I.; Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant. 2014, 151, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Pop, T.I.; Pamfil, D.; Bellini, C. Auxin control in the formation of adventitious roots. Not. Bot. Hortic. Agrobot. Cluj-Napoca 2011, 39, 307–316. [Google Scholar] [CrossRef]
- Lakehal, A.; Bellini, C. Control of adventitious root formation: Insight into synergistic and antagonistic hormonal interactions. Physiol. Plant. 2018, 165, 90–100. [Google Scholar] [CrossRef]
- Della Rovere, F.; Fattorini, L.; D’Angeli, S.; Veloccia, A.; Falasca, G.; Altamura, M.M. Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann. Bot. 2013, 112, 1395–1407. [Google Scholar] [CrossRef]
- Garrido, G.; Ramón Guerrero, J.; Angel Cano, E.; Acosta, M.; Sánchez-Bravo, J. Origin and basipetal transport of the IAA responsible for rooting of carnation cuttings. Physiol. Plant. 2002, 114, 303–312. [Google Scholar] [CrossRef]
- Abarca, D.; Diaz-Sala, C. Reprogramming adult cells during organ regeneration in forest species. Plant Signal Behav. 2009, 4, 793–795. [Google Scholar] [CrossRef]
- Ahkami, A.H.; Lischewski, S.; Haensch, K.-T.; Porfirova, S.; Hofmann, J.; Rolletschek, H.; Melzer, M.; Franken, P.; Hause, B.; Druege, U.; et al. Molecular physiology of adventitious root formation in Petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytol. 2008, 181, 613–625. [Google Scholar] [CrossRef]
- Abu-Abied, M.; Rogovoy, O.; Mordehaev, I.; Grumberg, M.; Elbaum, R.; Wasteneys, G.O.; Sadot, E. Dissecting the contribution of microtubule behaviour in adventitious root induction. J. Exp. Bot. 2015, 66, 2813–2824. [Google Scholar] [CrossRef] [PubMed]
- Alaguero-Cordovilla, A.; Sánchez-García, A.B.; Ibáñez, S.; Albacete, A.; Cano, A.; Acosta, M.; Pérez-Pérez, J.M. An auxin-mediated regulatory framework for wound-induced adventitious root formation in tomato shoot explants. Plant Cell Environ. 2021, 44, 1642–1662. [Google Scholar] [CrossRef] [PubMed]
- Lane, W.D. Regeneration of apple plants from shoot meristem tips. Plant Sci. Lett. 1978, 13, 281–285. [Google Scholar] [CrossRef]
- Corrêa, L.; Paim, D.C.; Schwambach, J.; Fett-Neto, A.G. Carbohydrates as regulatory factors on the rooting of Eucalyptus saligna Smith and Eucalyptus globulus Labill. Plant Growth Regul. 2005, 45, 63–73. [Google Scholar] [CrossRef]
- Ricci, A.; Rolli, E. Chemicals as adjuvants in auxin induced adventitious rooting. In Auxins: Structure, Biosynthesis and Functions; Andrew, H.K., Michelle, D.F., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2012; pp. 79–92. ISBN 978-1-62100-504-9. [Google Scholar]
- Sarropoulou, V.; Dimassi-Theriou, K.; Therios, I. L-arginine impact on cherry rootstock rooting and biochemical characteristics in tissue culture. Turk. J. Agric. For. 2014, 38, 887–897. [Google Scholar] [CrossRef]
- Ricci, A.; Carra, A.; Torelli, A.; Maggiali, C.A.; Morini, G.; Branca, C. Cytokinin-like activity of N,N′-diphenylureas. N,N′-bis-(2,3-methylenedioxyphenyl)urea and N,N′-bis-(3,4-methylenedioxyphenyl)urea enhance adventitious root formation in apple rootstock M26 (Malus pumila Mill.). Plant Sci. 2001, 160, 1055–1065. [Google Scholar] [CrossRef]
- Ricci, A.; Incerti, M.; Rolli, E.; Vicini, P.; Morini, G.; Comini, M.; Branca, C. Diheteroarylurea derivatives as adventitious rooting adjuvants in mung bean shoots and M26 apple rootstock. Plant Growth Regul. 2006, 50, 201–209. [Google Scholar] [CrossRef]
- Ricci, A.; Rolli, E.; Dramis, L.; Diaz-Sala, C. N,N′-bis-(2,3-methylenedioxyphenyl)urea and N,N′-bis-(3,4-methylenedioxyphenyl)urea enhance adventitious rooting in Pinus radiata and affect expression of genes induced during adventitious rooting in the presence of exogenous auxin. Plant Sci. 2008, 175, 356–363. [Google Scholar] [CrossRef]
