Unlocking the Multifaceted Mechanisms of Bud Outgrowth: Advances in Understanding Shoot Branching
Abstract
1. Introduction
2. Mechanisms Regulating Shoot Branching
2.1. Internal Inputs Determine Bud Outgrowth
2.1.1. TB1/BRC1 Acts as a Key Integrator of Branching
2.1.2. SQUAMOSA Binding Proteins Inhibit Bud Outgrowth
2.1.3. Auxin Indirectly Inhibits Sustained Bud Outgrowth
2.1.4. Strigolactones Have an Inhibitory Effect on Bud Outgrowth
2.1.5. Other Phytohormones Regulate Tillering/Branching
2.1.6. Phytohormones Interact Influencing Bud Outgrowth
2.1.7. Sugars Play an Essential Role in Bud Release
2.2. Effects of Environmental Inputs on Bud Outgrowth
2.2.1. Light Plays a Critical Role in Bud Outgrowth
2.2.2. Impacts of Nutrients on Bud Outgrowth
2.2.3. Water Availability Influences Bud Outgrowth
2.2.4. Effects of Temperature on Tillering
2.2.5. Biotic Stresses Impact Tiller Bud Outgrowth
3. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bowman, J.L.; Eshed, Y. Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 2000, 5, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef]
- Springer, N. Shaping a better rice plant. Nat. Genet. 2010, 42, 475–476. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J. Molecular Basis of Plant Architecture. Annu. Rev. Plant Biol. 2008, 59, 253–279. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef]
- Wang, M.; Le Moigne, M.-A.; Bertheloot, J.; Crespel, L.; Perez-Garcia, M.-D.; Ogé, L.; Demotes-Mainard, S.; Hamama, L.; Davière, J.-M.; Sakr, S. BRANCHED1: A key hub of shoot branching. Front. Plant Sci. 2019, 10, 76. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Chin-Atkins, A.N.; Wilson, I.W.; Chapple, R.; Dennis, E.S.; Chaudhury, A. The Arabidopsis AMP1 Gene Encodes a Putative Glutamate Carboxypeptidase. Plant Cell 2001, 13, 2115–2125. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 441–445. [Google Scholar] [CrossRef]
- Lo, S.-F.; Yang, S.-Y.; Chen, K.-T.; Hsing, Y.-I.; Zeevaart, J.A.D.; Chen, L.-J.; Yu, S.-M. A Novel Class of Gibberellin 2-Oxidases Control Semidwarfism, Tillering, and Root Development in Rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef] [PubMed]
- Bayer, E.M.; Smith, R.S.; Mandel, T.; Nakayama, N.; Sauer, M.; Prusinkiewicz, P.; Kuhlemeier, C. Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 2009, 23, 373–384. [Google Scholar] [CrossRef]
- Lin, H.; Wang, R.; Qian, Q.; Yan, M.; Meng, X.; Fu, Z.; Yan, C.; Jiang, B.; Su, Z.; Li, J.; et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef] [PubMed]
- Finlayson, S.A.; Krishnareddy, S.R.; Kebrom, T.H.; Casal, J.J. Phytochrome Regulation of Branching in Arabidopsis. Plant Physiol. 2010, 152, 1914–1927. [Google Scholar] [CrossRef] [PubMed]
- González-Grandío, E.; Pajoro, A.; Franco-Zorrilla, J.M.; Tarancón, C.; Immink, R.G.H.; Cubas, P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP/cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA 2017, 114, E245–E254. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, S.; Zhu, W.; Jia, K.; Yang, H.; Wang, X. Strigolactone/MAX2-Induced Degradation of Brassinosteroid Transcriptional Effector BES1 Regulates Shoot Branching. Dev. Cell 2013, 27, 681–688. [Google Scholar] [CrossRef]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M. Transporters involved in source to sink partitioning of amino acids and ureides: Opportunities for crop improvement. J. Exp. Bot. 2014, 65, 1865–1878. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Zhang, X.; He, J.; Yu, H.; Wang, Y.; Shi, B.; Han, Y.; Wang, G.; Feng, X.; Zhang, C.; et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014, 10, 755. [Google Scholar] [CrossRef]
- Wang, Q.; Kohlen, W.; Rossmann, S.; Vernoux, T.; Theres, K. Auxin Depletion from the Leaf Axil Conditions Competence for Axillary Meristem Formation in Arabidopsis and Tomato. Plant Cell 2014, 26, 2068–2079. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Shi, B.; Yu, T.; Qi, J.; Meyerowitz, E.M.; Jiao, Y. The Stem Cell Niche in Leaf Axils Is Established by Auxin and Cytokinin in Arabidopsis. Plant Cell 2014, 26, 2055–2067. [Google Scholar] [CrossRef]
- Zhang, Q.Q.; Wang, J.G.; Wang, L.Y.; Wang, J.F.; Wang, Q.; Yu, P.; Bai, M.Y.; Fan, M. Gibberellin repression of axillary bud formation in Arabidopsis by modulation of DELLA-SPL9 complex activity. J. Integr. Plant Biol. 2020, 62, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Igari, K.; Endo, S.; Hibara, K.; Aida, M.; Sakakibara, H.; Kawasaki, T.; Tasaka, M. Constitutive activation of a CC-NB-LRR protein alters morphogenesis through the cytokinin pathway in Arabidopsis. Plant J. Cell Mol. Biol. 2008, 55, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.H.; Chen, L.; Guo, H.F.; Feng, R.; Lou, Q.J.; Rashid, M.A.R.; Zhu, X.Y.; Qing, D.J.; Liang, H.F.; Gao, L.J.; et al. Systematic Analysis of NB-ARC Gene Family in Rice and Functional Characterization of GNP12. Front Genet 2022, 13, 887217. [Google Scholar] [CrossRef]
- Stafstrom, J.P.; Sussex, I.M. Expression of a Ribosomal Protein Gene in Axillary Buds of Pea Seedlings. Plant Physiol. 1992, 100, 1494–1502. [Google Scholar] [CrossRef]
