Abstract
Verticillium wilt is a soil-borne fungal disease that affects olive trees (Olea europaea) and poses a serious threat to their cultivation. The causal agent of this disease is Verticillium dahliae, a pathogen that is difficult to control with conventional methods. Therefore, there is a need to explore alternative strategies for the management of Verticillium wilt. In this study, we aimed to isolate and characterize actinobacteria from the rhizosphere of olive trees that could act as potential biocontrol agents against V. dahliae. We selected a Streptomyces sp. OR6 strain based on its in vitro antifungal activity and its ability to suppress the pathogen growth in soil samples. We identified the main active compound produced by this strain as albocycline, a macrolide polyketide with known antibacterial properties and some antifungal activity. Albocycline was able to efficiently suppress the germination of conidiospores. To our knowledge, this is the first report of albocycline as an effective agent against V. dahliae. Our results suggest that Streptomyces sp. OR6, or other albocycline-producing strains, could be used as a promising tool for the biological control of Verticillium wilt.
1. Introduction
Verticillium is a genus of ascomycete fungi that cause vascular wilt diseases in various plant hosts. These fungi can persist in the soil for up to 14 years by producing resting structures such as microsclerotia, chlamydospores, or resting mycelia [1]. Verticillium wilt is one of the most destructive fungal diseases worldwide, affecting many economically important crops such as alfalfa, almond, pistachio, peach [2,3], cotton, lettuce, potato, strawberry, ornamental plants [1], hops [4], and olive [3,5], among others. Although different species of Verticillium cause wilt, Verticillium dahliae is the most widespread and virulent, due to its broad host range and geographic distribution.
Olive (Olea europaea L.) is one of the earliest domesticated and cultivated tree species, and it has a significant historical, social, and economic value in the Mediterranean Basin as the main source of olive oil, which is a key component of the Mediterranean diet [5]. However, olive production is threatened by Verticillium wilt, a soil-borne fungal disease caused by V. dahliae, which has become a severe problem for olive growers in recent years [6]. Spain is the world’s leading producer of olive oil, accounting for more than half of the global production [7]. Most of the Spanish olive oil (83%) is produced in Andalucía, a region in southern Spain, where the average disease incidence has reached 20.4% [8]. However, this figure may be underestimated due to the lack of updated data [9].
Olive Verticillium wilt (OVW) is a devastating disease that has no effective control measure available. Therefore, an integrated control strategy is the most feasible approach to manage this disease. An integrated control strategy would involve pre-cultivation measures, such as strict hygiene practices during plant propagation in nurseries, heat treatment for the sanitation of olive plants, planting in disease-free soils, proper water treatment and management to prevent the spread of infective propagules, the use of organic amendments, or the development of new control methods based on novel fungicides or biological control agents. Moreover, the development of selection and breeding programs for plant material with higher intrinsic resistance to the disease would also be beneficial [5,10]. However, the lack of effective fungicides to eradicate the pathogen from the plant vascular system, and the current policies in Europe to limit the use of chemical fungicides in agriculture, are encouraging the development of new strategies to control fungal pathogens, such as the use of new and effective biocontrol agents [11].
V. dahliae is a soil-borne fungal pathogen that infects plants through their root system. Therefore, it is important to develop biocontrol agents (BCAs) that can inhibit its growth in soil before it reaches the roots. Ideally, these BCAs should colonize the rhizosphere and form a protective barrier against the pathogen [11]. Several fungal BCAs have been tested for their potential to control olive Verticillium wilt, a disease caused by V. dahliae. These include Fusarium oxisporum [12,13], Trichoderma asperellum [14], Mucor sp., Rhizopus sp., Phoma sp. [15], and entomopathogenic fungi such as Beauveria bassiana and Metarhizium brunneum [16]. These fungi have shown some ability to reduce the inoculum density of V. dahliae in soil and the severity of disease symptoms in plants. They may also produce secondary metabolites with antifungal activity (AF) against V. dahliae [16]. In addition, a consortium of arbuscular mycorrhizal fungi (Rhizolive) has demonstrated some efficacy in alleviating the symptoms of OVW [17,18].
Several bacterial strains have also been evaluated as potential BCAs against OVW. One of them is Pseudomonas fluorescens PICF7, which can suppress the growth of V. dahliae by competing for the niche and nutrients, producing antibacterial compounds, and inducing plant resistance [19]. Other Pseudomonas strains, such as Pseudomonas sp. PIC25, P. indica PIC105, and Pseudomonas sp. PIC141, isolated from healthy olive plants (cv. Picual), have shown in vitro antagonism against V. dahliae, with P. indica PIC105 being the most effective in reducing disease symptoms in plants [20]. Moreover, some members of the Bacillales order, such as Paenibacillus alvei K165 [21] and Bacillus velezensis [22], have demonstrated their ability to decrease the severity of Verticillium wilt in plants.
Streptomyces spp. are a promising alternative for the biocontrol of soil-borne fungal pathogens [23]. These bacteria belong to the order Streptomycetales, which comprises about 600 species of diverse and prolific producers of antibiotics [24]. Streptomyces spp. have been reported to control various plant diseases, including those caused by soil-borne fungi [25,26]. They are also abundant in different types of soils, accounting for about 10% of the total soil microbiome [27]. However, no data are available on the biocontrol of OVW by Streptomyces spp., despite the fact that some strains have shown AF activity against Verticillium spp. affecting other crops (such as cotton, potato, strawberry, and tomato) [28]. Therefore, the aim of this study was to evaluate the AF activity of Streptomyces spp. isolated from the rhizosphere of olive trees against V. dahliae and their potential to inhibit the pathogen growth in test soils.
2. Results
2.1. Isolation and Selection of Culturable Streptomycetes Associated with the Olive Rhizosphere and Their Antifungal Activity
A total of 96 putative different strains were obtained from rhizosphere soil after selection according to their different morphological and cultural characteristics. A bioassay-based in vitro screening showed that 32.3% (31 out of 96) of the isolates exhibited some degree of AF activity against the fungal pathogen V. dahliae V937I strain.
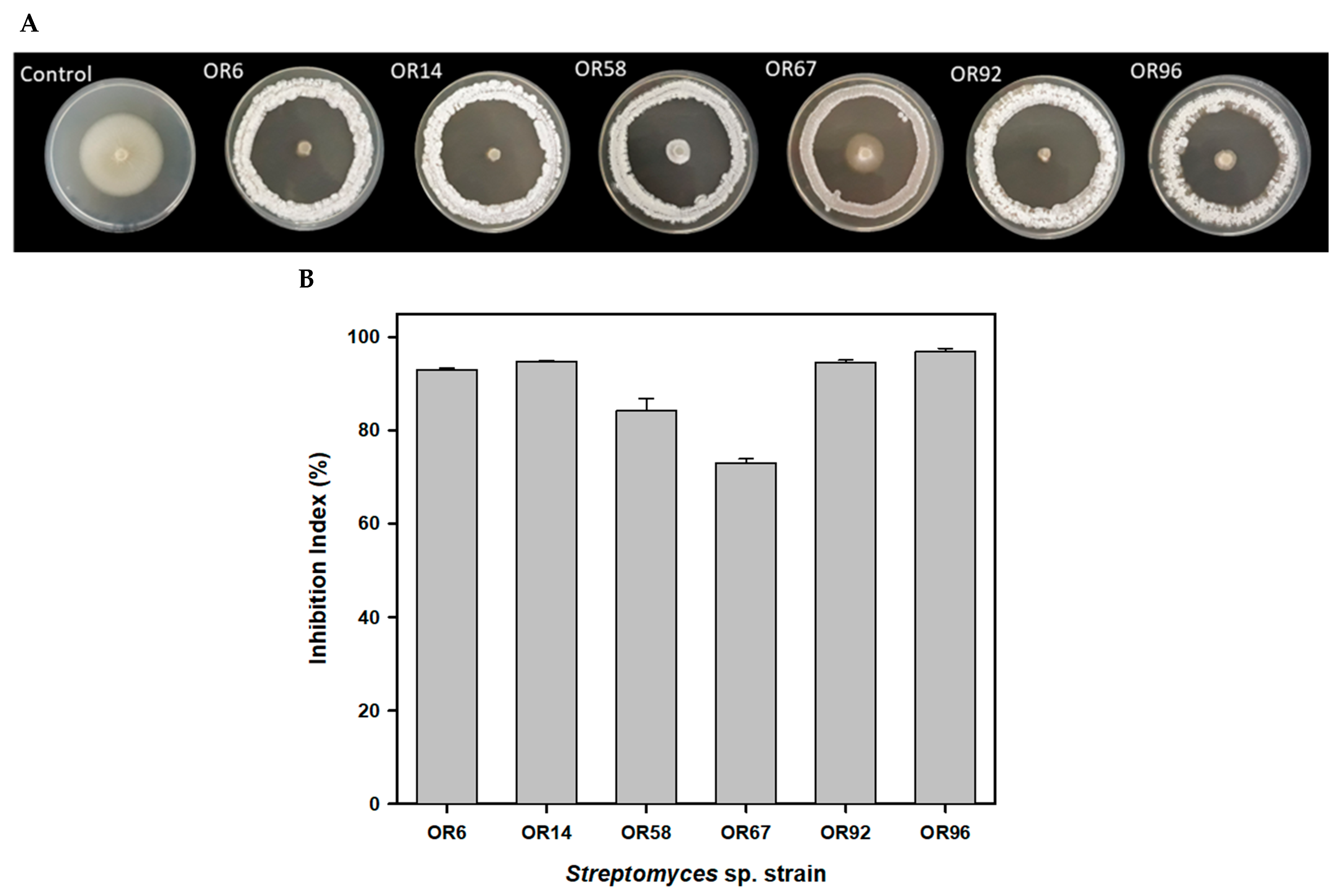
The six strains showing the highest Inhibition Index (I index), ranging from 73.07% for OR67 to 96.99% for OR96, were tested individually and selected for further studies (Figure 1).
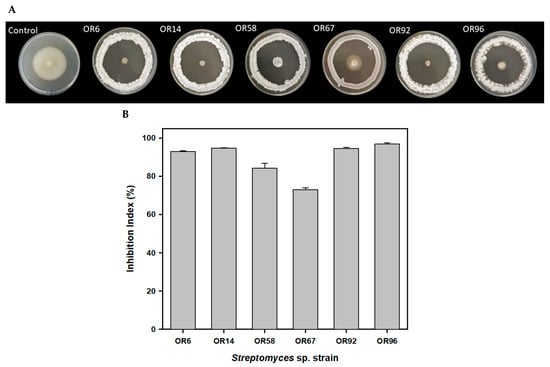
Figure 1.
Antifungal activity of the 6 best rhizosphere isolates against the fungal phytopathogen V. dahliae V937I as detected by a bioassay-based in vitro screening (A); inhibition index of the 6 best isolates (B).