- Brunoni, F.; Rolli, E.; Dramis, L.; Incerti, M.; Abarca, D.; Pizarro, A.; Diaz-Sala, C.; Ricci, A. Adventitious rooting adjuvant activity of 1,3-di(benzo[d]oxazol-5-yl)urea and 1,3-di(benzo[d]oxazol-6-yl)urea: New insights and perspectives. Plant Cell Tissue Org. Cult. 2014, 118, 111–124. [Google Scholar] [CrossRef]
- Ricci, A.; Rolli, E.; Brunoni, F.; Dramis, L.; Sacco, E.; Fattorini, L.; Ruffoni, B.; Diaz-Sala, C.; Altamura, M.M. 1,3-di(benzo[d]oxazol-5-yl)urea acts either as adventitious rooting adjuvant or xylogenesis enhancer in carob and pine microcuttings depending on the presence/absence of exogenous indole-3-butyric acid. Plant Cell Tissue Org. Cult. 2016, 126, 411–427. [Google Scholar] [CrossRef]
- Ricci, A.; Rolli, E. Some urea derivatives positively affect adventitious root formation: Old concepts and the state of the art. Plants 2020, 9, 321–328. [Google Scholar]
- Vielba, J.M.; Vidal, N.; Ricci, A.; Castro, R.; Covelo, P.; Carmen San-Jose, M.; Sanchez, C. Effects of auxin and urea derivatives on adventitious rooting in chestnut and oak microshoots. Isr. J. Plant Sci. 2020, 67, 52–68. [Google Scholar] [CrossRef]
- Michniewicz, M.; Brewer, P.B.; Friml, J. Polar auxin transport and asymmetric auxin distribution. Arab. Book 2007, 5, e0108. [Google Scholar]
- Dhonukshe, P.; Grigoriev, I.; Fischer, R.; Tominaga, M.; Robinson, D.G.; Hašek, J.; Paciorek, T.; Petrášek, J.; Seifertová, D.; Tejos, R.; et al. Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc. Natl. Acad. Sci. USA 2008, 105, 4489–4494. [Google Scholar] [CrossRef]
- Klíma, P.; Laňková, M.; Zažímalová, E. Inhibitors of plant hormone transport. Protoplasma 2016, 253, 1391–1404. [Google Scholar] [CrossRef]
- Teale, W.; Palme, K. Naphthylphthalamic acid and the mechanism of polar auxin transport. J. Exp. Bot. 2018, 69, 303–312. [Google Scholar] [CrossRef]
- Abas, L.; Kolb, M.; Stadlmann, J.; Janacek, D.P.; Lukic, K.; Schwechheimer, C.; Sazanov, L.A.; Mach, L.; Friml, J.; Hammes, U.Z. Naphthylphthalamic acid associates with and inhibits PIN auxin transporters. Proc. Natl. Acad. Sci. USA 2021, 118, e2020857118. [Google Scholar] [CrossRef]
- Grones, P.; Friml, J. ABP1: Finally docking. Mol. Plant 2015, 8, 356–358. [Google Scholar] [CrossRef]
- Grones, P.; Friml, J. Auxin transporters and binding proteins at a glance. J. Cell Sci. 2015, 128, 1–7. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signalling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef]
- Tromas, A.; Braun, N.; Muller, P.; Khodus, T.; Paponov, I.A.; Palme, K.; Ljung, K.; Lee, J.Y.; Benfey, P.; Murray, J.A.; et al. The AUXIN BINDING PROTEIN 1 is required for differential auxin responses mediating root growth. PLoS ONE 2009, 4, e6648. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, Y.; Dai, X.; Estelle, M.; Zhao, Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc. Natl. Acad. Sci. USA 2015, 112, 2275–2280. [Google Scholar] [CrossRef] [PubMed]
- Habets, M.E.J.; Offringa, R. Auxin binding protein 1: A red herring after all? Mol. Plant 2015, 8, 1131–1134. [Google Scholar] [CrossRef]
- Steffens, B.; Feckler, C.; Palme, K.; Christian, M.; Böttger, M.; Lüthen, H. The auxin signal for protoplast swelling is perceived by extracellular ABP1. Plant, J. 2001, 27, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Paque, S.; Mouille, G.; Grandont, L.; Alabadí, D.; Gaertner, C.; Goyallon, A.; Muller, P.; Primard-Brisset, C.; Sormani, R.; Blázquez, M.A.; et al. AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in Arabidopsis. Plant Cell 2014, 26, 280–295. [Google Scholar] [CrossRef]