- Devitt, M.L.; Stafstrom, J.P. Cell cycle regulation during growth-dormancy cycles in pea axillary buds. Plant Mol. Biol. 1995, 29, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Dun, E.A.; Ferguson, B.J.; Beveridge, C.A. Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiol. 2006, 142, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Waldie, T.; Hayward, A.; Beveridge, C.A. Axillary bud outgrowth in herbaceous shoots: How do strigolactones fit into the picture? Plant Mol. Biol. 2010, 73, 27–36. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J. Branching in rice. Curr. Opin. Plant Biol. 2011, 14, 94–99. [Google Scholar] [CrossRef]
- Hubbard, L.; McSteen, P.; Doebley, J.; Hake, S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 2002, 162, 1927–1935. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Grandío, E.G.; Serra, F.; Marcel, F.; Rodríguez-Buey, M.L.; Schmitz, G.; Theres, K.; Bendahmane, A.; Dopazo, H.; Cubas, P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 2011, 67, 701–714. [Google Scholar] [CrossRef]
- Braun, N.; de Saint Germain, A.; Pillot, J.-P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; Le Signor, C.; Bouteiller, N. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 2012, 158, 225–238. [Google Scholar] [CrossRef]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2015, 5, 741–755. [Google Scholar] [CrossRef]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef]
- Lincoln, C.; Britton, J.H.; Estelle, M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 1990, 2, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Coneva, V.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Nambara, E.; Bi, Y.M.; Rothstein, S.J. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 2015, 83, 913–925. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, H.; Li, B.; Huang, J.; Liu, X.; Zhou, Y.; Mou, Z.; Li, J. Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell 2006, 18, 308–320. [Google Scholar] [CrossRef]
- Jia, W.; Li, B.; Li, S.; Liang, Y.; Wu, X.; Ma, M.; Wang, J.; Gao, J.; Cai, Y.; Zhang, Y.; et al. Mitogen-Activated Protein Kinase Cascade MKK7-MPK6 Plays Important Roles in Plant Development and Regulates Shoot Branching by Phosphorylating PIN1 in Arabidopsis. PLoS Biol. 2016, 14, e1002550. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant Development Is Regulated by a Family of Auxin Receptor F Box Proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The Path from β-Carotene to Carlactone, a Strigolactone-Like Plant Hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Booker, J.; Auldridge, M.; Wills, S.; McCarty, D.; Klee, H.; Leyser, O. MAX3/CCD7 Is a Carotenoid Cleavage Dioxygenase Required for the Synthesis of a Novel Plant Signaling Molecule. Curr. Biol. 2004, 14, 1232–1238. [Google Scholar] [CrossRef]
- Sorefan, K. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003, 17, 1469–1474. [Google Scholar] [CrossRef]
- Booker, J.; Sieberer, T.; Wright, W.; Williamson, L.; Willett, B.; Stirnberg, P.; Turnbull, C.; Srinivasan, M.; Goddard, P.; Leyser, O. MAX1 Encodes a Cytochrome P450 Family Member that Acts Downstream of MAX3/4 to Produce a Carotenoid-Derived Branch-Inhibiting Hormone. Dev. Cell 2005, 8, 443–449. [Google Scholar] [CrossRef]
- Smith, S.M.; Li, J. Signalling and responses to strigolactones and karrikins. Curr. Opin. Plant Biol. 2014, 21, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, B.; Jiang, L.; Liu, X.; Li, X.; Lu, Z.; Meng, X.; Wang, Y.; Smith, S.M.; Li, J. Strigolactone Signaling in Arabidopsis Regulates Shoot Development by Targeting D53-Like SMXL Repressor Proteins for Ubiquitination and Degradation. Plant Cell 2015, 27, 3128–3142. [Google Scholar] [CrossRef] [PubMed]
- Soundappan, I.; Bennett, T.; Morffy, N.; Liang, Y.; Stanga, J.P.; Abbas, A.; Leyser, O.; Nelson, D.C. SMAX1-LIKE/D53 Family Members Enable Distinct MAX2-Dependent Responses to Strigolactones and Karrikins in Arabidopsis. Plant Cell 2015, 27, 3143–3159. [Google Scholar] [CrossRef] [PubMed]
- Medford, J.I.; Horgan, R.; El-Sawi, Z.; Klee, H.J. Alterations of Endogenous Cytokinins in Transgenic Plants Using a Chimeric Isopentenyl Transferase Gene. Plant Cell 1989, 1, 403–413. [Google Scholar] [CrossRef]
- Tantikanjana, T.; Yong, J.W.H.; Letham, D.S.; Griffith, M.; Hussain, M.; Ljung, K.; Sandberg, G.; Sundaresan, V. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 2001, 15, 1577–1588. [Google Scholar] [CrossRef] [PubMed]
- Shimizu-Sato, S.; Tanaka, M.; Mori, H. Auxin–cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 2009, 69, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.K.; Holalu, S.V.; Casal, J.J.; Finlayson, S.A. Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol. 2013, 163, 1047–1058. [Google Scholar] [CrossRef]
- Yao, C.; Finlayson, S.A. Abscisic Acid Is a General Negative Regulator of Arabidopsis Axillary Bud Growth. Plant Physiol. 2015, 169, 611–626. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A New Class of Transcription Factors Mediates Brassinosteroid-Regulated Gene Expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Koumoto, T.; Shimada, H.; Kusano, H.; She, K.-C.; Iwamoto, M.; Takano, M. Rice monoculm mutation moc2, which inhibits outgrowth of the second tillers, is ascribed to lack of a fructose-1, 6-bisphosphatase. Plant Biotechnol. 2013, 30, 47–56. [Google Scholar] [CrossRef]
