The Effects of Accompanying Ryegrass on Bayberry Trees by Change of Soil Property, Rhizosphere Microbial Community Structure, and Metabolites
Abstract
1. Introduction
2. Results and Discussion
2.1. Effects of Accompanying Ryegrass on Vegetative Growth and Fruit Quality of Bayberry
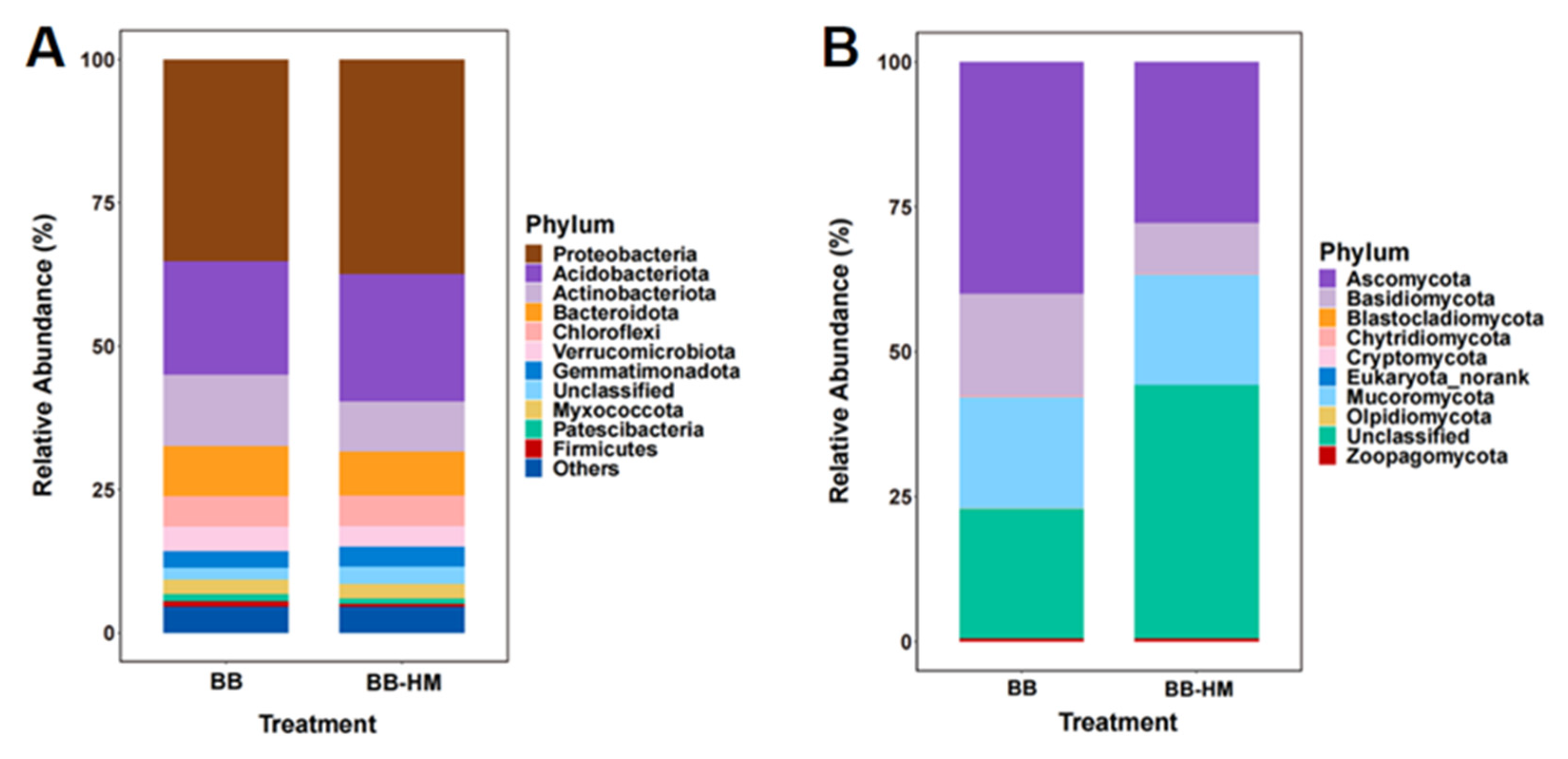
2.2. Change in Soil Microbial Community Composition
2.3. Change in Microbial Community Diversity
2.4. Soil Properties Related to Microbial Communities
2.4.1. Soil Physical and Chemical Properties
2.4.2. RDA of Soil Properties and Microbial Communities
2.5. Change in Rhizosphere Soil Metabolites Composition
2.6. Analysis of Soil Secondary Metabolites Differential Content
2.7. Correlation Analysis of Soil Microorganisms and Metabolites
3. Materials and Methods
3.1. Field Trial and Sampling
3.2. Measurement of Vegetative Growth Parameters
3.3. Measurement of Fruit Parameters with Economic Impact
3.4. Soil Sample Collection
3.5. Soil Genome Sequencing
3.6. Soil Physical and Chemical Properties
3.7. Gas Chromatography-Mass Spectrometry (GC-MS) Metabolomics Analysis
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, C.D.; Huang, H.Z.; Xu, C.J.; Li, X.; Chen, K.S. Biological activities of extracts from Chinese bayberry (Myrica rubra Sieb. et Zucc.): A review. Plant Foods Hum. Nutr. 2013, 68, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.Y.; Yu, H.Y.; Zhang, S.W.; Liang, S.M.; Zheng, X.L.; Zhang, S.J.; Yao, P.; Zheng, H.K.; Qi, X.J. Genome sequencing provides insights into the evolution and antioxidant activity of Chinese bayberry. BMC Genom. 2019, 20, 458. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.L.; Zhang, S.L.; Chen, D.M. Red bayberry (Myrica rubra Sieb. & Zucc.): A valuable evergreen tree fruit for tropical and subtropical areas. Front. Trop. Fruit Res. 1992, 321, 112–121. [Google Scholar]
- Mathew, I.; Shimelis, H.; Mutema, M.; Minasny, B.; Chaplot, V. Crops for increasing soil organic carbon stocks—A global meta analysis. Geoderma 2020, 367, 114230. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, F.; Tang, L.; Zheng, Y.; Li, Y.; Christie, P.; Li, L. Wheat powdery mildew and foliar N concentrations as influenced by N fertilization and belowground interactions with intercropped faba bean. Plant Soil 2007, 291, 1–13. [Google Scholar] [CrossRef]