- Gelová, Z.; Gallei, M.; Pernisová, M.; Brunoud, G.; Zhang, X.; Glanc, M.; Li, L.; Michalko, J.; Pavlovičová, Z.; Verstraeten, I.; et al. Developmental roles of Auxin Binding Protein 1 in Arabidopsis thaliana. Plant Sci. 2021, 303, 110750–110767. [Google Scholar] [CrossRef]
- Friml, J.; Gallei, M.; Gelová, Z.; Johnson, A.; Mazur, E.; Monzer, A.; Rodriguez, L.; Roosjen, M.; Verstraeten, I.; Živanović, B.D. ABP1–TMK auxin perception for global phosphorylation and auxin canalization. Nature 2022, 609, 575–597. [Google Scholar] [CrossRef]
- Xu, T.; Wen, M.; Nagawa, S.; Fu, Y.; Chen, J.-G.; Wu, M.-J.; Perrot-Rechenmann, C.; Friml, J.; Jones, A.M.; Yang, Z. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 2010, 143, 99–110. [Google Scholar] [CrossRef]
- Xu, T.; Dai, N.; Chen, J.; Nagawa, S.; Cao, M.; Li, H.; Zhou, Z.; Chen, X.; De Rycke, R.; Rakusová, H.; et al. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 2014, 343, 1025–1028. [Google Scholar] [CrossRef]
- Woo, E.J.; Marshall, J.; Bauly, J.; Chen, J.G.; Venis, M.; Napier, R.M.; Pickersgill, R.W. Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO J. 2002, 21, 2877–2885. [Google Scholar] [CrossRef]
- Kopečný, D.; Briozzo, P.; Popelková, H.; Šebela, M.; Končitíková, R.; Spíchal, L.; Nisler, J.; Madzak, C.; Frébort, I.; Laloue, M.; et al. Phenyl- and benzylurea cytokinins as competitive inhibitors of cytokinin oxidase/dehydrogenase: A structural study. Biochimie 2010, 92, 1052–1062. [Google Scholar] [CrossRef] [PubMed]
- Bae, E.; Bingman, C.A.; Bitto, E.; Aceti, D.J.; Phillips, G.N. Crystal structure of Arabidopsis thaliana cytokinin dehydrogenase. Proteins Struct. Funct. Genet. 2008, 70, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Brunoni, F.; Rolli, E.; Polverini, E.; Spíchal, L.; Ricci, A. The adjuvant activity of two urea derivatives on cytokinins: An example of serendipitous dual effect. Plant Growth Regul. 2021, 95, 169–190. [Google Scholar] [CrossRef]
- Diekmann, W.; Venis, M.A.; Robinson, D.G. Auxins induce clustering of the auxin-binding protein at the surface of maize coleoptile protoplasts. Proc. Natl. Acad. Sci. USA 1995, 92, 3425–3429. [Google Scholar] [CrossRef] [PubMed]
- Bertoša, B.; Kojić-Prodić, B.; Wade, R.C.; Tomić, S. Mechanism of auxin interaction with Auxin Binding Protein (ABP1): A molecular dynamics simulation study. Biophys. J. 2008, 94, 27–37. [Google Scholar] [CrossRef]
- Nisler, J.; Kopečný, D.; Končitíková, R.; Zatloukal, M.; Bazgier, V.; Berka, K.; Zalabák, D.; Briozzo, P.; Strnad, M.; Spíchal, L. Novel thidiazuron-derived inhibitors of cytokinin oxidase/dehydrogenase. Plant Mol. Biol. 2016, 92, 235–248. [Google Scholar] [CrossRef]
- Kopečný, D.; Končitíková, R.; Popelka, H.; Briozzo, P.; Vigouroux, A.; Kopečná, M.; Zalabák, D.; Šebela, M.; Skopalová, J.; Frébort, I.; et al. Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site. FEBS J. 2016, 283, 361–377. [Google Scholar] [CrossRef]
- Cheng, Y.; Prusoff, W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [PubMed]
- Murthy, B.N.S.; Murch, S.J.; Saxena, P.K. Thidiazuron: A potent regulator of in vitro plant morphogenesis. Vitr. Cell. Dev. Biol. Plant 1998, 34, 267–275. [Google Scholar] [CrossRef]
- Ricci, A.; Carra, A.; Rolli, E.; Bertoletti, C.; Branca, C. N,N′-bis-(2,3-methylenedioxyphenyl)urea and N,N′-bis-(3,4-methylenedioxyphenyl)urea cooperate with auxin in enhancing root formation of M26 apple (Malus pumila Mill.) stem slices. Plant Growth Regul. 2003, 40, 207–212. [Google Scholar] [CrossRef]