- Huang, W.; Bai, G.; Wang, J.; Zhu, W.; Zeng, Q.; Lu, K.; Sun, S.; Fang, Z. Two splicing variants of OsNPF7.7 regulate shoot branching and nitrogen utilization efficiency in rice. Front. Plant Sci. 2018, 9, 300. [Google Scholar] [CrossRef]
- He, X.; Qu, B.; Li, W.; Zhao, X.; Teng, W.; Ma, W.; Ren, Y.; Li, B.; Li, Z.; Tong, Y. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 2015, 169, 1991–2005. [Google Scholar] [CrossRef]
- Guo, S.; Xu, Y.; Liu, H.; Mao, Z.; Zhang, C.; Ma, Y.; Zhang, Q.; Meng, Z.; Chong, K. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 2013, 4, 1566. [Google Scholar] [CrossRef]
- Bai, M.Y.; Zhang, L.Y.; Gampala, S.S.; Zhu, S.W.; Song, W.Y.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef]
- Qiao, S.; Sun, S.; Wang, L.; Wu, Z.; Li, C.; Li, X.; Wang, T.; Leng, L.; Tian, W.; Lu, T.; et al. The RLA1/SMOS1 Transcription Factor Functions with OsBZR1 to Regulate Brassinosteroid Signaling and Rice Architecture. Plant Cell 2017, 29, 292–309. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef]
- Doebley, J.; Stec, A.; Gustus, C. teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics 1995, 141, 333–346. [Google Scholar] [CrossRef]
- Doebley, J. The genetics of maize evolution. Annu. Rev. Genet. 2004, 38, 37–59. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 1997, 9, 1607–1619. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Minakuchi, K.; Kameoka, H.; Yasuno, N.; Umehara, M.; Luo, L.; Kobayashi, K.; Hanada, A.; Ueno, K.; Asami, T.; Yamaguchi, S.; et al. FINE CULM1 (FC1) Works Downstream of Strigolactones to Inhibit the Outgrowth of Axillary Buds in Rice. Plant Cell Physiol. 2010, 51, 1127–1135. [Google Scholar] [CrossRef]
- González-Grandío, E.; Poza-Carrión, C.; Sorzano, C.O.S.; Cubas, P. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 2013, 25, 834–850. [Google Scholar] [CrossRef]
- Seale, M.; Bennett, T.; Leyser, O. BRC1 expression regulates bud activation potential, but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 2017, 144, 1661–1673. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of Plant MicroRNA Targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific Effects of MicroRNAs on the Plant Transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Gruber, M.Y.; Yu, B.; Gao, M.-J.; Khachatourians, G.G.; Hegedus, D.D.; Parkin, I.A.; Hannoufa, A. Arabidopsis mutant sk156 reveals complex regulation of SPL15 in a miR156-controlled gene network. BMC Plant Biol. 2012, 12, 169. [Google Scholar] [CrossRef]
- Matthes, M.S.; Best, N.B.; Robil, J.M.; Malcomber, S.; Gallavotti, A.; McSteen, P. Auxin EvoDevo: Conservation and Diversification of Genes Regulating Auxin Biosynthesis, Transport, and Signaling. Mol. Plant 2019, 12, 298–320. [Google Scholar] [CrossRef]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef]
- Zenser, N.; Ellsmore, A.; Leasure, C.; Callis, J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 11795–11800. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Wang, X.-J.; Hagen, G.; Guilfoyle, T.J. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef]
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 2001, 414, 271–276. [Google Scholar] [CrossRef]
- Park, J.-Y.; Kim, H.-J.; Kim, J. Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J. 2002, 32, 669–683. [Google Scholar] [CrossRef]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef]
- Wu, K.; Chen, A.; Pan, Z.-Q. Conjugation of Nedd8 to CUL1 Enhances the Ability of the ROC1-CUL1 Complex to Promote Ubiquitin Polymerization. J. Biol. Chem. 2000, 275, 32317–32324. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.C.; Dharmasiri, S.; Hellmann, H.; Walker, L.; Gray, W.M.; Estelle, M. AXR1-ECR1–Dependent Conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 Is Required for Auxin Response. Plant Cell 2002, 14, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Prasad, T.; Li, X.; Abdel-Rahman, A.; Hosokawa, Z.; Cloud, N.; Lamotte, C.; Cline, M. Does auxin play a role in the release of apical dominance by shoot inversion in Ipomoea nil? Ann. Bot. 1993, 71, 223–229. [Google Scholar] [CrossRef]
- Li, C.J.; Bangerth, F. Autoinhibition of indoleacetic acid transport in the shoots of two-branched pea (Pisum sativum) plants and its relationship to correlative dominance. Physiol. Plant. 1999, 106, 415–420. [Google Scholar] [CrossRef]
- Domagalska, M.A.; Leyser, O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011, 12, 211–221. [Google Scholar] [CrossRef]
- Barbier, F.; Péron, T.; Lecerf, M.; Perez-Garcia, M.-D.; Barrière, Q.; Rolčík, J.; Boutet-Mercey, S.; Citerne, S.; Lemoine, R.; Porcheron, B. Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J. Exp. Bot. 2015, 66, 2569–2582. [Google Scholar] [CrossRef]
- Bennett, T.; Hines, G.; Leyser, O. Canalization: What the flux? Trends Genet. 2014, 30, 41–48. [Google Scholar] [CrossRef]
- Shinohara, N.; Taylor, C.; Leyser, O. Strigolactone Can Promote or Inhibit Shoot Branching by Triggering Rapid Depletion of the Auxin Efflux Protein PIN1 from the Plasma Membrane. PLoS Biol. 2013, 11, e1001474. [Google Scholar] [CrossRef]