- Zhou, W.J.; Zhang, Y.Z.; Wang, K.R.; Li, H.S.; Hao, Y.J.; Liu, X. Plant phosphorus uptake in a soybean-citrus intercropping system in the red soil hilly region of South China. Pedosphere 2009, 19, 244–250. [Google Scholar] [CrossRef]
- Loranger-Merciris, G.; Barthes, L.; Gastine, A.; Leadley, P. Rapid effects of plant species diversity and identity on soil microbial communities in experimental grassland ecosystems. Soil Biol. Biochem. 2006, 38, 2336–2343. [Google Scholar] [CrossRef]
- Song, Y.N.; Zhang, F.S.; Marschner, P.; Fan, F.L.; Gao, H.M.; Bao, X.G.; Sun, J.H.; Li, L. Effect of intercropping on crop yield and chemical and microbiological properties in rhizosphere of wheat (Triticum aestivum L.), maize (Zea mays L.), and faba bean (Vicia faba L.). Biol. Fertil. Soils 2007, 43, 565–574. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, F.; Zhou, X. Allelopathic effects of wheat, soybean and oat residues on cucumber and Fusarium oxysporum f.sp cucumerinum Owen. Allelopath. J. 2010, 25, 107–114. [Google Scholar]
- Li, Q.; Wu, F.; Yang, Y.; Wang, X. Effects of rotation and interplanting on soil bacterial communities and cucumber yield. Acta Agric. Scand. Sect. B-Soil Plant Sci. 2009, 59, 431–439. [Google Scholar] [CrossRef]
- Liu, Y.; Zhan, L.; Xu, C.; Jiang, H.; Zhu, C.; Sun, L.; Sun, C.; Li, X. α-Glucosidase inhibitors from Chinese bayberry (Morella rubra Sieb. et Zucc.) fruit: Molecular docking and interaction mechanism of flavonols with different B-ring hydroxylations. RSC Adv. 2020, 10, 29347–29361. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Cai, Y.; Sun, M.; Wang, G.; Corke, H. Anthocyanins, flavonols, and free radical scavenging activity of Chinese bayberry (Myrica rubra) extracts and their color properties and stability. J. Agric. Food Chem. 2005, 53, 2327–2332. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Chung, D.; Himmel, M.E.; Bomble, Y.J.; Westpheling, J. Heterologous expression of family 10 xylanases from Acidothermus cellulolyticus enhances the exoproteome of Caldicellulosiruptor bescii and growth on xylan substrates. Biotechnol. Biofuels 2016, 9, 176. [Google Scholar] [CrossRef][Green Version]
- Mhedbi-Hajri, N.; Jacques, M.A.; Koebnik, R. Adhesion mechanisms of plant-pathogenic Xanthomonadaceae. Adv. Exp. Med. Biol. 2011, 715, 71–89. [Google Scholar]