2.2. Molecular Identification of the Selected Isolates by Partial Sequencing of 16S rRNA and Multilocus Sequence Analysis (MLSA)
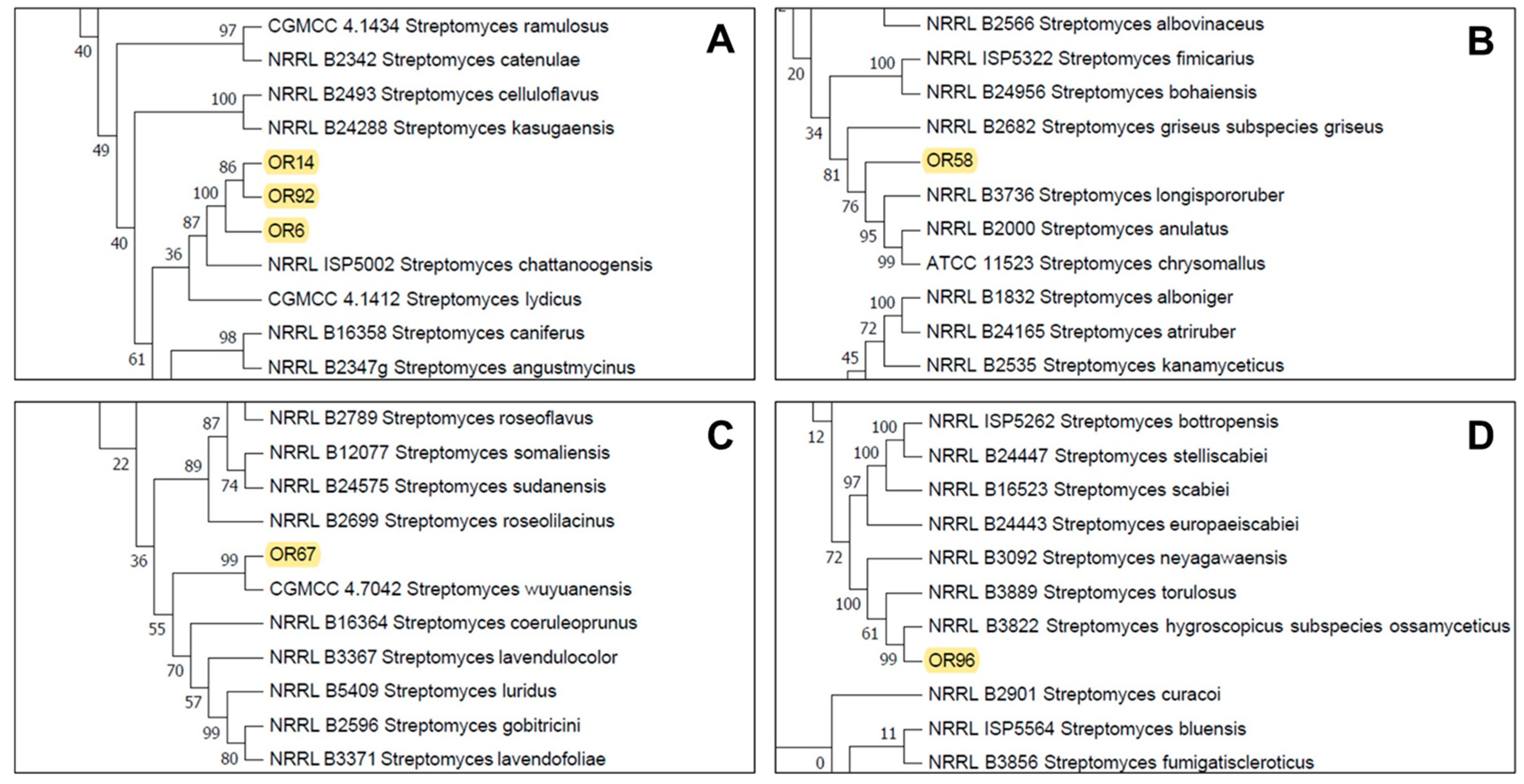
The partial sequencing of 16S rRNA identified the isolates as members of the Streptomyces sp. genus and they were initially designated as Streptomyces sp. OR6, OR14, OR58, OR67, OR92, and OR96. To further characterize and accurately identify them, we performed a multilocus sequence analysis (MLSA) of five housekeeping genes (atpD, gyrB, recA, rpoB, and trpB). MLSA (Supplementary Material, Figure S1) confirmed that isolates OR6, OR14, and OR92 belonged to the same species (sharing an MLSA evolutionary distance of ≤0.007; Table 1). These strains are closely related to Streptomyces chattanoogensis (Figure 2A), showing MLSA distances ranging between 0.043–0.045 with this species (Table 1). Strain OR58 is related to members of a subclade that included Streptomyces longispororuber and Streptomyces anulatus (Figure 2B), although its MLSA distance with these 2 species (0.013 and 0.014 respectively) clearly indicated that this isolate could represent a putative new species. Strain OR67 was placed in a subclade including Streptomyces wuyuanensis (Figure 2C), but its MLSA distance with this strain was 0.047, suggesting that OR67 could be a putative novel species. Similarly, strain OR96 was placed in a subclade that included Streptomyces hygroscopicus subsp. ossamyceticus (Figure 2D), but its MLSA distance with this strain was 0.018, implying that OR96 could be a putative novel species within that subclade.

Table 1.
Identification of the 6 rhizosphere selected isolates by MLSA analysis and MLSA distances from strains phylogenetically near to our isolates.
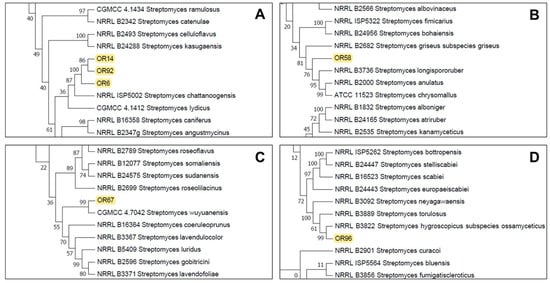
Figure 2.
Partial details of the Neighbor-Joining tree generated from the MLSA analysis showing the location of the selected strains Streptomyces sp. OR14, OR92, OR6 (A), Streptomyces sp. OR58 (B), Streptomyces sp. OR67 (C), and Streptomyces sp. OR96 (D). See Supplementary Materials, Figure S1 for the complete tree.
2.3. Strain-Specific Patterns of Inhibition in Streptomyces–Streptomyces Interaction Bioassays
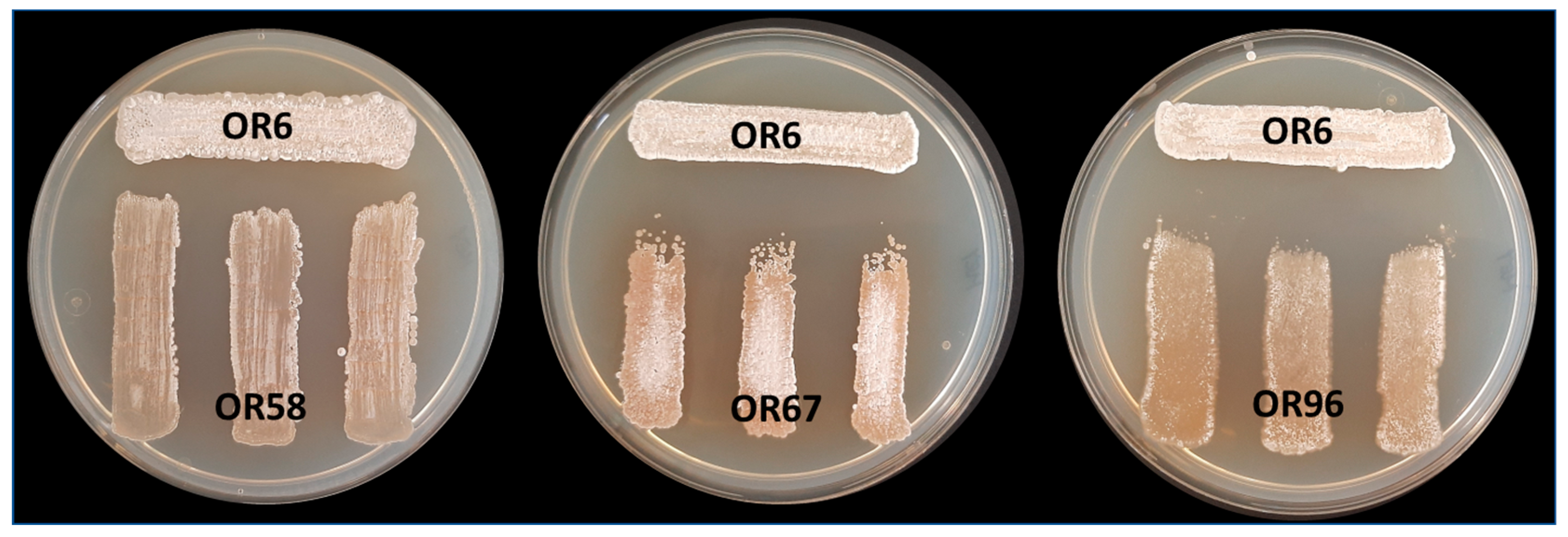
To examine the potential negative interactions among the selected Streptomyces sp. strains, we performed co-culture bioassays to evaluate the effects of four strains (OR6, OR58, OR67, and OR96) on each other. Based on the MLSA analysis, strains OR6, OR14, and OR92 belonged to the same species, so we chose strain OR6 from these strains for further studies. The main goal of this analysis was to detect any possible competition or other types of negative interactions between the selected strains as a preliminary step for designing a mixture of isolates that could have higher AF activity than each individual strain in the mixture. We observed that strain OR6 inhibited the growth of the other strains to some extent (Figure 3), with inhibition rates ranging from 27.33 (±2.83)% with OR67 to 20.00 (±2.24)% with OR96, and 10.44 (±0.88)% with OR58 (Table 2).
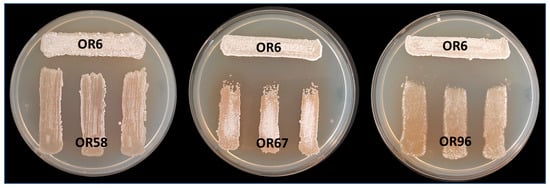
Figure 3.
Analysis of cross interactions between the four different selected rhizosphere Streptomyces sp. strains after MLSA analysis. In this case, strain OR6 was acting as “tester strain” and strains OR58, OR67, and OR96 were acting as “receiver strains”. Each strain was challenged with each other on a Petri dish co-culture bioassay 3 times (n = 9). Similar assays were carried out using strains OR58, OR67, and OR96 as receivers.

Table 2.
Inhibition index (Ii) between the 4 best rhizosphere strains selected according their antifungal activity against V. dahliae V937I. Each strain was challenged with each other on a Petri dish co-culture bioassay 3 times (n = 9).
When we used OR58 and OR67 as tester strains, they showed inhibition indexes that were always ≤8.00%, indicating low levels of inhibition. In contrast, strain OR96 did not exhibit any noticeable signs of growth inhibition or negative interactions with the other three strains in the test (Table 2).
2.4. Analysis of AF Activity Due to Volatile Organic Compounds (VOCs)
The capability of OR6, OR58, OR67, and OR96 to inhibit the growth of V. dahliae due to the putative production of VOCs was tested by using a classic double-dish chamber test. In all the cases, the AF activity due to VOC production was very low with an inhibition index of 20.54% ± 11.43 (OR6), 11.11% ± 8.57 (OR58), and 23.25% ± 6.39 (OR96), whereas no AF activity mediated by VOCs was detected for OR67.