- Brewer, P.B.; Dun, E.A.; Ferguson, B.J.; Rameau, C.; Beveridge, C.A. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 2009, 150, 482–493. [Google Scholar] [CrossRef]
- Hayward, A.; Stirnberg, P.; Beveridge, C.; Leyser, O. Interactions between Auxin and Strigolactone in Shoot Branching Control. Plant Physiol. 2009, 151, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Dun, E.A.; de Saint Germain, A.; Rameau, C.; Beveridge, C.A. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 2012, 158, 487–498. [Google Scholar] [CrossRef]
- Shimizu, S.; Mori, H. Analysis of Cycles of Dormancy and Growth in Pea Axillary Buds Based on mRNA Accumulation Patterns of Cell Cycle-Related Genes. Plant Cell Physiol. 1998, 39, 255–262. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Brutnell, T.P.; Finlayson, S.A. Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 2009, 33, 48–58. [Google Scholar] [CrossRef]
- Cook, C.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef]
- Akiyama, K.; Matsuzaki, K.-I.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Napoli, C.; Ruehle, J. New mutations affecting meristem growth and potential in Petunia hybrida Vilm. J. Hered. 1996, 87, 371–377. [Google Scholar] [CrossRef]
- Beveridge, C.A.; Symons, G.M.; Murfet, I.C.; Ross, J.J.; Rameau, C. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal (s). Plant Physiol. 1997, 115, 1251. [Google Scholar] [CrossRef]
- Beveridge, C.A. Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul. 2000, 32, 193–203. [Google Scholar] [CrossRef]
- Beveridge, C.A. Axillary bud outgrowth: Sending a message. Curr. Opin. Plant Biol. 2006, 9, 35–40. [Google Scholar] [CrossRef]
- Turnbull, C.G.N.; Booker, J.P.; Leyser, H.M.O. Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 2002, 32, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Turnbull, C.G.N.; Beveridge, C.A. Long-Distance Signaling and the Control of Branching in therms1 Mutant of Pea. Plant Physiol. 2001, 126, 203–209. [Google Scholar] [CrossRef]
- Morris, S.E.; Turnbull, C.G.; Murfet, I.C.; Beveridge, C.A. Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol. 2001, 126, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Sado, A.; Tanaka, K.; Kisugi, T.; Asami, K.; Ota, S.; Kim, H.I.; Yoneyama, K.; Xie, X.; Ohnishi, T.; et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. 2014, 111, 18084–18089. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wang, Y.; Li, J. Action of strigolactones in plants. Enzymes 2014, 35, 57–84. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.A. Transport of exogenous auxin in two-branched dwarf pea seedlings (Pisum sativum L.). Planta 1977, 136, 91–96. [Google Scholar] [CrossRef]
- CLINE, M.G. Exogenous auxin effects on lateral bud outgrowth in decapitated shoots. Ann. Bot. 1996, 78, 255–266. [Google Scholar] [CrossRef]
- El-Showk, S.; Ruonala, R.; Helariutta, Y. Crossing paths: Cytokinin signalling and crosstalk. Development 2013, 140, 1373–1383. [Google Scholar] [CrossRef]
- Tanaka, M.; Takei, K.; Kojima, M.; Sakakibara, H.; Mori, H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006, 45, 1028–1036. [Google Scholar] [CrossRef]
- Turnbull, C.G.; Raymond, M.A.; Dodd, I.C.; Morris, S.E. Rapid increases in cytokinin concentration in lateral buds of chickpea (Cicer arietinum L.) during release of apical dominance. Planta 1997, 202, 271–276. [Google Scholar] [CrossRef]
- Li, C.; Bangerth, F. The possible role of cytokinins, ethylene and indoleacetic acid in apical dominance. In Progress in Plant Growth Regulation. Current Plant Science and Biotechnology in Agriculture; Springer: Dordrecht, The Netherlands, 1992; pp. 431–436. [Google Scholar] [CrossRef]
- Chatfield, S.P.; Stirnberg, P.; Forde, B.G.; Leyser, O. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 2000, 24, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Beveridge, C.A. Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol. 2009, 149, 1929–1944. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.J.; Schreiber, B. Role and function of cytokinin oxidase in plants. Plant Growth Regul. 1997, 23, 123–134. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Mak, P.Y.A.; Martinez, E.C.; Sun, T.-P. The New RGA Locus Encodes a Negative Regulator of Gibberellin Response in Arabidopsis thaliana. Genetics 1997, 146, 1087–1099. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.-Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 Accumulates in the Nucleus in Response to Brassinosteroids to Regulate Gene Expression and Promote Stem Elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef]
- Arite, T.; Iwata, H.; Ohshima, K.; Maekawa, M.; Nakajima, M.; Kojima, M.; Sakakibara, H.; Kyozuka, J. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007, 51, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Zhang, S.; Zhang, W.; Li, G.; Chen, Z.; Zhai, W.; Zhao, X.; Pan, X.; Xie, Q.; Zhu, L. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006, 48, 687–698. [Google Scholar] [CrossRef]
- Johnson, X.; Brcich, T.; Dun, E.A.; Goussot, M.; Haurogné, K.; Beveridge, C.A.; Rameau, C. Branching Genes Are Conserved across Species. Genes Controlling a Novel Signal in Pea Are Coregulated by Other Long-Distance Signals. Plant Physiol. 2006, 142, 1014–1026. [Google Scholar] [CrossRef]