- Zhao, Y.; Wu, F.; Yang, W.; Tan, B.; He, W. Variations in bacterial communities during foliar litter decomposition in the winter and growing seasons in an alpine forest of the eastern Tibetan Plateau. Can. J. Microbiol. 2016, 62, 35–48. [Google Scholar] [CrossRef]
- Sreevidya, M.; Gopalakrishnan, S.; Kudapa, H.; Varshney, R.K. Exploring plant growth-promotion actinomycetes from vermicompost and rhizosphere soil for yield enhancement in chickpea. Braz. J. Microbiol. 2016, 47, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Angelini, P.; Rubini, A.; Gigante, D.; Reale, L.; Pagiotti, R.; Venanzoni, R. The endophytic fungal communities associated with the leaves and roots of the common reed (Phragmites australis) in Lake Trasimeno (Perugia, Italy) in declining and healthy stands. Fungal Ecol. 2012, 5, 683–693. [Google Scholar] [CrossRef]
- Kellner, H.; Zak, D.R.; Vandenbol, M. Fungi unearthed: Transcripts encoding lignocellulolytic and chitinolytic enzymes in forest soil. PLoS ONE 2010, 5, e10971. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Cheng, L.; Han, M.; Yang, L.-m.; Li, Y.; Sun, Z.; Zhang, T. Changes in the physiological characteristics and baicalin biosynthesis metabolism of Scutellaria baicalensis Georgi under drought stress. Ind. Crops Prod. 2018, 122, 473–482. [Google Scholar] [CrossRef]
- White, R.A.; Rivas-Ubach, A.; Borkum, M.I.; Köberl, M.; Bilbao, A.; Colby, S.M.; Hoyt, D.W.; Bingol, K.; Kim, Y.-M.; Wendler, J.P.; et al. The state of rhizospheric science in the era of multi-omics: A practical guide to omics technologies. Rhizosphere 2017, 3, 212–221. [Google Scholar] [CrossRef]
- Good, A.G.; Muench, D.G. Long-Term Anaerobic Metabolism in Root Tissue (Metabolic Products of Pyruvate Metabolism). Plant Physiol. 1993, 101, 1163–1168. [Google Scholar] [CrossRef]
- Sogi, D.S.; Siddiq, M.; Dolan, K.D. Total phenolics, carotenoids and antioxidant properties of Tommy Atkin mango cubes as affected by drying techniques. LWT—Food Sci. Technol. 2015, 62, 564–568. [Google Scholar] [CrossRef]
- Jiang, Y.; Nie, W.J. Chemical properties in fruits of mulberry species from the Xinjiang province of China. Food Chem. 2015, 174, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Qu, B.; Liu, Y.; Sun, X.; Li, S.; Wang, X.; Xiong, K.; Yun, B.; Zhang, H. Effect of various mulches on soil physico-chemical properties and tree growth (Sophora japonica) in urban tree pits. PLoS ONE 2019, 14, e0210777. [Google Scholar] [CrossRef]