2.5. Analysis of the Antifungal Activity of the Selected Strains in Small-Scale Soil Tests
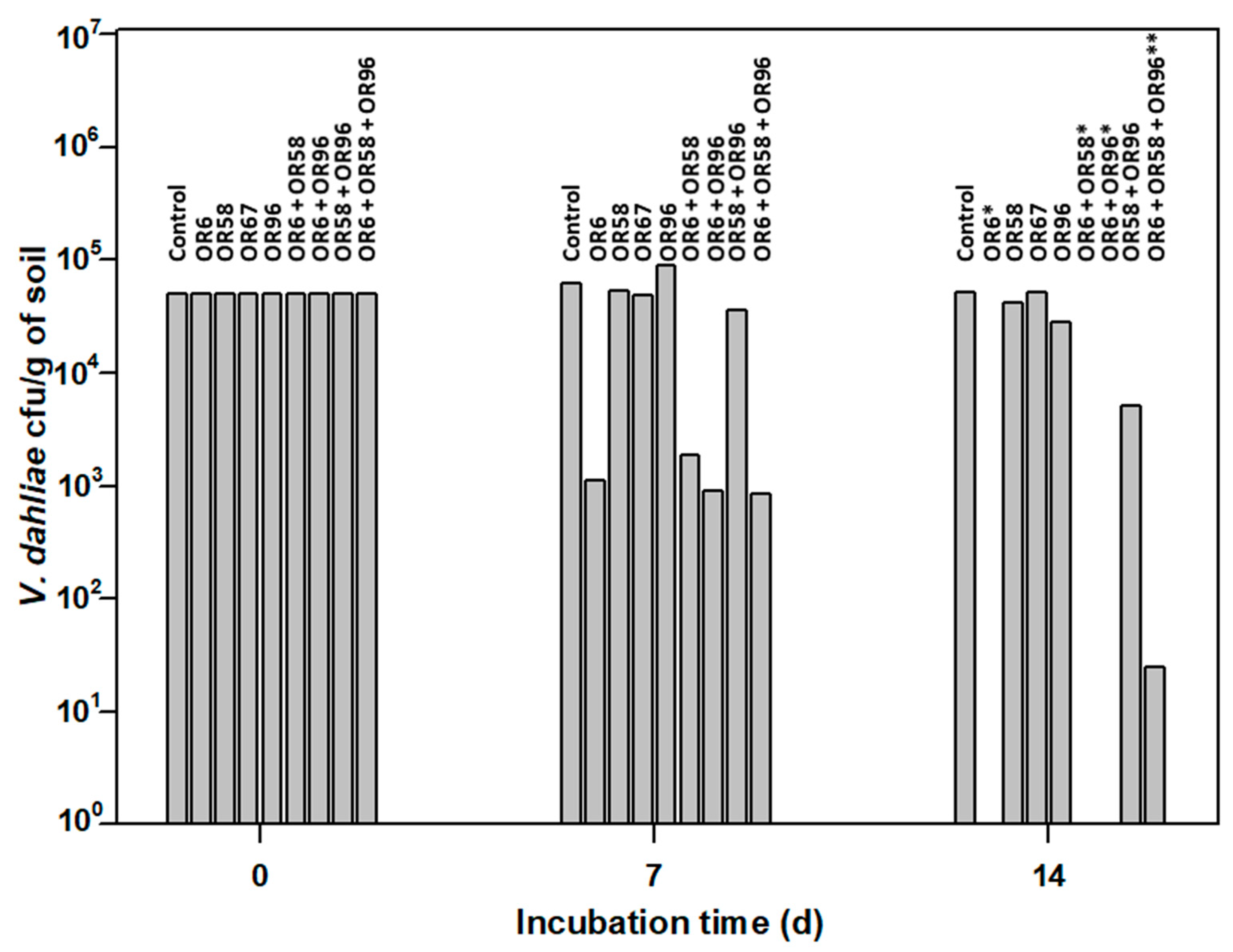
Many microbial isolates show good AF activity in in vitro assays, but it is unclear whether they can maintain the same activity when inoculated into soils. Therefore, we designed a small-scale in vitro assay to test the AF activity of the selected strains OR6, OR58, OR67, and OR96 against the pathogen under soil conditions. We inoculated these strains into sterile soil and measured their effects on the pathogen growth and survival (Figure 4). Sterile soil was used in this kind of experiment in order to avoid putative interferences by other microorganisms presents in the original soil sample. In this way we could be sure that the inhibitory effect observed on the pathogen was exerted by the microorganism(s) used in each individual assay.
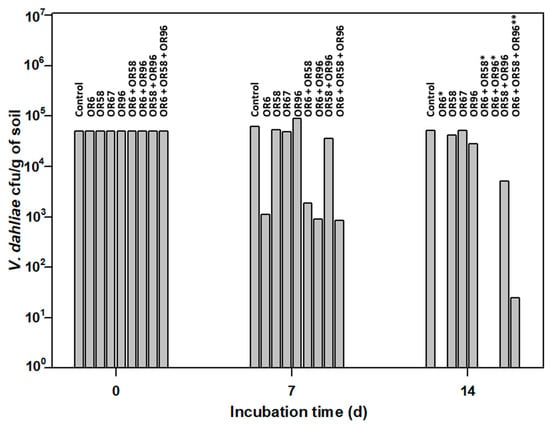
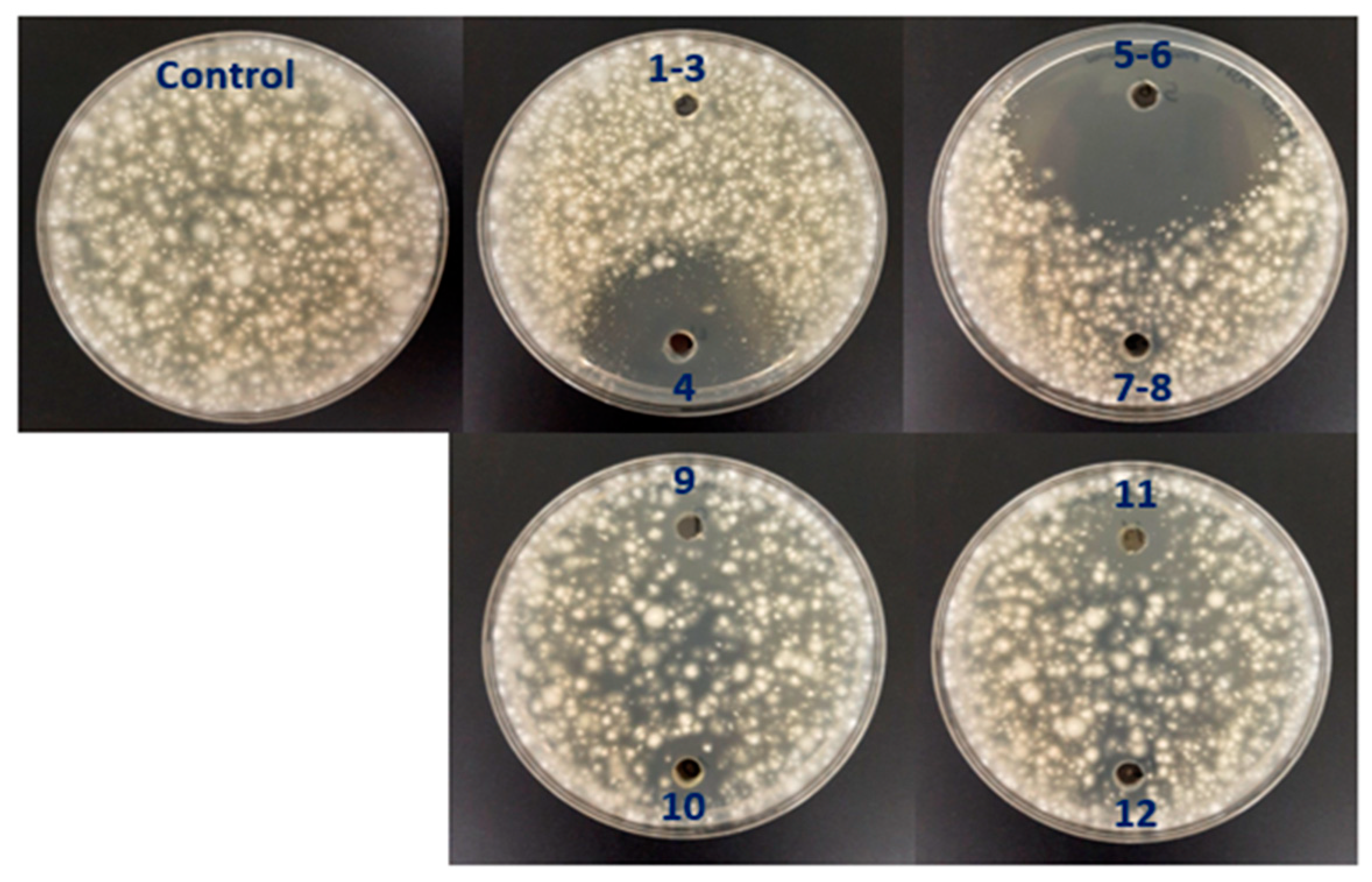
Figure 4.
Small-scale in vitro soil assay to test the antifungal activity of the selected strains OR6, OR58, OR67, and OR96 against V. dahliae. * No viable V. dahliae was detected at 14 days (d) of the experiment. ** An average of 25 ± 3 Colony Forming Units (cfu) of V. dahliae per g of soil were detected at 14 d of the experiment. The values shown are the mean of two independent experiments performed in triplicate.
The soil used In this study had a loamy texture, as per the USDA standard classification. The pH of the soil was 7.98, and it had an organic matter content of 6.59%. The total nitrogen content was 0.32%. These were the most characteristic aspects of the soil, and no other relevant data were available.
The pathogen was viable in this assay, as the average number of cfu/g of soil detected after 14 days of incubation was 51,500, close to the 50,000 cfu/g of soil added at the beginning of the experiment.
As shown in Figure 4, OR6 exhibited strong AF activity. After 7 days of incubation, only 1100 cfu/g of soil of the pathogen were detected, and its efficacy at 14 days was close to 100%, as less than 10 cfu/g of soil were detected in each replicate. In contrast, OR67 had no effect on the pathogen number at 7 and 14 days of incubation. Isolate OR58 did not affect the pathogen survival at 7 days of incubation but showed a slight reduction (16.0% on average) in pathogen viability after 14 days of incubation. Similarly, isolate OR96 did not inhibit the pathogen growth at 7 days of incubation but reduced it by 44% on average at 14 days of incubation. When we applied strain OR6 in co-culture with strains OR58, OR67, or OR96, their effectiveness did not decrease in small-scale soil tests, despite the negative interactions observed above (Table 2). After 14 days of incubation, the viability of V. dahliae in OR6–OR58, OR6–OR67, or OR6–OR96 co-cultures was always close to zero, as we detected less than 25 cfu/g of soil on average.
These results indicated that isolate OR6 was the most promising for controlling V. dahliae in soils, as its AF activity strongly affected the pathogen survival in soil. Therefore, we decided to identify the chemical compound responsible for the strong AF activity observed.
2.6. Identification of Albocycline as the Main Antifungal Compound Produced by OR6
We cultivated strain OR6 in ISP2 medium in buffled Erlenmeyer flasks for 72 h at 28 °C and 200 rpm to achieve optimal AF activity in liquid medium [30]. Next, we performed an activity-guided vacuum flash chromatography (VFC) on the crude extract (fermentation broth). Thus, after further filtration and evaporation of the solvents under vacuum, 2.28 g of extract were obtained. A 2 mg sample of this extract was analyzed by LC/MS and checked for the presence of AF activity by plate bioassay. After VFC, 12 fractions were obtained and AF activity was tested by performing a plate bioassay. Fractions 4 and 5–6 showed the highest AF activity (Figure 5).
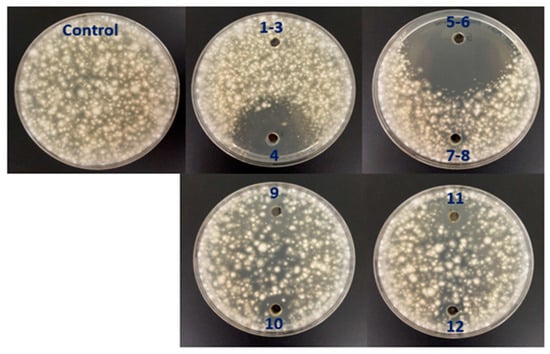
Figure 5.
Analysis of antifungal activity against V.dahliae in the 12 fractions obtained by vacuum flash chromatography (VFC) during the fractionation of a crude extract (fermentation broth) obtained from a liquid culture of Streptomyces sp. OR6. Note that most of the total antifungal activity is concentrated in fractions 4 and 5–6.