- Foo, E.; Bullier, E.; Goussot, M.; Foucher, F.; Rameau, C.; Beveridge, C.A. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 2005, 17, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, K.; Sorefan, K.; Ward, S.; Leyser, O. Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J. 2005, 44, 569–580. [Google Scholar] [CrossRef]
- Beveridge, C.A.; Dun, E.A.; Rameau, C. Pea has its tendrils in branching discoveries spanning a century from auxin to strigolactones. Plant Physiol. 2009, 151, 985–990. [Google Scholar] [CrossRef]
- Bennett, T.; Leyser, O. Something on the Side: Axillary Meristems and Plant Development. Plant Mol. Biol. 2006, 60, 843–854. [Google Scholar] [CrossRef]
- Prusinkiewicz, P.; Crawford, S.; Smith, R.S.; Ljung, K.; Bennett, T.; Ongaro, V.; Leyser, O. Control of bud activation by an auxin transport switch. Proc. Natl. Acad. Sci. USA 2009, 106, 17431–17436. [Google Scholar] [CrossRef]
- Crawford, S.; Shinohara, N.; Sieberer, T.; Williamson, L.; George, G.; Hepworth, J.; Müller, D.; Domagalska, M.A.; Leyser, O. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 2010, 137, 2905–2913. [Google Scholar] [CrossRef]
- Bangerth, F. Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta 1994, 194, 439–442. [Google Scholar] [CrossRef]
- Nordström, A.; Tarkowski, P.; Tarkowska, D.; Norbaek, R.; Åstot, C.; Dolezal, K.; Sandberg, G. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin–cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 2004, 101, 8039–8044. [Google Scholar] [CrossRef]
- Li, C.J.; Guevara, E.; Herrera, J.; Bangerth, F. Effect of apex excision and replacement by 1-naphthylacetic acid on cytokinin concentration and apical dominance in pea plants. Physiol. Plant. 1995, 94, 465–469. [Google Scholar] [CrossRef]
- Bangerth, F.; Li, C.-J.; Gruber, J. Mutual interaction of auxin and cytokinins in regulating correlative dominance. Plant Growth Regul. 2000, 32, 205–217. [Google Scholar] [CrossRef]
- To, J.P.C.; Haberer, G.; Ferreira, F.J.; DeruèRe, J.; Mason, M.G.; Schaller, G.E.; Alonso, J.M.; Ecker, J.R.; Kieber, J.J. Type-A Arabidopsis Response Regulators Are Partially Redundant Negative Regulators of Cytokinin Signaling. Plant Cell 2004, 16, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Vysotskaya, L.B.; Korobova, A.V.; Veselov, S.Y.; Dodd, I.C.; Kudoyarova, G.R. ABA mediation of shoot cytokinin oxidase activity: Assessing its impacts on cytokinin status and biomass allocation of nutrient-deprived durum wheat. Funct. Plant Biol. 2009, 36, 66. [Google Scholar] [CrossRef]
- Chang, H.; Jones, M.L.; Banowetz, G.M.; Clark, D.G. Overproduction of Cytokinins in Petunia Flowers Transformed with PSAG12-IPT Delays Corolla Senescence and Decreases Sensitivity to Ethylene. Plant Physiol. 2003, 132, 2174–2183. [Google Scholar] [CrossRef]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef]
- Smeekens, S.; Ma, J.; Hanson, J.; Rolland, F. Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 2010, 13, 273–278. [Google Scholar] [CrossRef]
- Wind, J.; Smeekens, S.; Hanson, J. Sucrose: Metabolite and signaling molecule. Phytochemistry 2010, 71, 1610–1614. [Google Scholar] [CrossRef]
- Barbier, F.F.; Lunn, J.E.; Beveridge, C.A. Ready, steady, go! A sugar hit starts the race to shoot branching. Curr. Opin. Plant Biol. 2015, 25, 39–45. [Google Scholar] [CrossRef]
- Morris, S.E.; Cox, M.C.; Ross, J.J.; Krisantini, S.; Beveridge, C.A. Auxin dynamics after decapitation are not correlated with the initial growth of axillary buds. Plant Physiol. 2005, 138, 1665–1672. [Google Scholar] [CrossRef]
- Renton, M.; Hanan, J.; Ferguson, B.J.; Beveridge, C.A. Models of long-distance transport: How is carrier-dependent auxin transport regulated in the stem? New Phytol. 2012, 194, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Kebrom, T.H.; Chandler, P.M.; Swain, S.M.; King, R.W.; Richards, R.A.; Spielmeyer, W. Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol. 2012, 160, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Rabot, A.; Henry, C.; Ben Baaziz, K.; Mortreau, E.; Azri, W.; Lothier, J.; Hamama, L.; Boummaza, R.; Leduc, N.; Pelleschi-Travier, S.; et al. Insight into the Role of Sugars in Bud Burst Under Light in the Rose. Plant Cell Physiol. 2012, 53, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Kebrom, T.H.; Mullet, J.E. Photosynthetic leaf area modulates tiller bud outgrowth in sorghum. Plant Cell Environ. 2015, 38, 1471–1478. [Google Scholar] [CrossRef]
- Brewer, P.B.; Koltai, H.; Beveridge, C.A. Diverse Roles of Strigolactones in Plant Development. Mol. Plant 2013, 6, 18–28. [Google Scholar] [CrossRef]
- Ongaro, V.; Leyser, O. Hormonal control of shoot branching. J. Exp. Bot. 2007, 59, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Kebrom, T.H.; Burson, B.L.; Finlayson, S.A. Phytochrome B Represses Teosinte Branched1 Expression and Induces Sorghum Axillary Bud Outgrowth in Response to Light Signals. Plant Physiol. 2006, 140, 1109–1117. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, Y.; Chen, G.; Zhang, A.; Yang, S.; Shang, L.; Wang, D.; Ruan, B.; Liu, C.; Jiang, H.; et al. The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 2019, 10, 5207. [Google Scholar] [CrossRef]