- Sukumar, P.; Maloney, G.S.; Muday, G.K. Localized Induction of the ATP-Binding Cassette B19 Auxin Transporter Enhances Adventitious Root Formation in Arabidopsis. Plant Physiol. 2013, 162, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Pernisová, M.; Klíma, P.; Horák, J.; Válková, M.; Malbeck, J.; Souček, P.; Reichman, P.; Hoyerová, K.; Dubová, J.; Friml, J.; et al. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl. Acad. Sci. USA 2009, 106, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Bustillo-Avendaño, E.; Ibáñez, S.; Sanz, O.; Sousa Barros, J.A.; Gude, I.; Perianez Rodriguez, J.; Micol, J.L.; Del Pozo, J.C.; Moreno-Risueno, M.A.; Pérez-Pérez, J.M. Regulation of hormonal control, cell reprogramming and patterning during de novo root organogenesis. Plant Physiol. 2018, 176, 1709–1727. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.Y.; Skoog, F. The use of dimethylsulfoxide as a solvent in the tobacco bioassay for cytokinins. Plant Physiol. 1970, 45, 537–538. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Van der Krieken, W.M.; Breteler, H.; Visser, M.H.M.; Mavridou, D. The role of the conversion of IBA into IAA on root regeneration in apple: Introduction of a test system. Plant Cell Rep. 1993, 12, 203–206. [Google Scholar] [CrossRef]
- De Klerk, G.J.; Caillat, E. Rooting responses of stem-disks excised from the same “M9 Jork” microcutting. Adv. Hort. Sci. 1994, 8, 15–18. [Google Scholar]
- Berlyn, G.P.; Miksche, J.P. Botanical Microtechnique and Cytochemistry; Iowa State University Press: Ames, IA, USA, 1976; ISBN 0-8138-0220-2. [Google Scholar]
- Morris, R.O.; Bilyeu, K.D.; Laskey, J.G.; Cheikh, N.N. Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem. Biophys. Res. Commun. 1999, 255, 328–333. [Google Scholar] [CrossRef]
- Bilyeu, K.D.; Cole, J.L.; Laskey, J.G.; Riekhof, W.R.; Esparza, T.J.; Kramer, M.D.; Morris, R.O. Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant physiol. 2001, 125, 378–386. [Google Scholar] [CrossRef]
- Zalabak, D.; Galuszka, P.; Mrizova, K.; Podlesakova, K.; Gu, R.; Frebortova, J. Biochemical characterization of the maize cytokinin dehydrogenase family and cytokinin profiling in developing maize plantlets in relation to the expression of cytokinin dehydrogenase genes. Plant physiol. Biochem. 2014, 74, 283–293. [Google Scholar] [CrossRef]
- Schmulling, T.; Werner, T.; Riefler, M.; Krupkova, E.; Bartrina y Manns, I. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 2003, 116, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- van Aalten, D.M.; Bywater, R.; Findlay, J.B.; Hendlich, M.; Hooft, R.W.; Vriend, G. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aided. Mol. Des. 1996, 10, 255–262. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Halgren, T. Merck Molecular Force Field. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricci, A.; Polverini, E.; Bruno, S.; Dramis, L.; Ceresini, D.; Scarano, A.; Diaz-Sala, C. New Insights into the Enhancement of Adventitious Root Formation Using N,N′-Bis(2,3-methylenedioxyphenyl)urea. Plants 2023, 12, 3610. https://doi.org/10.3390/plants12203610
Ricci A, Polverini E, Bruno S, Dramis L, Ceresini D, Scarano A, Diaz-Sala C. New Insights into the Enhancement of Adventitious Root Formation Using N,N′-Bis(2,3-methylenedioxyphenyl)urea. Plants. 2023; 12(20):3610. https://doi.org/10.3390/plants12203610
Chicago/Turabian StyleRicci, Ada, Eugenia Polverini, Stefano Bruno, Lucia Dramis, Daniela Ceresini, Antonio Scarano, and Carmen Diaz-Sala. 2023. "New Insights into the Enhancement of Adventitious Root Formation Using N,N′-Bis(2,3-methylenedioxyphenyl)urea" Plants 12, no. 20: 3610. https://doi.org/10.3390/plants12203610
APA StyleRicci, A., Polverini, E., Bruno, S., Dramis, L., Ceresini, D., Scarano, A., & Diaz-Sala, C. (2023). New Insights into the Enhancement of Adventitious Root Formation Using N,N′-Bis(2,3-methylenedioxyphenyl)urea. Plants, 12(20), 3610. https://doi.org/10.3390/plants12203610