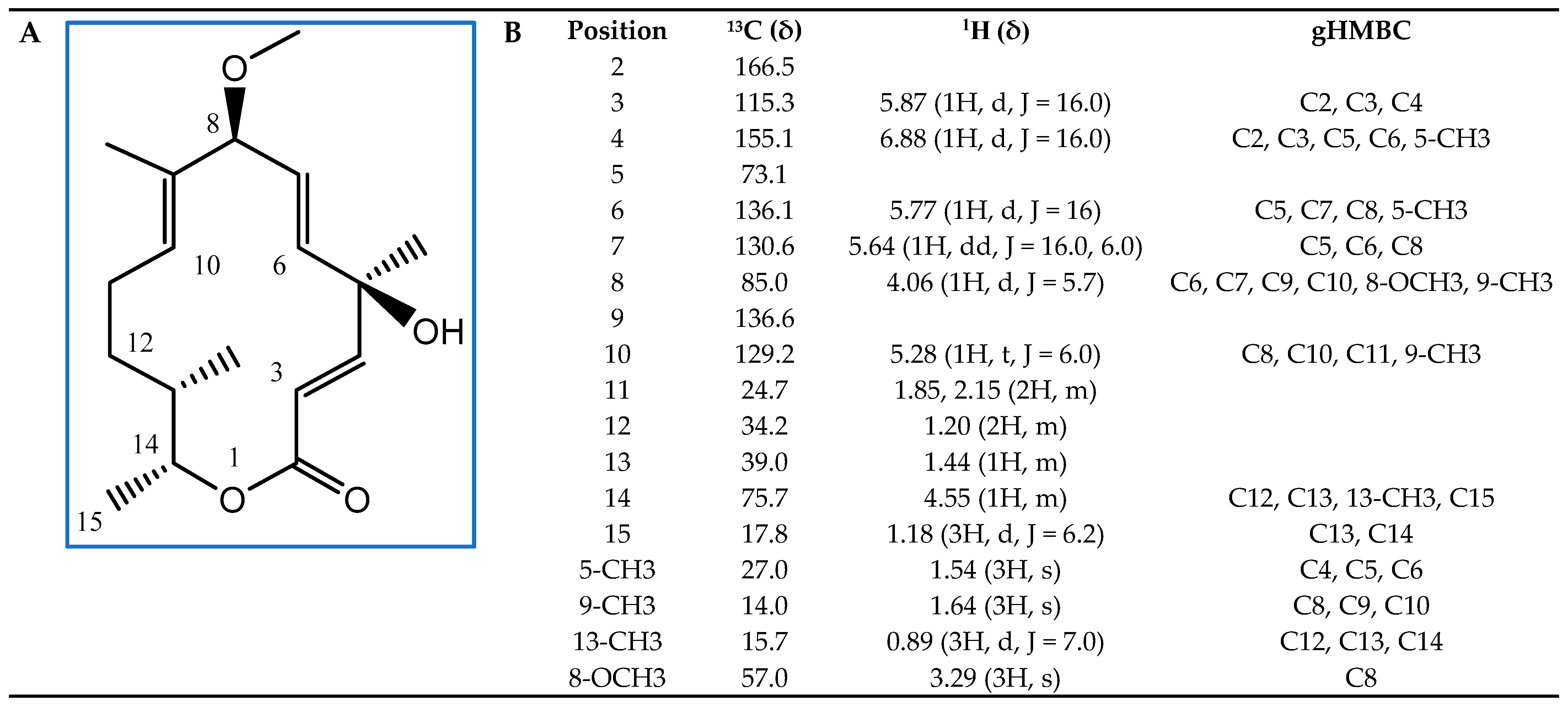
Next, we subjected these active fractions to liquid chromatography–mass spectrometry (LC/MS) and detected a major compound (Supplementary Materials, Figure S2). We determined its structure by nuclear magnetic resonance (NMR) spectroscopy (Figure 6). This analysis allowed us to identify the compound with AF activity as the 14-membered macrolide albocycline (also known as cineromycin-β-methyl ester or ingramycin). Previous studies have reported that albocycline inhibits the nicotinate [31] and peptidoglycan biosynthesis in prokaryotic microorganisms [32], whereas it also inhibits prolyl endopeptidases in eukaryotic cells [33], and exhibits both antibacterial and AF activity against various microorganisms [31,32,33,34,35,36,37,38]. Albocycline ESI-TOF-MS analysis showed an ion peak at m/z 309.2207 [M + H]+ (Supplementary Materials, Figure S2). The molecular formula was determined as C18H28O4 based on the HR ESI-TOF-MS and NMR spectral data. Extensive NMR experiments (1H NMR, 13C NMR, 1H-1H COSY, gHSQC, and gHMBC) indicated that albocycline is a 14-membered macrolide (Figure 6A). The spectroscopic data are in line with those reported in the literature [39,40]. 1H-NMR and 13C-NMR data are shown in Figure 6B.
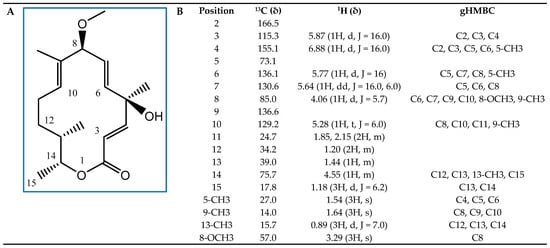
Figure 6.
Structural elucidation of albocycline as the main antifungal compound produced by isolate OR6. (A) Chemical structure with carbon positions labeled; (B) 13C, 1H, and Heteronuclear Multiple Bond Connectivity (gHMBC) NMR spectral data of albocycline (δ (ppm), JHH (Hz); CDCl3).
2.7. Effect of Albocycline on the Germination of V. dahliae Conidiospores and Sclerotia and Calculation of MIC Value
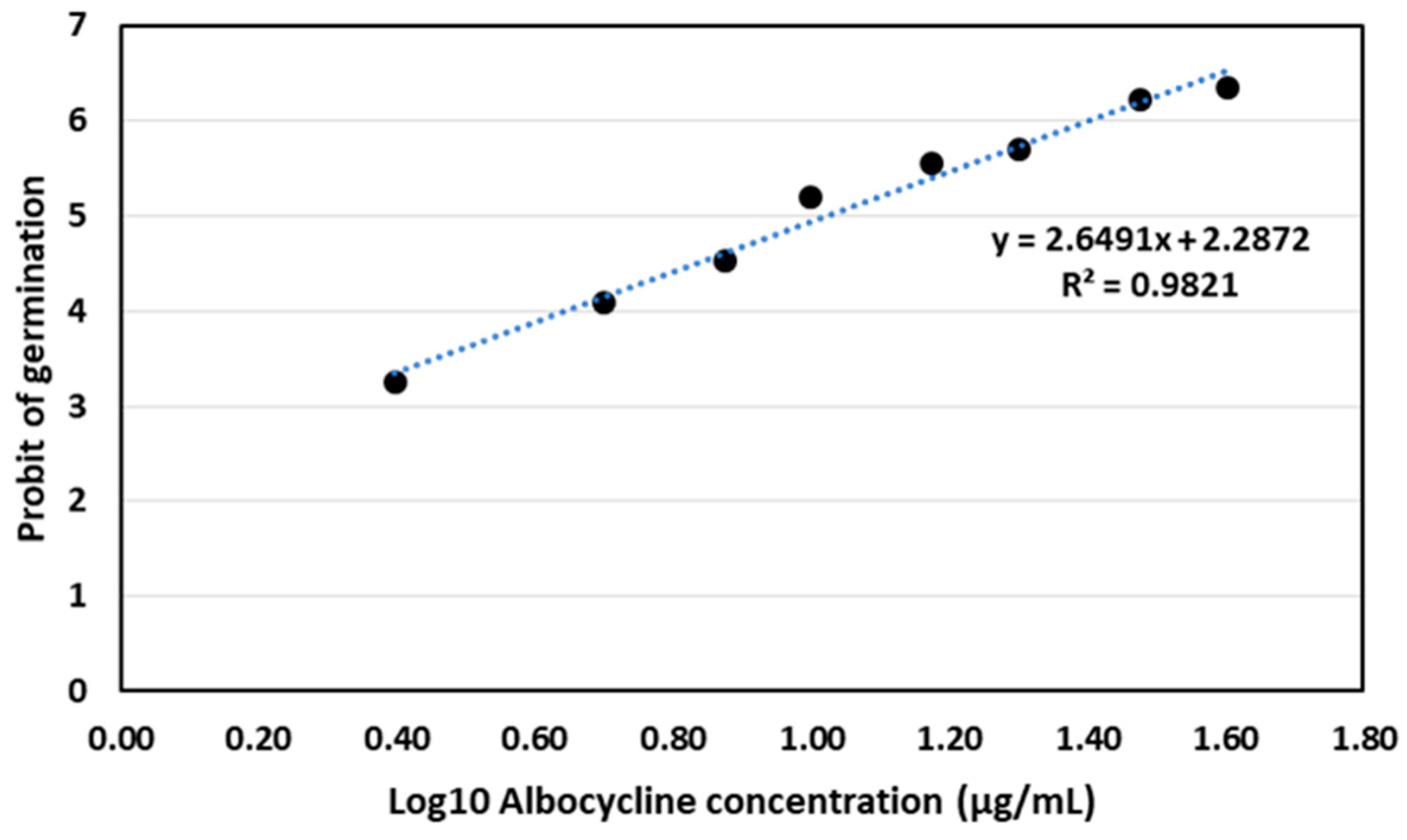
Albocycline was found to be effective in inhibiting the germination of conidiospores of V. dahliae V937I in the tested range of concentrations of 2.5–40 μg/mL. The Probit analysis of the germination data yielded a regression curve of y = 2.6491x + 2.2872 (R2 = 0.9821), from which we calculated the LC50 of albocycline to be 10.57 µg/mL (Figure 7).
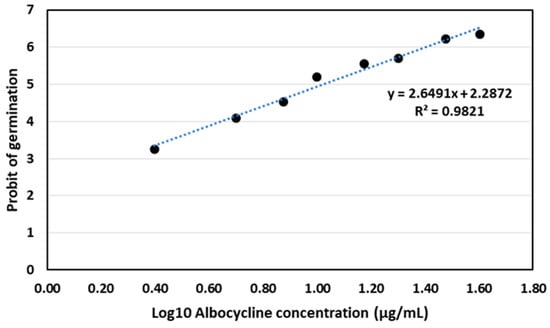
Figure 7.
Probit analysis for germination of conidiospores of V. dahliae V937I strain after treatment with albocycline. The data represented are the average of 3 independent experiments made by duplicate.
However, albocycline did not affect the germination of V. dahliae sclerotia at concentrations between 2.5 and 40 µg/mL. Microscopic observation revealed hyphal development from the germinated sclerotia, but the subsequent mycelial growth was inhibited by the AF activity of albocycline used in the assays carried out. The MIC value was calculated by standard procedures and determined to be between 5–6 µg.
3. Discussion
The productivity of agriculture worldwide is threatened by numerous fungal pathogenic pests that affect all crops [41,42,43]. Soil-borne fungal pathogens are particularly damaging and worrying because they spend much of their life cycle in the soil, where they can remain viable for long periods of time due to the production of different forms of resistance [5,44]. Once in contact with the sensitive plant system, they can infect it by penetrating through the root system itself and exerting their pathological activity. In many cases, by the time the symptoms of the disease are visible, the possibilities for control are practically non-existent [11]. Therefore, it is necessary to look for alternatives that allow for their control and propagation in the soil to reduce either their virulence or degree of infectivity.
Traditionally, the control of phytopathogenic fungi has been developed through the use of chemically synthesized pesticides. However, in the most advanced countries, such as those of the European Union, the medium-term policy is to ban their use and replace them with natural control methods that are non-toxic to humans and animals and are respectful of the environment. In this context, the development of BCAs emerges as a powerful tool for the control of soil-borne fungal pathogens. Although numerous scientific studies have analyzed the possible role of BCAs for the control of soil-borne fungal pathogens, relatively few have identified the substance(s) responsible for the observed AF activity.