- Wu, K.; Wang, S.; Song, W.; Zhang, J.; Wang, Y.; Liu, Q.; Yu, J.; Ye, Y.; Li, S.; Chen, J.; et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 2020, 367, eaaz2046. [Google Scholar] [CrossRef]
- Sun, H.; Qian, Q.; Wu, K.; Luo, J.; Wang, S.; Zhang, C.; Ma, Y.; Liu, Q.; Huang, X.; Yuan, Q.; et al. Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 2014, 46, 652–656. [Google Scholar] [CrossRef]
- Li, X.; Xia, K.; Liang, Z.; Chen, K.; Gao, C.; Zhang, M. MicroRNA393 is involved in nitrogen-promoted rice tillering through regulation of auxin signal transduction in axillary buds. Sci. Rep. 2016, 6, 32158. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Jiang, Z.; Wang, W.; Xu, R.; Wang, Q.; Zhang, Z.; Li, A.; Liang, Y.; Ou, S.; et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 2021, 590, 600–605. [Google Scholar] [CrossRef]
- Fioreze, S.L.; Castoldi, G.; Pivetta, L.A.; Pivetta, L.G.; Fernandes, D.M.; Büll, L.T. Tillering of two wheat genotypes as affected by phosphorus levels. Acta Sci. Agron. 2012, 34, 331–338. [Google Scholar] [CrossRef]
- Li, X.R.; Sun, J.; Albinsky, D.; Zarrabian, D.; Hull, R.; Lee, T.; Jarratt-Barnham, E.; Chiu, C.H.; Jacobsen, A.; Soumpourou, E.; et al. Nutrient regulation of lipochitooligosaccharide recognition in plants via NSP1 and NSP2. Nat. Commun. 2022, 13, 6421. [Google Scholar] [CrossRef]
- Chérel, I. Regulation of K+ channel activities in plants: From physiological to molecular aspects. J. Exp. Bot. 2004, 55, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, S.; Guo, H.; Wang, S.; Xu, L.; Li, C.; Qian, Q.; Chen, F.; Geisler, M.; Qi, Y.; et al. OsABCB14 functions in auxin transport and iron homeostasis in rice (Oryza sativa L.). Plant J. Cell Mol. Biol. 2014, 79, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Feng, H.; Hu, Q.; Qu, H.; Chen, A.; Yu, L.; Xu, G. Improving rice tolerance to potassium deficiency by enhancing OsHAK16p:WOX11-controlled root development. Plant Biotechnol. J. 2015, 13, 833–848. [Google Scholar] [CrossRef]
- Kang, J.; Li, J.; Gao, S.; Tian, C.; Zha, X. Overexpression of the leucine-rich receptor-like kinase gene LRK 2 increases drought tolerance and tiller number in rice. Plant Biotechnol. J. 2017, 15, 1175–1185. [Google Scholar] [CrossRef]
- Weng, X.; Wang, L.; Wang, J.; Hu, Y.; Du, H.; Xu, C.; Xing, Y.; Li, X.; Xiao, J.; Zhang, Q. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 2014, 164, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Cai, W. RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). J. Exp. Bot. 2014, 65, 3277–3287. [Google Scholar] [CrossRef]
- Evers, J.B.; Vos, J.; Andrieu, B.; Struik, P.C. Cessation of Tillering in Spring Wheat in Relation to Light Interception and Red: Far-red Ratio. Ann. Bot. 2006, 97, 649–658. [Google Scholar] [CrossRef]
- Girault, T.; Bergougnoux, V.; Combes, D.; Viemont, J.-D.; Leduc, N. Light controls shoot meristem organogenic activity and leaf primordia growth during bud burst in Rosa sp. Plant Cell Environ. 2008, 31, 1534–1544. [Google Scholar] [CrossRef]
- Stirnberg, P.; Van De Sande, K.; Leyser, H.M.O. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002, 129, 1131–1141. [Google Scholar] [CrossRef]
- Beveridge, C.A.; Weller, J.L.; Singer, S.R.; Hofer, J.M.I. Axillary Meristem Development. Budding Relationships between Networks Controlling Flowering, Branching, and Photoperiod Responsiveness. Plant Physiol. 2003, 131, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Kebrom, T.H.; Brutnell, T.P. The molecular analysis of the shade avoidance syndrome in the grasses has begun. J. Exp. Bot. 2007, 58, 3079–3089. [Google Scholar] [CrossRef]
- Ballaré, C.L.; Scopel, A.L.; Sánchez, R.A. Far-red radiation reflected from adjacent leaves: An early signal of competition in plant canopies. Science 1990, 247, 329–332. [Google Scholar] [CrossRef]
- Smith, H.; Whitelam, G.C. The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environ. 1997, 20, 840–844. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, Y.; Xu, G. How does nitrogen shape plant architecture? J. Exp. Bot. 2020, 71, 4415–4427. [Google Scholar] [CrossRef]
- Luo, Z.; Janssen, B.J.; Snowden, K.C. The molecular and genetic regulation of shoot branching. Plant Physiol. 2021, 187, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Qi, S.; Wang, Y. Nitrate signaling and use efficiency in crops. Plant Commun. 2022, 3, 100353. [Google Scholar] [CrossRef] [PubMed]
- Duan, E.; Wang, Y.; Li, X.; Lin, Q.; Zhang, T.; Wang, Y.; Zhou, C.; Zhang, H.; Jiang, L.; Wang, J.; et al. OsSHI1 Regulates Plant Architecture Through Modulating the Transcriptional Activity of IPA1 in Rice. Plant Cell 2019, 31, 1026–1042. [Google Scholar] [CrossRef] [PubMed]
- Drummond, R.S.M.; Janssen, B.J.; Luo, Z.; Oplaat, C.; Ledger, S.E.; Wohlers, M.W.; Snowden, K.C. Environmental Control of Branching in Petunia. Plant Physiol. 2015, 168, 735–751. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Xie, X.; Kim, H.I.; Kisugi, T.; Nomura, T.; Sekimoto, H.; Yokota, T.; Yoneyama, K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 2012, 235, 1197–1207. [Google Scholar] [CrossRef]
- Kamada-Nobusada, T.; Makita, N.; Kojima, M.; Sakakibara, H. Nitrogen-Dependent Regulation of De Novo Cytokinin Biosynthesis in Rice: The Role of Glutamine Metabolism as an Additional Signal. Plant Cell Physiol. 2013, 54, 1881–1893. [Google Scholar] [CrossRef]
- Yoneyama, K.; Xie, X.; Kisugi, T.; Nomura, T.; Yoneyama, K. Nitrogen and phosphorus fertilization negatively affects strigolactone production and exudation in sorghum. Planta 2013, 238, 885–894. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Puga, M.I.; Rojas-Triana, M.; Martinez-Hevia, I.; Diaz, S.; Poza-Carrión, C.; Miñambres, M.; Leyva, A. Plant adaptation to low phosphorus availability: Core signaling, crosstalks, and applied implications. Mol. Plant 2022, 15, 104–124. [Google Scholar] [CrossRef]
- Yuan, K.; Zhang, H.; Yu, C.; Luo, N.; Yan, J.; Zheng, S.; Hu, Q.; Zhang, D.; Kou, L.; Meng, X.; et al. Low phosphorus promotes NSP1-NSP2 heterodimerization to enhance strigolactone biosynthesis and regulate shoot and root architectures in rice. Mol. Plant 2023. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Magome, H.; Takeda-Kamiya, N.; Yamaguchi, S. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 2010, 51, 1118–1126. [Google Scholar] [CrossRef]
- Yang, T.; Feng, H.; Zhang, S.; Xiao, H.; Hu, Q.; Chen, G.; Xuan, W.; Moran, N.; Murphy, A.; Yu, L.; et al. The Potassium Transporter OsHAK5 Alters Rice Architecture via ATP-Dependent Transmembrane Auxin Fluxes. Plant Commun. 2020, 1, 100052. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.; Mishra, S.S.; Behera, P.K. Drought tolerance in rice: Focus on recent mechanisms and approaches. Rice Sci. 2021, 28, 119–132. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, V.V.; Babu, M.S.; Basava, R.K.; Tripura Venkata, V.G.N.; Mangrauthia, S.K.; Voleti, S.R.; Neelamraju, S. Trait and Marker Associations in Oryza nivara and O. rufipogon Derived Rice Lines under Two Different Heat Stress Conditions. Front. Plant Sci. 2017, 8, 1819. [Google Scholar] [CrossRef]
- Harsant, J.; Pavlovic, L.; Chiu, G.; Sultmanis, S.; Sage, T.L. High temperature stress and its effect on pollen development and morphological components of harvest index in the C3 model grass Brachypodium distachyon. J. Exp. Bot. 2013, 64, 2971–2983. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, F.; Cao, H.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 2012, 7, e48445. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, Y.; Xu, S.; Zhang, Z.; Xu, Y.; Zhang, J.; Chong, K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018, 218, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.H.; Lee, S.S.; Park, H.J.; Lyu, J.I.; Chong, W.S.; Liu, J.R.; Kim, B.G.; Ahn, J.C.; Cho, H.S. Overexpression of OsCYP19-4 increases tolerance to cold stress and enhances grain yield in rice (Oryza sativa). J. Exp. Bot. 2016, 67, 69–82. [Google Scholar] [CrossRef]
- Clarke, P.R.; Zhang, C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 2008, 9, 464–477. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, K.; Li, D.; Yan, J.; Zhang, W. Enhanced Cold Tolerance and Tillering in Switchgrass (Panicum virgatum L.) by Heterologous Expression of Osa-miR393a. Plant Cell Physiol. 2017, 58, 2226–2240. [Google Scholar] [CrossRef]
- Holbrook-Smith, D.; Toh, S.; Tsuchiya, Y.; McCourt, P. Small-molecule antagonists of germination of the parasitic plant Striga hermonthica. Nat. Chem. Biol. 2016, 12, 724–729. [Google Scholar] [CrossRef]
- Cardoso, C.; Zhang, Y.; Jamil, M.; Hepworth, J.; Charnikhova, T.; Dimkpa, S.O.; Meharg, C.; Wright, M.H.; Liu, J.; Meng, X.; et al. Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proc. Natl. Acad. Sci. USA 2014, 111, 2379–2384. [Google Scholar] [CrossRef] [PubMed]
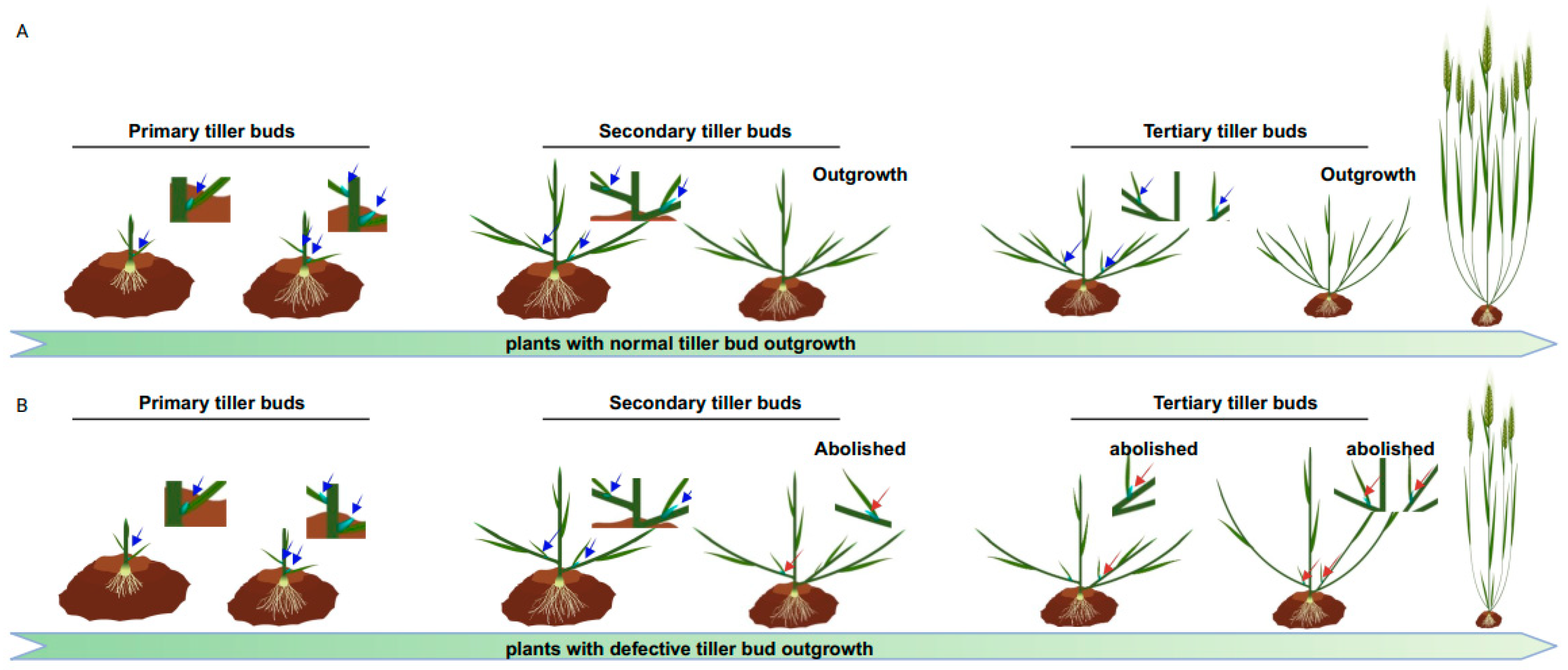
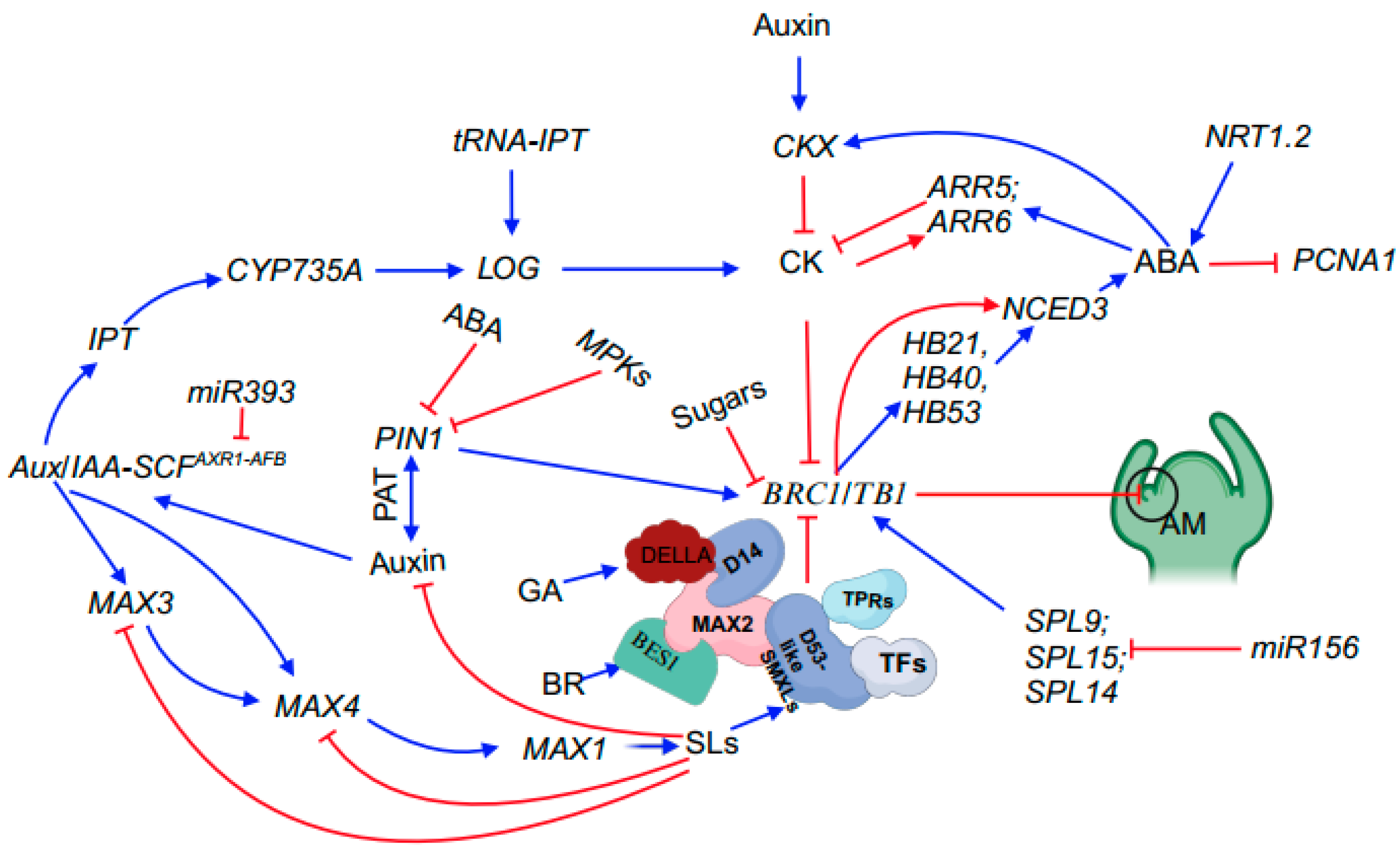
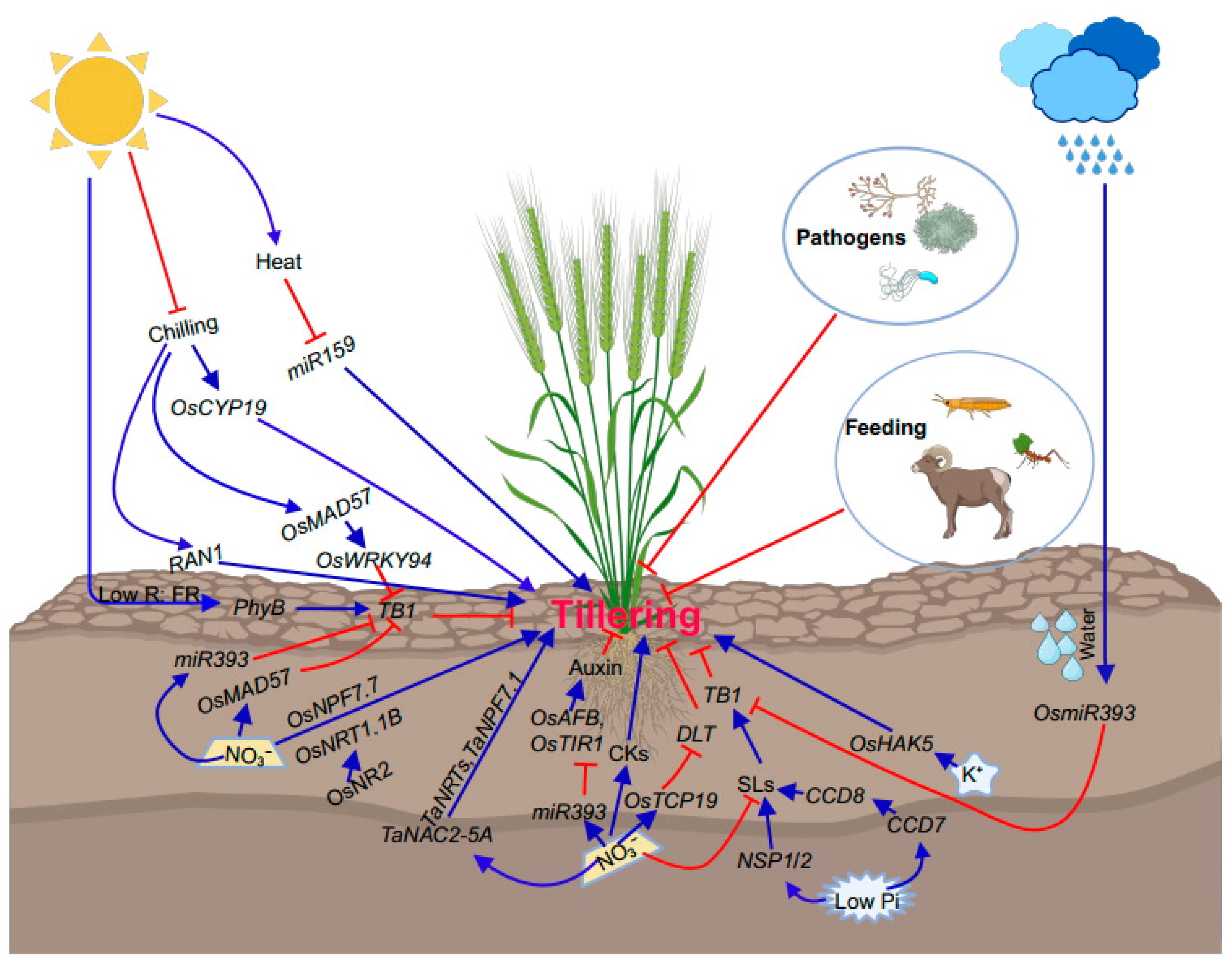
| Gene Names | Accession Numbers | Reported Species (Homolog) | Functional Annotation | References |
|---|---|---|---|---|
| OsTB1 | Os03g0706500 | Arabidopsis, rice, maize, pea, tomato | Transcription factor TCP family | [6,29,30,31] |
| OsSPL14 (IPA1), OsSPL15 | Os08g0509600, Os08g0513700 | Rice, Arabidopsis | SQUAMOSA promoter binding protein-like transcription factors | [5,32,33] |
| AXR1 | AT1G05180 | Arabidopsis | a subunit of the RUB1 activating enzyme | [34] |
| YUCCA | AT4G32540 | Arabidopsis | A flavin monooxygenase-like enzyme, auxin biosynthesis | [35] |
| PIN1 | Os02g0743400 | Rice | an auxin transporter | [36] |
| OsPIN5b | Os09g0505400 | Rice | an auxin transporter | [37] |
| MKK7 | AT1G18350 | Arabidopsis | MAP kinase kinase7 | [38] |
| MKK6 | AT5G56580 | Arabidopsis | MAP kinase kinase 6 | [39] |
| TIR1 | Os05g0150500, AT1G72930 | Rice, Arabidopsis | auxin receptor | [9,40] |
| IAA12 | AT1G04550 | Arabidopsis | an auxin-responsive gene | [41] |
| AFB2 | Os04g0395600 | Rice | Auxin signaling f-box 2 | [42] |
| RUB1 | AT1G31340 | Arabidopsis | A ubiquitin-related protein | [9] |
| D27 | Os11g0587000 | Rice | An iron-containing protein | [12,43] |
| CCD7/MAX3 | AT2G44990, Os04g0550600 | Arabidopsis, rice | carotenoid cleavage dioxygenases | [44] |
| CCD8/MAX4 | AT4G32810, Os01g0746400 | Arabidopsis, rice | carotenoid cleavage dioxygenases | [45] |