The genus Streptomyces has broad biotechnological potential and is a promising candidate for the biocontrol of phytopathogenic microorganisms. The efficacy of some species of this genus in plant protection and their continued presence in the intensely competitive rhizosphere is due to their great potential to produce a wide variety of soluble bioactive secondary metabolites, including up to 8700 different antibiotics produced by actinomycetes [45], and volatile organic compounds. The capacity of Streptomyces as a BCA is based on different mechanisms such as induction resistance or priming plants, nutrient competition, hyperparasitism, or antibiosis [46,47].
Unfortunately, the potential effectiveness of a biological control agent (BCA) in the field is often limited by the fact that its ability to produce an antimicrobial compound in vitro does not necessarily correlate with its in situ antagonism [46]. In our study, we demonstrate that the potent antifungal activity of isolate OR6 is not only exerted in vitro but can also be detected in soil assays. This finding overcomes the aforementioned limitation, at least in soils with characteristics similar to those of the soil used in our assay. However, other isolates (OR58, OR67, and OR96) that exhibited good antifungal activity in vitro were ineffective in controlling the fungal pathogen in the soil assays we carried out. This data suggests that these isolates may not be well adapted to the characteristics of the soil used in our tests or may be unable to produce antifungal compounds when grown in soil. It is also possible that the soil contains substances or chemical compounds that inhibit antifungal production. We previously tested the AF activity of the OR6 isolate due to the production of VOCs and found it to be very limited. Therefore, it is highly probable that the potent AF activity detected in the soil test is due to the production of diffusible compounds.
Although a large number of reports dealing with Streptomyces isolates selected as BCAs to manage fungal phytopathogens can be found in the scientific literature, studies that identify the main compound(s) with AF activity are scarce. The identification of a chemical compound responsible for the AF properties of a particular strain is of great interest in order to develop formulations with a broad spectrum of antimicrobial action, promote plant growth, increase shelf life, and suppress diseases under field conditions. These characteristics would be highly attractive for technological exploitation and commercialization [47]. In our case, we were able to conclude that the AF activity of Streptomyces sp. OR6 was mostly due to an AF compound identified as albocycline.
Albocycline is a 14-membered macrolactone (macrolide) that is naturally produced by several Streptomyces strains. It was first isolated by Tanabe Seiyaku Company and The Upjohn company in 1967 and initially named as ingramycin [48]. From a structural point of view, its exact structure was elucidated by X-ray crystallography by Thomas and Chidester in 1982 [40]. Interestingly, albocycline is a 14-membered macrolactone with four stereogenic centers and three alkenes, in such a way that its skeleton is different from those of other representative 14-membered macrolide antibiotics such as erythromycin and oleandomycin. Moreover, albocycline lacks any carbohydrate moiety [49].
The production capacity of albocycline has been detected in several Streptomyces strains, including S. maizeus [32], S. brunneogriseus, S. roseocinereus, S. roseochromogenes var. albocyclini [50], and Streptomyces sp. AR10, a strain phylogenetically close to S. lanatus [37]. Albocycline production has also been reported by a Streptomyces sp. isolated from pady soil and for an S. sparsus strain isolated from deep-sea sediment samples from the Bay of Bengal [36]. Albocycline production has also been detected in other actinobacteria such as Propionicimonas sp. ENT-18 [35]. Its production capacity is expanded with the albocycline producer Streptomyces sp. OR6 that, according to the MLSA analysis carried out, is a putative new species phylogenetically close to S. chattanoogensis, which does not appear in the bibliography as an albocycline producer.
Initially, albocycline was characterized as a potent antibacterial compound that inhibits the in vitro growth of a variety of Gram-positive and Gram-negative bacteria [48]. It is also active against methicillin-resistant Staphylococcus aureus (MRSA) and is equipotent with vancomycin [38]. The antibacterial properties of albocycline are due to its capability to inhibit peptidoglycan biosynthesis [38], although the exact molecular target remains to be elucidated [32]. Albocycline is also able to inhibit nicotinate biosynthesis in Bacillus subtilis cells [31].
Interestingly, more recently the AF activity of albocycline against Candida albicans [36] and the fungal phytopathogen Sclerotinia sclerotiorum [35] has been reported. Albocycline has also been reported to display AF activity in vitro against the fungal phytopathogen Rhizoctonia solani, being able to suppress Rhizoctonia dumping-off of cucumber in infection control assays [37]. Our findings indicate that albocycline is also active against V. dahliae, which causes Verticillium wilt in many different crops, inhibiting both conidiospore germination and mycelial growth. This is important since it adds another important fungal phytopathogen to the list of fungi sensitive to its AF action and reinforces a possible future use in the control of these pathologies.
Interestingly, it has been reported that a low concentration of albocycline (1.6 μg/sclerotia) was able to inhibit the sclerotia germination in S. sclerotiorum [35]. In addition, Zucchi and colleagues had previously reported that an ethyl acetate extract of Propionicimonas sp. ENT-18 was able to induce severe histological abnormalities in the sclerotia of S. sclerotiorum, affecting both the cell structure of the medullae and rind cell wall [34]. Although albocycline was identified as the main AF compound of this ethyl acetate extract, it cannot be concluded that albocycline is the cause of the observed abnormalities in the sclerotia structure [35]. In our case, albocycline was unable to inhibit sclerotia germination in the tested concentration range. This different behavior of albocycline in relation to its ability to inhibit sclerotia germination observed in V. dahliae and S. sclerotiorum could be due to structural or compositional differences in the wall of both sclerotia.
Unfortunately, there is no clear evidence about how albocycline can exert its AF activity, although we do know that it is a potent inhibitor of human prolyl endopeptidases, enzymes that are involved in the degradation of neuronal peptides containing proline residues [33]. Interestingly, albocycline lacks toxicity to mice and humans [31,32,51], which is a positive aspect that must be taken into account in a possible future application in the field.
Finally, we would like to emphasize that semi-synthetic albocycline analogs can be obtained by functionalization at three specific sites: the C2-C3 enone, the tertiary carbinol at C4, and the allylic C16 methyl group. One of the semi-synthetic C4 ester analogs obtained was twice as potent as albocycline, although only its antibacterial activity was tested [52]. Additionally, it has been reported that albocycline can be biomodified by Streptomyces venezuelae. However, the modification carried out consisted of a reduction in albocycline to yield a 2,3-dihydroalbocycline derivative that was antimicrobially inactive [53]. Both types of studies are very interesting since they allow us to visualize that it is possible to modify the structure of albocycline to generate similar compounds, either by chemical modification or biotransformation, that could result in new derivatives with greater AF activity.
4. Materials and Methods
4.1. Isolation of Culturable Streptomycetes Strains from the Rhizosphere of Olive Trees
Root-adjacent soil (rhizosphere) samples were collected from three adult olive trees that exhibited visual symptoms compatible with Verticillium wilt in a plot belonging to the company Río Lacarón in La Garrovilla, Spain, at 215 m above sea level (38°55′08.1″ N 6°31′27.7″ W). A visual inspection indicated that approximately 13% of the trees in that plot exhibited Verticillium wilt visual symptoms. Sampling of rhizosphere soil was made after digging to access the root system of the plants. After leaving the root system exposed, rhizosphere soil in direct physical contact with the roots was collected with a small sterile spatula. Soil samples were transferred to sterile Falcon tubes (50 mL), kept in an icebox, and preserved at 4 °C until processing. We isolated culturable actinobacteria from 1 g of soil samples by suspending them in 10 mL of Solution I (0.5% SDS; 6.0% yeast extract), homogenizing them using a vortex, and incubating them at 40 °C for 15 min. We made tenfold serial dilutions in sterile water and inoculated 0.1-mL aliquots of each dilution on starch-casein agar (SCA) [54] and International Streptomyces Project 2 (ISP2) [30] agar media, supplemented with 100 μg/mL pimaricin and 50 μg/mL nalidixic acid (Sigma-Aldrich, St. Louis, MO, USA) to prevent fungal and Gram-negative bacterial growth, respectively. We incubated the plates at 30 °C for 5 to 7 days. We selected different isolates based on their morphological and cultural characteristics, such as colony properties, presence/absence of aerial mycelia, spore mass color, distinctive reverse colony color, and production of diffusible pigments. We routinely cultivated and maintained the isolates on MEY (Maltose Yeast Extract) medium [55] at 4 °C. We maintained spore-producing isolates as spore suspensions at −20 °C in glycerol (40%).
4.2. Preliminary Identification of Strains by 16S rRNA Sequencing
Those strains exhibiting higher AF activity were identified by partial sequencing of 16S rRNA. Briefly, genomic DNA extraction was performed as described by Hopwood et al. [56]. The 16S rRNA genes were amplified using the oligonucleotides 27F and 1492R [57]. Isolates were identified by comparing them to corresponding sequences of the type strain found on the EzTaxon-e database [58]. (http://www.ezbiocloud.net/eztaxon/identify (accessed on 9 January 2023)). Sequence alignments were performed using the MEGA v7.0 software (http://www.megasoftware.net/ (accessed on 10 January 2023)). Evolutionary distance was calculated using the Kimura two-parameter (K2P) model for nucleotide sequences [59].
4.3. Identification by Multilocus Sequence Analysis (MLSA)
MLSA analyses were carried out by using five housekeeping genes: atpD (ATP synthase F1, β-subunit), gyrB (DNA gyrase B subunit), recA (recombinase A), rpoB (RNA polymerase, β-subunit), and trpB (tryptophan synthase, β-subunit) [29]. We amplified the housekeeping genes using the primers and conditions described by Guo [60] and Rong [61]. The GenBank accession numbers of DNA sequences from the housekeeping genes are listed in Table 3. A phylogenetic tree was constructed from a concatenation of the five housekeeping genes. All the sequences were concatenated by joining them head to tail. DNA sequences were manually trimmed at the same position before being aligned using MEGA 7.0 software with sequences from type strains obtained from the ARS Microbial Genomic Sequence Database server (https://data.nal.usda.gov/dataset/ars-microbial-genomic-sequence-database-server (accessed on 19 January 2023)). The phylogenetic tree was constructed using the maximum likelihood method with the Kimura two-parameter model [59]. MLSA evolutionary distances were calculated using MEGA 7.0 by calculation of the K2P distance. Strain pairs having ≤0.007 MLSA evolutionary distance were considered conspecific based on the guideline empirically determined by Rong and Huang in 2012 [29].

Table 3.
GenBank accession numbers of DNA sequences corresponding to partially sequenced genes used in the MLSA analysis of the different rhizosphere Streptomyces sp. strains tested.
4.4. In Vitro Selection of Isolates by Their AF Activity in Plate Assays
We tested all the selected isolates for AF activity using an in vitro AF assay as described previously [62]. Briefly, we inoculated the isolates on potato dextrose agar (PDA) plates in a 1.0 cm2 area (four isolates per plate, 1 cm from the edge of the plates). An agar plug (0.5 cm) containing V. dahliae V937I strain [63] was deposited in the center of the plates. Then, they were incubated for up to 12 days at 25 °C and we measured the growth inhibition zones. In order to quantify the AF activity of the best isolates, they were individually tested on MEY agar plates forming a circle at 1 cm from the edge of the plate. An agar plug containing V. dahliae was deposited in the center of the plate (Figure 1A). Plates were incubated at 25 °C up to 10 days. The inhibition index (I index) was calculated as follows: I index (%) = [(Rc − R)/Rc] × 100, where R is the radius of the fungal colony in the presence of the bacterial colony, and Rc is the maximum radius of the fungal colony (control). We performed this assay in triplicate for each actinobacteria tested.
4.5. Analysis of Antifungal Activity of Volatile Organic Compounds (VOCs)
The production of VOCs with putative AF activity was tested by using the double-dish chamber test [64]. Briefly, two sterile bottom dishes of Petri dish (9 cm in diameter) were used. One dish contained Potato Dextrose Agar (PDA) medium (Sigma-Aldrich) that was inoculated with a 9 mm diameter agar plug containing V. dahliae. Another dish contained a Streptomyces sp. culture grown in MEY agar medium [55] at 30 °C until a good sporulation was obtained (normally 3–5 days) depending on the strain tested. The two plates were faced so that the plate inoculated with the fungus was placed inverted on the plate inoculated with the bacteria. Both dishes were sealed with Parafilm and incubated at 25 °C for 14 days. Then, the inhibition index was calculated as indicated above.
4.6. Analysis of AF Activity in Liquid Culture Media and in Fractions Obtained during Albocycline Purification Process
We grew 100-mL liquid cultures of the OR6 strain in four different media: ISP2 [30], SPG [65], TSB, and yeast extract malt extract (YEME) [56]. We used indented 500 mL Erlenmeyer flasks containing 100 mL of liquid media. Each flask was inoculated with 10 agar plugs (0.5 cm diameter) of Streptomyces sp. OR6 grown on SCA plates until they produced a good amount of spores. To determine the best media for AF production, as well as the growth rate and the time of maximum AF activity, we took samples of 1 mL of culture every 24 h. The samples were centrifuged to remove cells and we tested the supernatant for AF activity using an in vitro AF plate assay against V. dahliae V937I strain [63] on PDA agar plates containing 0.8% of agar (w/v). We embedded 5.0 × 104 fungal spores/mL in the agar media before plating. Then, we placed culture supernatant samples (60 µL) in a well on the agar plate at 1 cm from the edge of the plate. We incubated the plates at 25 °C for 4 days and measured the inhibition halos.
Plate assays to detect the presence of AF activity in the different fractions obtained during the albocycline purification protocol (see Section 4.11) were conducted as described above. Fractions (60 μL) were deposited in a well (5 mm diameter) made in the center of PDA plates containing 0.8% agar.
4.7. Confrontation of Rhizosphere Streptomyces sp. Isolates with Each Other
In order to rule out the possible existence of any negative interaction, the influence of the six selected Streptomyces sp. isolates upon each other was tested pair-wise in a bioassay following the protocol described by Schrey et al. [66]. Streptomyces liquid cultures were grown for three days in ISP2 medium. From the tester strain, 40 μL of this suspension culture were applied on the lower part of an SCA Petri dish, forming a line. After sporulation of the tester strain began, three parallel lines of the receiver strains were applied perpendicularly to the tester line. For each Streptomyces sp. pair, three tester and nine receiver lines were applied. The impact of the tester strain on the growth and formation of the receiver strain’s substrate mycelium and sporulation was recorded at the time point of the onset of sporulation in the control cultures.
4.8. Analysis of AF Activity in Small-Scale Soil Tests
To evaluate the AF activity in soil, we conducted small-scale tests in 50 mL Falcon tubes containing 5 g of sterile soil. Spores of each particular strain tested were obtained by growing in MEY agar plates at 28 °C and up to 6–8 days. Spores were recovered in glycerol (20%) and later were quantified by performing tenfold serial dilutions and plating on MEY plates. We inoculated the soil with 500 µL of a bacterial suspension supplemented with starch (1%) and casein (0.5%) to achieve a final concentration of 107 cfu/g of soil. We homogenized the soil samples by vortexing and manually with a spatula to obtain a uniform distribution of the inoculum. We incubated the tubes at 30 °C for 48 h. Then, we added 105 spores/g of soil of V. dahliae to each tube and homogenized them again as before. We also included a non-inoculated negative control and a positive control inoculated only with the fungal pathogen in the study. We incubated the tubes at 25 °C. Next, we collected samples of 100 mg of soil at 7 and 14 days after the bacterial inoculation and made tenfold serial dilutions in sterile water. We inoculated 0.1 mL aliquots of each dilution on PDA plates supplemented with 250 μg/mL of chloramphenicol to quantify fungal growth, and on SCA plates supplemented with 100 μg/mL pimaricin and 50 μg/mL nalidixic acid to quantify the presence of the tested actinobacteria.
4.9. Soil Physicochemical Properties
Physicochemical properties of the soil used for the small-scale soil test were determined in the Laboratory of Instrumental Techniques (University of León, León, Spain). Soil samples were taken at 20–30 cm deep, after removing the 5 cm of superficial soil. Five samples were taken and combined for analysis. Soil texture and mechanical analysis were performed according to the standard sedimentation method with particle size distribution in a combination of sieving and sedimentation techniques (ISO 11277:2020). The organic carbon was determined by Walkley–Black wet oxidation. The pH and EC were measured in the supernatants of soil: water 1:2.5 suspensions following 25 min of shaking and 5 min of soil settlement, and using, respectively, a micro-pH 2001 pH meter and a conductivity meter 522. Soil nitrogen was determined by the Kjeldahl method [67], which determines all soil nitrogen except that in nitrates [68]. Soil phosphorus available for vines was determined by ultraviolet–visible spectroscopy after treatment with the Olsen–Watanabe extractant. Exchangeable potassium was determined by extraction with successive aliquots of 1 M ammonium acetate (NH4C2H3O2) followed by determination by atomic absorption spectrometry (AAS; Unicam SOLAAR 969). The micronutrients (Fe, Cu, Mn, and Zn) were determined using atomic absorption spectrometry (AAS) in an air-acetylene flame [69]. Magnesium was analyzed with the CaCl2 method [70] determined by AAS. Available boron was measured according to the standard NF X31-122, and humus content according to the ISO 14235:1999. The total carbonates and active carbonates were determined with the Bernard calcimeter method, after extraction with ammonium oxalate 0.2 N for the last ones.
4.10. Obtaining Crude Extracts and HPLC Analysis
We obtained crude extracts of the OR6 strain in the selected media for antifungals production as described by Das et al. [71], with minor modifications. We mixed the cell-free culture supernatant vigorously with ethyl acetate in a 1:1 (v/v) ratio for 30 min and separated the organic layer. We evaporated the ethyl acetate extract in a vacuum concentrator CentriVap (Labconco, Kansas City, MO, USA) and dissolved the residue in 80% methanol to a final concentration of 1 mg/mL. We tested samples of 60 µL of the crude extract for antifungal activity using the antifungal bioassay described above. We filtered the rest of the sample by 0.45 µm pore Corning® Costar® Spin-X® centrifuge tube filters (Merck KGaA, Darmstadt, Germany) and stored them at −20 °C until analysis. We analyzed the crude extracts by high performance liquid chromatography (HPLC) following the chromatographic method described by Awla et al. [72] using an Agilent 1200 Series Gradient HPLC System (Agilent Technologies, Santa Clara, CA, USA) equipped with a quaternary pump delivery system (G1311A), a preparative autosampler (G1329A), a diode array multi-wavelength detector (G7115A), and an analytical fraction collector (G1364F) equipped with an Autosampler Thermostat (G1330B). We injected samples of 10 µL and resolved them on an analytical Lichospher RP18 column (40 × 250 mm; 5 µm) (Teknokroma, San Cugat del Vallés, Spain).
4.11. Purification of Albocycline by Vacuum Flash Chromatography (VFC)
Albocycline was purified from a crude extract (fermentation broth) by vacuum flash chromatography (VFC) as follows. Streptomyces sp. OR6 was grown in 500-mL indented Erlenmeyer flasks containing 125 mL of ISP2 medium. Each flask was inoculated with 10 agar plugs (0.5 cm diameter) of the bacterial strain grown on SCA plates until a good sporulation had been obtained. Cultures were incubated at 30 °C and 150 rpm for 72 h. Fermentation broth (6 L) was shaken with the adsorption resin Amberlite XAD-1180 (Dupont, Mississauga, ON, Canada), and filtered using Radifil RW50 as filtration coadjuvant (Agrovin, Alcázar de San Juan, Spain). The exhausted broth was discarded and the mixture of resin and mycelium was extracted with 3L of EtOAc/MeOH (3:1). After further filtration and evaporation of the solvents under vacuum, 2.28 g of extract were obtained. The extract was analyzed by LCMS, and checked for the presence of AF activity by plate bioassay.
A 2 mg sample of the obtained extract was dissolved in 400 μL of MeOH. The sample was analyzed by HPLC (1290 Infinity II, Agilent) coupled to an Agilent 6230 time-of-flight LC/MS (LC/TOF) mass spectrometer. The chromatographic column used was an Agilent Zorbax Eclipse Plus, C18 RRHD (2.1 × 50 mm; 1.8 μm particle size), gradient system MeOH/H2O (0.1% formic acid), 20% to 100% over 8 min, UV detection 220 nm, and flow rate 0.6 mL/min. Once checked that the obtained extract exhibited AF activity, it was subjected to VFC (Vacuum Flash Chromatography) using silica gel (40–60 μm, 60 A) (Thermo ScientificTM, Waltham, MA USA) as a stationary phase and for the mobile phase a gradient of Hexane/EtOAc/MeOH of increasing polarity was used to obtain a total of 12 fractions. All the fractions were analyzed by LCMS, as described above, and tested for AF activity. Active fractions eluted with Hexane-EtOAc 1:1 and 3:7 showed the presence of a major compound in their LC/MS analysis (Supplementary Materials, Figure S2).
4.12. Structural Elucidation of Albocycline
The molecular formula of the major compound detected in the active fractions was determined based on the HR ESI-TOF-MS and NMR spectral data. Extensive NMR experiments (1H NMR, 13C NMR, 1H-1H COSY, gHSQC, and gHMBC) were carried out in order to elucidate its structure. 1H-NMR and 13C-NMR data were recorded on a Varian “Mercury 400” spectrometer (Agilent Technologies) at 400 and 100 MHz, respectively. gHMQC and gHMBC experiments were carried out using an inverse resonance probe. Chemical shifts are reported in ppm relative to solvent (CDCl3 δH 7.26, δC 77.0). MS data were recorded on an Agilent 6230 time-of-flight LC/MS (LC/TOF) mass spectrometer.
4.13. Analysis of the Ability of Albocycline to Inhibit Conidiospore Germination
The ability of albocycline to inhibit conidiospore germination was analyzed using a modified protocol of López-Moral [73]. Conidial suspensions were obtained from 12-day-old colonies of V. dahliae isolate V937I growing on PDA and adjusted to 1 × 106 conidia/mL. In parallel, a 5 mg/mL albocycline-concentrated solution in methanol was used to prepare more diluted 20, 40, 60, 80, 100, 120, 140, 160, 180, and 200 µg/mL solutions in sterile distilled water, in order to avoid a putative inhibitory effect of methanol on conidiospore germination. Subsequently, a 5 µL drop of the conidial suspension was placed in the center of a microscope coverslip; then, a 5 µL drop of the albocycline solution was mixed. The albocycline was evaluated at the following final concentrations: 2.5, 5, 7.5, 10, 15, 20, 30, and 40 µg/mL. A concentration of 0 µg/mL consisting of a 5 µL drop of the conidial suspension mixed with a 5 µL drop of sterile distilled water was used as a negative control. The coverslips were placed inside Petri dishes containing water agar and used as humid chambers. They were incubated at a temperature of 25 °C in the dark for a period of ten hours. After the incubation period, a 5 µL drop of 0.01% acid fuchsine in lactoglycerol (1:2:1 lactic acid:glycerol:water) was added to each coverslip to stop conidial germination. The coverslips were then mounted on a slide. The assay was performed in triplicate for each concentration and repeated twice. For each replicated coverslip, 100 randomly selected conidia were observed at a magnification of ×400 using a phase-contrast microscope (Olympus CX41), and germinated conidia were counted. Conidia were considered germinated when the germ tube was at least as long as the longitudinal axis of the conidia. The inhibition of conidial germination (RGI; %) was estimated with respect to the control using the following formula: RGI = [(Gcontrol − GAlb)/Gcontrol] × 100 where Gcontrol = percentage of germinated conidia after incubation in water and GAlb = percentage of germinated conidia after incubation in the albocycline solution. The data obtained were plotted to obtain a regression line and an equation from which the Inhibitory Concentration of albocycline causing a 50% growth inhibition (IC50) was calculated by Probit analysis of the mortality data.