| MAX1 | AT2G26170 | Arabidopsis | Belonging to the CYP711A cytochrome P450 family | [46] |
| MAX2/D3 | AT2G42620, Os06g0154200 | Arabidopsis, rice | Belonging to a member of the F-box leucine-rich repeat family | [46] |
| D14 | Os03g0203200, AT3G03990 | Rice, Arabidopsis | An alpha/beta hydrolase | [47] |
| D53 | Os11g0104300 | Rice | The substrate of SCF-D3 ubiquitin complex | [48] |
| SMXL6, SMXL7, SMXL8 | AT1G07200, AT2G29970, AT2G40130 | Arabidopsis | D53-like proteins | [48,49] |
| IPT | AT3G23630 | Arabidopsis | An isopentenyl transferase | [50] |
| SPS | AT1G16410 | Arabidopsis | Belonging to a member of CYP79F | [51] |
| AMP1 | AT3G54720 | Arabidopsis | A glutamate carboxypeptidase | [8] |
| PsCKX2 | LOC127082854 | Pea | Cytokinin dehydrogenase 6-like | [52] |
| NCED3 | AT3G14440 | Arabidopsis | A 9-cis-epoxycarotenoid dioxygenase | [53] |
| ABA2 | AT1G52340 | Arabidopsis | A cytosolic short-chain dehydrogenase | [54] |
| HB21, HB40, HB53 | AT2G02540, AT4G36740, AT5G66700 | Arabidopsis | Homeobox proteins | [14] |
| SLR1 | Os03g0707600 | Rice | A DELLA protein | [32] |
| BES1 | AT1G19350 | Arabidopsis | A transcription factor | [55] |
| MOC2 | Os01g0866400 | Rice | A cytosolic fructose 1,6-bisphosphatase | [56] |
| OsNPF7.7 | Os10g0579600 | Rice | One nitrate transporter | [57] |
| TaNAC2-5A | AY625683 | Wheat | A transcription factor | [58] |
| OsMADS57 | Os02g0731200 | Rice | A MADS transcription factor 57 | [59] |
| OsBZR1, BES1 | Os07g0580500, AT1G19350 | Rice, Arabidopsis | A key transcription factor involved in brassinosteroid (BS) signaling | [60] |
| DLT | Os06g0127800 | Rice | A GRAS protein | [61] |
| GSK2 | Os05g0207500 | Rice | A conserved glycogen synthase kinase 3-like kinase | [61] |
| RLA1 | Os05g0389000 | Rice | An APETALA2 (AP2) DNA binding domain protein | [62] |
| BRI1/D61 | Os01g0718300 | Rice | A BR receptor | [61] |
| Gene Names | Accession Numbers | Reported Species (Homolog) | Functional Annotation | References |
|---|---|---|---|---|
| PHYB | LOC8081072 | Sorghum bicolor | Phytochrome B | [149] |
| SbTB1 | LOC8062930 | Sorghum bicolor | Belonging to transcription factor of the TCP family | [149] |
| NCED3 | AT3G14440 | Arabidopsis | A 9-cis-epoxycarotenoid dioxygenase | [53] |
| ABA2 | AT1G52340 | Arabidopsis | A cytosolic short-chain dehydrogenase/reductase | [53] |
| OsNPF7.7 | Os10g0579600 | Rice | Belonging to the peptide transporter (PTR) gene family | [57] |
| OsNR2 | Os02g0770800 | Rice | NADH/NADPH-dependent NO3− reductase 2 | [150] |
| NGR5 | Os05g0389000 | Rice | One APETALA2-domain transcription factor | [151] |
| PRC2 | Os03g0108700 | Rice | A polycomb repressive complex 2-associated coiled-coil protein | [151] |
| D14 | Os03g0203200 | Rice | A strigolactone receptor | [151] |
| SPL14 | Os08g0509600 | Rice | A squamosa promoter-binding-like transcription activator | [151] |
| OsDEP1 | Os09g0441900 | Rice | One unknown phosphatidylethanolamine-binding protein (PEBP)-like domain protein | [152] |
| OsAFB2 | Os04g0395600 | Rice | An auxin receptor | [153] |
| OsTIR1 | Os05g0150500 | Rice | A F-Box auxin receptor protein | [153] |
| OsTCP19 | Os06g0226700 | Rice | A class-I TCP transcription factor | [154] |
| TaNAC2-5A | LOC606326 | Wheat | NAC domain-containing protein 2 | [58] |
| OsMADS57 | Os02g0731200 | Rice | A MADS-box transcription factor | [59] |
| OsPHR2 | Os07g0438800 | Rice | A MYB-CC family protein | [155] |
| NSP1 | Os03g0408600 | Rice | A GRAS-domain transcription factor | [156] |
| NSP2 | Os03g0263300 | Rice | A GRAS-domain transcription factor | [156] |
| OsHAK5 | Os01g0930400 | Rice | A potassium transporter | [157] |
| OsABCB14 | Os04g0459000 | Rice | An auxin transport | [158] |
| WOX11 | Os07g0684900 | Rice | A WUSCHEL-related homeobox protein | [159] |
| OsHAK16 | Os03g0575200 | Rice | A high-affinity potassium transporter | [159] |
| OsAUX1 | Os01g0856500 | Rice | An auxin transporter | [42] |
| LRK2 | Os02g0154000 | Rice | A leucine-rich repeat receptor-like kinase | [160] |
| GHD7 | Os07g0261200 | Rice | A CCT(CONSTANS, CONSTANS-LIKE, and TIMING OF CHLOROPHYLL A/B BINDING1) domain protein | [161] |
| OsRAN1 | Os01g0611100 | Rice | A small GTPase | [162] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Y.; Khourchi, S.; Li, S.; Du, Y.; Delaplace, P. Unlocking the Multifaceted Mechanisms of Bud Outgrowth: Advances in Understanding Shoot Branching. Plants 2023, 12, 3628. https://doi.org/10.3390/plants12203628
Yuan Y, Khourchi S, Li S, Du Y, Delaplace P. Unlocking the Multifaceted Mechanisms of Bud Outgrowth: Advances in Understanding Shoot Branching. Plants. 2023; 12(20):3628. https://doi.org/10.3390/plants12203628
Chicago/Turabian StyleYuan, Yundong, Said Khourchi, Shujia Li, Yanfang Du, and Pierre Delaplace. 2023. "Unlocking the Multifaceted Mechanisms of Bud Outgrowth: Advances in Understanding Shoot Branching" Plants 12, no. 20: 3628. https://doi.org/10.3390/plants12203628
APA StyleYuan, Y., Khourchi, S., Li, S., Du, Y., & Delaplace, P. (2023). Unlocking the Multifaceted Mechanisms of Bud Outgrowth: Advances in Understanding Shoot Branching. Plants, 12(20), 3628. https://doi.org/10.3390/plants12203628