The calculation of the Minimum Inhibitory Concentration (MIC) was carried out by performing a classic disk-diffusion test [74,75] in PDA plates. The amount of albocycline per disk ranged from 5 to 50 µg.
5. Conclusions
A Streptomyces sp. OR6 strain isolated from olive rhizosphere exhibited notable AF activity in vitro against the phytopathogenic fungus V. dahliae. AF activity was mainly exerted by the macrolide albocycline. This strain brings together a series of very interesting properties that could in the future result in a possible commercial use as a biopesticide, such as its use in the design of biofertilizers, including: (i) the produced albocycline is active against several of the most important phytopathogenic fungi affecting many crops including V. dahliae (this work), R. solani [33], and S. sclerotiorum [35]; (ii) the ability to show AF activity in soil assays; (iii) albocycline is able to strongly inhibit the germination of V. dahliae conidiospores; (iv) since it is a strain isolated from the rhizosphere of olive trees, it is assumed to have a good adaptation to colonize that microenvironment, in which the infection of the tree by the pathogen occurs, where it can exert its AF activity; (v) finally, it should be highlighted that, as previously described in the literature, albocycline does not exhibit toxicity against mice and humans, a positive aspect that should be taken into account in its possible future field application.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12203612/s1, Figure S1: Streptomyces phylogenetic tree inferred from concatenated partial sequences of the housekeeping genes (atpD, gyrB, recA, rpoB, and trpB) of rhizosphere isolates (OR6, OR14, OR58, OR67, OR92, and OR96) with type strains obtained from the ARS Microbial Genomic Sequence Database server; Figure S2: Analysis of antifungal activity against V. dahliae in the 12 fractions obtained by vacuum flash chromatography (VFC) during the fractionation of a crude extract (fermentation broth) obtained from a liquid culture of Streptomyces sp. OR6.
Author Contributions
Conceptualization, R.C. and J.J.R.C.; methodology, C.C.-P., R.C., J.M.S.-L. and A.I.; technical support, C.C.-P., R.C., J.M.S.-L. and A.I.; writing—original draft preparation, R.C., J.M.S.-L. and J.J.R.C.; writing—review and editing, C.C.-P., R.C., J.M.S.-L. and J.J.R.C.; funding acquisition, J.J.R.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Rio Lacarón S.L. (La Garrovilla, Badajoz, Spain). Carla Calvo-Peña (EDU/601/2020) was supported by a predoctoral contract from the Junta de Castilla y León and the European Social Fund. Ana Ibañez was supported by a “Margarita Salas” modality postdoctoral grant (Reference no.: UP2021-025) through the University of León awarded by the Spanish Ministry of Universities within the Recovery, Transformation and Resilience Plan (Modernization and digitalization of the Educational System), for which funding comes from the European Recovery Instrument European Union—NextGenerationEU.
Data Availability Statement
Not applicable.
Acknowledgments
We thank D. Diego Díaz García (Río Lacarón S.L.) for his valuable help in collecting samples in the field. We are also grateful to Jesús Mercado Blanco (Estación Experimental del Zaidín, Spanish National Research Council) for kindly providing the V. dahliae V937I strain.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Inderbitzin, P.; Subbarao, K.V. Verticillium Systematics and Evolution: How Confusion Impedes Verticillium Wilt Management and How to Resolve It. Phytopathology 2014, 104, 564–574. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, Pathogenicity, and Management of Verticillium Species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Keykhasaber, M.; Thomma, B.P.H.J.; Hiemstra, J.A. Verticillium Wilt Caused by Verticillium dahliae in Woody Plants with Emphasis on Olive and Shade Trees. Eur. J. Plant Pathol. 2018, 150, 21–37. [Google Scholar] [CrossRef]
- Gent, D.H.; Woods, J.L.; Putnam, M.L. New Outbreaks of Verticillium Wilt on Hop in Oregon Caused by Nonlethal Verticillium albo-atrum. Plant Manag. Netw. 2012, 13, 14. [Google Scholar] [CrossRef]
- López-Escudero, F.J.; Mercado-Blanco, J. Verticillium Wilt of Olive: A Case Study to Implement an Integrated Strategy to Control a Soil-Borne Pathogen. Plant Soil 2011, 344, 1–50. [Google Scholar] [CrossRef]
- Castro, D.; Torres, M.; Sampedro, I.; Martínez-Checa, F.; Torres, B.; Béjar, V. Biological Control of Verticillium Wilt on Olive Trees by the Salt-Tolerant Strain Bacillus velezensis XT1. Microorganisms 2020, 8, 1080. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). FAOSTAT Database, Crops Processed, Data for Olive Oil; FAO: Rome, Italy, 2014. [Google Scholar]
- Ministerio de Agricultura. Alimentación y Medio Ambiente (MAGRAMA). Avances de Superficies y Producciones de Cultivos. Available online: https://www.mapa.gob.es/es/estadistica/temas/estadisticas-agrarias/agricultura/avances-superficies-producciones-agricolas/2012-2015.aspx (accessed on 10 July 2020).
- López-Escudero, F.J.; Mercado-Blanco, J.; Roca, J.M.; Valverde-Corredor, A.; Blanco-López, M.A. Verticillium Wilt of Olive in the Guadalquivir Valley (Southern Spain): Relations with Some Agronomical Factors and Spread of Verticillium dahliae. Phytopathol. Mediterr. 2010, 49, 370–380. [Google Scholar] [CrossRef]
- Montes-Osuna, N.; Mercado-Blanco, J. Verticillium Wilt of Olive and Its Control: What Did We Learn during the Last Decade? Plants 2020, 9, 735. [Google Scholar] [CrossRef]
- Coque, J.J.R.; Álvarez-Pérez, J.M.; Cobos, R.; González-García, S.; Ibáñez, A.M.; Diez Galán, A.; Calvo-Peña, C. Advances in the Control of Phytopathogenic Fungi That Infect Crops through Their Root System. In Advances in Applied Microbiology; Gadd, G.M., Sariaslani, S., Eds.; Academic Press Inc.: Cambridge, MA, USA, 2020; Volume 111, pp. 123–170. [Google Scholar]
- Mulero-Aparicio, A.; Agustí-Brisach, C.; Varo, Á.; López-Escudero, F.J.; Trapero, A. A Non-Pathogenic Strain of Fusarium oxysporum as a Potential Biocontrol Agent against Verticillium Wilt of Olive. Biol. Control 2019, 139, 104045. [Google Scholar] [CrossRef]
- Mulero-Aparicio, A.; Cernava, T.; Turrà, D.; Schaefer, A.; di Pietro, A.; López-Escudero, F.J.; Trapero, A.; Berg, G. The Role of Volatile Organic Compounds and Rhizosphere Competence in Mode of Action of the Non-Pathogenic Fusarium oxysporum FO12 toward Verticillium Wilt. Front. Microbiol. 2019, 10, 1808. [Google Scholar] [CrossRef]
- Carrero-Carrón, I.; Trapero-Casas, J.L.; Olivares-García, C.; Monte, E.; Hermosa, R.; Jiménez-Díaz, R.M. Trichoderma asperellum Is Effective for Biocontrol of Verticillium Wilt in Olive Caused by the Defoliating Pathotype of Verticillium dahliae. Crop Prot. 2016, 88, 45–52. [Google Scholar] [CrossRef]
- Varo, A.; Raya-Ortega, M.C.; Trapero, A. Selection and Evaluation of Micro-Organisms for Biocontrol of Verticillium dahliae in Olive. J. Appl. Microbiol. 2016, 121, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Tovar, M.D.; Garrido-Jurado, I.; Quesada-Moraga, E.; Raya-Ortega, M.C.; Trapero-Casas, A. Metarhizium brunneum and Beauveria Bassiana Release Secondary Metabolites with Antagonistic Activity against Verticillium dahliae and Phytophthora megasperma Olive Pathogens. Crop Prot. 2017, 100, 186–195. [Google Scholar] [CrossRef]
- Boutaj, H.; Meddich, A.; Wahbi, S.; Moukhli, A.; Alaoui-Talibi, Z.; Douira, A.; Filali-Maltouf, A.; Modafar, C. Effect of Arbuscular Mycorrhizal Fungi on Verticillium Wilt Development of Olive Trees Caused by Verticillium dahliae. Res. J. Biotechnol. 2019, 14, 79–88. [Google Scholar]
- Boutaj, H.; Chakhchar, A.; Meddich, A.; Wahbi, S.; El Alaoui-Talibi, Z.; Douira, A.; Filali-Maltouf, A.; El Modafar, C. Bioprotection of Olive Tree from Verticillium Wilt by Autochthonous Endomycorrhizal Fungi. J. Plant Dis. Prot. 2020, 127, 349–357. [Google Scholar] [CrossRef]
- Gómez-Lama Cabanás, C.; Sesmero, R.; Valverde-Corredor, A.; López-Escudero, F.J.; Mercado-Blanco, J. A Split-Root System to Assess Biocontrol Effectiveness and Defense-Related Genetic Responses in Above-Ground Tissues during the Tripartite Interaction Verticillium dahliae-Olive-Pseudomonas Fluorescens PICF7 in Roots. Plant Soil 2017, 417, 433–452. [Google Scholar] [CrossRef]
- Gómez-Lama Cabanás, C.; Legarda, G.; Ruano-Rosa, D.; Pizarro-Tobías, P.; Valverde-Corredor, A.; Niqui, J.L.; Triviño, J.C.; Roca, A.; Mercado-Blanco, J. Indigenous pseudomonas spp. Strains from the Olive (Olea europaea L.) Rhizosphere as Effective Biocontrol Agents against Verticillium dahliae: From the Host Roots to the Bacterial Genomes. Front. Microbiol. 2018, 9, 277. [Google Scholar] [CrossRef]
- Markakis, E.A.; Tjamos, S.E.; Antoniou, P.P.; Paplomatas, E.J.; Tjamos, E.C. Biological Control of Verticillium Wilt of Olive by Paenibacillus alvei, Strain K165. BioControl 2016, 61, 293–303. [Google Scholar] [CrossRef]
- Cheffi Azabou, M.; Gharbi, Y.; Medhioub, I.; Ennouri, K.; Barham, H.; Tounsi, S.; Triki, M.A. The Endophytic Strain Bacillus velezensis OEE1: An Efficient Biocontrol Agent against Verticillium Wilt of Olive and a Potential Plant Growth Promoting Bacteria. Biol. Control 2020, 142, 104168. [Google Scholar] [CrossRef]
- Bubici, G. Streptomyces spp. as Biocontrol Agents against Fusarium Species. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2018, 13, 1–15. [Google Scholar] [CrossRef]
- Labeda, D.P.; Goodfellow, M.; Brown, R.; Ward, A.C.; Lanoot, B.; Vanncanneyt, M.; Swings, J.; Kim, S.B.; Liu, Z.; Chun, J.; et al. Phylogenetic Study of the Species within the Family Streptomycetaceae. Antonie Van Leeuwenhoek 2012, 101, 73–104. [Google Scholar] [CrossRef] [PubMed]
- Van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; Van Wezel, G.P. Chemical Ecology of Antibiotic Production by Actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef] [PubMed]
- Vurukonda, S.S.K.P.; Giovanardi, D.; Stefani, E. Plant Growth Promoting and Biocontrol Activity of Streptomyces spp. as Endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.H. Identifying the Dominant Soil Bacterial Taxa in Libraries of 16S RRNA and 16S RRNA Genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Deketelaere, S.; Tyvaert, L.; França, S.C.; Höfte, M. Desirable Traits of a Good Biocontrol Agent against Verticillium Wilt. Front. Microbiol. 2017, 8, 1186. [Google Scholar] [CrossRef]
- Rong, X.; Huang, Y. Taxonomic Evaluation of the Streptomyces Hygroscopicus Clade Using Multilocus Sequence Analysis and DNA-DNA Hybridization, Validating the MLSA Scheme for Systematics of the Whole Genus. Syst. Appl. Microbiol. 2012, 35, 7–18. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for the Characterization of Streptomyces Species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Reusser, F. Mode of Action of Albocycline, an Inhibitor of Nicotinate Biosynthesis. J. Bacteriol. 1969, 100, 11–13. [Google Scholar] [CrossRef]
- Liang, H.; Zhou, G.; Ge, Y.; D’Ambrosio, E.A.; Eidem, T.M.; Blanchard, C.; Shehatou, C.; Chatare, V.K.; Dunman, P.M.; Valentine, A.M.; et al. Elucidating the Inhibition of Peptidoglycan Biosynthesis in Staphylococcus aureus by Albocycline, a Macrolactone Isolated from Streptomyces maizeus. Bioorg. Med. Chem. 2018, 26, 3453–3460. [Google Scholar] [CrossRef]
- Christner, C.; Kullertz, G.; Fischer, G.; Zerlin, M.; Grabley, S.; Thiericke, R.; Taddei, A.; Zeeck, A. Albocycline- and Carbomycin-Type Macrolides, Inhibitors of Human Prolyl Endopeptidases. J. Antibiot. 1998, 51, 368–371. [Google Scholar] [CrossRef][Green Version]
- Zucchi, T.D.; Almeida, L.G.; Dossi, F.C.A.; Cônsoli, F.L. Secondary Metabolites Produced by Propionicimonas sp. (ENT-18) Induce Histological Abnormalities in the Sclerotia of Sclerotinia Sclerotiorum. BioControl 2010, 55, 811–819. [Google Scholar] [CrossRef]
- Zucchi, T.D.; Almeida, L.G.; Moraes, L.A.B.; Cônsoli, F.L. Albocycline, the Main Bioactive Compound from Propionicimonas sp. ENT-18 against Sclerotinia Sclerotiorum. Ind. Crops Prod. 2014, 52, 264–268. [Google Scholar] [CrossRef]
- Gu, C.Z.; Yuan, S.H.; Jing, L.; Qiao, Y.J.; Song, Y.Y.; Abdalla Elzaki, M.E.; Yang, C.R.; Zhang, Y.J.; Zeng, R. Sen Albocycline-Type Macrolides with Antibacterial Activities from Streptomyces sp. 4205. Chem. Biodivers. 2019, 16, e1800344. [Google Scholar] [CrossRef]
- Ohike, T.; Matsukawa, T.; Okanami, M.; Kajiyama, S.; Ano, T. Biological Control Potential of Streptomyces sp. AR10 Producing Albocycline Isolated from Soil around Ant Nest. J. Agric. Sci. 2018, 10, 54. [Google Scholar] [CrossRef]
- Koyama, N.; Yotsumoto, M.; Onaka, H.; Tomoda, H. New Structural Scaffold 14-Membered Macrocyclic Lactone Ring for Selective Inhibitors of Cell Wall Peptidoglycan Biosynthesis in Staphylococcus aureus. J. Antibiot. 2013, 66, 303–304. [Google Scholar] [CrossRef]
- Nagahama, N.; Takamori, I.; Kotera, K.; Suzuki, M. Studies on an Antibiotic, Albocycline. Chem. Pharm. Bull. 1971, 19, 619–654. [Google Scholar] [CrossRef]
- Thomas, R.C.; Chidester, C.G. Albocycline: Structure Determination by X-ray Crystallography. J. Antibiot. 1982, 35, 1658–1664. [Google Scholar] [CrossRef]
- Stukenbrock, E.; Gurr, S. Address the Growing Urgency of Fungal Disease in Crops. Nature 2023, 617, 31–34. [Google Scholar] [CrossRef]
- Bebber, D.P.; Holmes, T.; Gurr, S.J. The Global Spread of Crop Pests and Pathogens. Glob. Ecol. Biogeogr. 2014, 23, 1398–1407. [Google Scholar] [CrossRef]
- Almeida, F.; Rodrigues, M.L.; Coelho, C. The Still Underestimated Problem of Fungal Diseases Worldwide. Front. Microbiol. 2019, 10, 214. [Google Scholar] [CrossRef]
- O’Sullivan, C.A.; Belt, K.; Thatcher, L.F. Tackling Control of a Cosmopolitan Phytopathogen: Sclerotinia. Front. Plant Sci. 2021, 12, 707509. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of Action of Microbial Biological Control Agents against Plant Diseases: Relevance beyond Efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed]
- Pacios-Michelena, S.; Aguilar González, C.N.; Alvarez-Perez, O.B.; Rodriguez-Herrera, R.; Chávez-González, M.; Arredondo Valdés, R.; Ascacio Valdés, J.A.; Govea Salas, M.; Ilyina, A. Application of Streptomyces Antimicrobial Compounds for the Control of Phytopathogens. Front. Sustain. Food Syst. 2021, 5, 696518. [Google Scholar] [CrossRef]
- Nagahama, N.; Suzuki, M.; Awataguchi, S.; Okuda, T. Studies on a New Antibiotic, Albocycline. I. Isolation, Purification and Properties. J. Antibiot. 1967, 20, 261–266. [Google Scholar]
- Harada, K.I.; Nishida, F.; Takagi, H.; Suzuki, M.; Iwashita, T. Studies on an Antibiotic, Albocycline VII. Minor Components of Albocycline. J. Antibiot. 1984, 37, 1187–1197. [Google Scholar] [CrossRef]
- Furumai, T.; Nagahama, N.; Okuda, T. Studies on a New Antibiotic, Albocycline. II. Taxonomic Studies on Albocycline-Producing Strains. J. Antibiot. 1968, 21, 85–90. [Google Scholar] [CrossRef]
- Miyairi, N.; Takashima, M.; Shimizu, K.; Sakai, H. Studies on New Antibiotics, Cineromycins A and B. J. Antibiot. 1966, 19, 56–62. [Google Scholar]
- Daher, S.S.; Franklin, K.P.; Scherzi, T.; Dunman, P.M.; Andrade, R.B. Synthesis and Biological Evaluation of Semi-Synthetic Albocycline Analogs. Bioorg. Med. Chem. Lett. 2020, 30, 127509. [Google Scholar] [CrossRef]
- Slechta, L.; Cialdella, J.; Hoeksema, H. Biomodification of Albocycline by Streptomyces venezuelae. J. Antibiot. 1978, 31, 319–323. [Google Scholar] [CrossRef]
- Küster, E.; Williams, S. Selection of Media for Isolation of Streptomycetes. Nature 1964, 202, 928–929. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Bibb, M.; Buttner, M.; Chater, K.; Hopwood, D. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000; ISBN 0708406238. [Google Scholar]
- Hopwood, D.A.; Bibb, M.J.; Chater, K.F.; Bruton, C.J.; Kieser, H.M.; Lydiate, D.; Smith, C.P.; Ward, J.M.; Schrempf, H. Genetic Manipulation of Streptomyces: A Laboratory Manual; The John Innes Institute: Norwich, UK, 1985. [Google Scholar]
- Lane, D. 16S/23S RRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A Prokaryotic 16S RRNA Gene Sequence Database with Phylotypes That Represent Uncultured Species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.P.; Zheng, W.; Rong, X.Y.; Huang, Y. A Multilocus Phylogeny of the Streptomyces griseus 16S RRNA Gene Clade: Use of Multilocus Sequence Analysis for Streptomycete Systematics. Int. J. Syst. Evol. Microbiol. 2008, 58, 149–159. [Google Scholar] [CrossRef]
- Rong, X.; Guo, Y.; Huang, Y. Proposal to Reclassify the Streptomyces albidoflavus Clade on the Basis of Multilocus Sequence Analysis and DNA-DNA Hybridization, and Taxonomic Elucidation of Streptomyces griseus subsp. solvifaciens. Syst. Appl. Microbiol. 2009, 32, 314–322. [Google Scholar] [CrossRef]
- Álvarez-Pérez, J.M.; González-García, S.; Cobos, R.; Olego, M.Á.; Ibañez, A.; Díez-Galán, A.; Garzón-Jimeno, E.; Coque, J.J.R. Use of Endophytic and Rhizosphere Actinobacteria from Grapevine Plants to Reduce Nursery Fungal Graft Infections That Lead to Young Grapevine Decline. Appl. Environ. Microbiol. 2017, 83, e01564-17. [Google Scholar] [CrossRef]
- Collado-Romero, M.; Mercado-Blanco, J.; Olivares-García, C.; Valverde-Corredor, A.; Jiménez-Díaz, R.M. Molecular Variability within and among Verticillium dahliae Vegetative Compatibility Groups Determined by Fluorescent Amplified Fragment Length Polymorphism and Polymerase Chain Reaction Markers. Phytopathology 2006, 96, 485–495. [Google Scholar] [CrossRef]
- Senthilkumar, M.; Amaresan, N.; Sankaranarayanan, A. Antagonistic Activity of Volatile Compound of bacteria against Phytopathogens: Dual Plate Assay. In Plant-Microbe Interactions. Springer Protocols Handbooks; Humana: New York, NY, USA, 2021; pp. 171–173. ISBN 978-1-0716-1079-4. [Google Scholar]
- Martin, J.F.; McDaniel, L.E. Biosynthesis of Candicidin by Phosphate-Limited Resting Cells of Streptomyces griseus. Eur. J. Appl. Microbiol. 1976, 3, 135–144. [Google Scholar] [CrossRef]
- Schrey, S.D.; Erkenbrack, E.; Früh, E.; Fengler, S.; Hommel, K.; Horlacher, N.; Schulz, D.; Ecke, M.; Kulik, A.; Fiedler, H.P.; et al. Production of Fungal and Bacterial Growth Modulating Secondary Metabolites Is Widespread among Mycorrhiza-Associated Streptomycetes. BMC Microbiol. 2012, 12, 164. [Google Scholar] [CrossRef]
- Tan, K.H. Soil Sampling, Preparation, and Analysis, 2nd ed.; CRC Press: New York, NY, USA, 2005; ISBN 9780429178993. [Google Scholar]
- Batjes, N.H. Total Carbon and Nitrogen in the Soils of the World. Eur. J. Soil Sci. 1996, 47, 151–163. [Google Scholar] [CrossRef]
- Ostrowska, A.; Gawlinski, S.; Szczubialka, Z. Methods for Analysis and Evaluation of Soil and Plant Properties. IOŚ Warszawa 1991, 334, 158–167. [Google Scholar]
- Houba, V.J.G.; Temminghoff, E.J.M.; Gaikhorst, G.A.; van Vark, W. Soil Analysis Procedures Using 0.01 M Calcium Chloride as Extraction Reagent. Commun. Soil Sci. Plant Anal. 2000, 31, 1299–1396. [Google Scholar] [CrossRef]
- Das, R.; Romi, W.; Das, R.; Sharma, H.K.; Thakur, D. Antimicrobial Potentiality of Actinobacteria Isolated from Two Microbiologically Unexplored Forest Ecosystems of Northeast India. BMC Microbiol. 2018, 18, 71. [Google Scholar] [CrossRef] [PubMed]
- Awla, H.K.; Rashid, T.S. HPLC Fractionation: A Comparative Analysis of Anti-Fungal Compounds from Different Streptomyces Isolates Inhibiting Colletotrichum acutatum. Biocatal Agric. Biotechnol. 2020, 27, 101688. [Google Scholar] [CrossRef]
- López-Moral, A.; Agustí-Brisach, C.; Leiva-Egea, F.M.; Trapero, A. Influence of Cultivar and Biocontrol Treatments on the Effect of Olive Stem Extracts on the Viability of Verticillium dahliae Conidia. Plants 2022, 11, 554. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.M.; Sherrys, J.C.; Turck, M. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Tenover, F.C. Antimicrobial Susceptibility Testing. In Encyclopedia of Microbiology; Academic Press: Cambridge, MA, USA, 2019; pp. 166–175. ISBN 9780128117378. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).