- Ma, M.; Zhou, J.; Ongena, M.; Liu, W.; Wei, D.; Zhao, B.; Guan, D.; Jiang, X.; Li, J. Effect of long-term fertilization strategies on bacterial community composition in a 35-year field experiment of Chinese Mollisols. AMB Express 2018, 8, 20. [Google Scholar] [CrossRef]
- Baran, A.; Mierzwa-Hersztek, M.; Gondek, K.; Tarnawski, M.; Szara, M.; Gorczyca, O.; Koniarz, T. The influence of the quantity and quality of sediment organic matter on the potential mobility and toxicity of trace elements in bottom sediment. Environ. Geochem. Health 2019, 41, 2893–2910. [Google Scholar] [CrossRef]
- Ijaz, M.U.; Ahmed, M.I.; Zou, X.; Hussain, M.; Zhang, M.; Zhao, F.; Xu, X.; Zhou, G.; Li, C. Beef, casein, and soy proteins differentially affect lipid metabolism, triglycerides accumulation and gut microbiota of high-fat diet-fed C57BL/6J mice. Front. Microbiol. 2018, 9, 2200. [Google Scholar] [CrossRef]





| Parameters | Value | Parameters | Value |
|---|---|---|---|
| Twig length (mm) | Twig diameter (mm) | ||
| BB-HM | 48.60 ± 8.16 | BB-HM | 2.11 ± 0.32 |
| BB | 46.70 ± 9.45 | BB | 2.20 ± 0.39 |
| Leaf length (mm) | Leaf width (mm) | ||
| BB-HM | 96.50 ± 0.92 | BB-HM | 29.10 ± 0.25 |
| BB | 96.80 ± 1.03 | BB | 29.90 ± 0.36 |
| Leaf thickness (mm) | Chlorophyll/(SPAD) | ||
| BB-HM | 0.97 ± 0.03 | BB-HM | 43.45 ± 2.69 |
| BB | 0.98 ± 0.06 | BB | 42.89 ± 3.71 |
| Parameters | Value | Parameters | Value |
|---|---|---|---|
| Single fruit weight (g) | Titratable sugar (mg/g) | ||
| BB-HM | 47.08 ± 3.40 | BB-HM | 109.83 ± 0.41 * |
| BB | 48.95 ± 4.01 | BB | 107.40 ± 0.42 |
| Vitamin C (mg/100 g) | Titratable acid (%) | ||
| BB-HM | 9.66 ± 0.54 * | BB-HM | 0.97 ± 0.08 |
| BB | 7.52 ± 1.05 | BB | 1.06 ± 0.07 * |
| Titratable flavone (mg/g) | |||
| BB-HM | 1.60 ± 0.01 * | ||
| BB | 1.28 ± 0.01 |
| Parameters | Value | Parameters | Value |
|---|---|---|---|
| pH | Available phosphorus (mg/kg) | ||
| BB-HM | 4.59 ± 0.13 | BB-HM | 68.11 ± 1.35 |
| BB | 5.34 ± 0.19 * | BB | 100.21 ± 4.50 * |
| Organic matter (%) | Exchangeable aluminum (cmol/kg) | ||
| BB-HM | 63.76 ± 1.35 | BB-HM | 2.99 ± 0.30 * |
| BB | 72.07 ± 3.19 * | BB | 0.14 ± 0.02 |
| Available nitrogen (mg/kg) | Available kalium (mg/kg) | ||
| BB-HM | 145.58 ± 3.58 | BB-HM | 255.17 ± 8.38 |
| BB | 169.67 ± 8.40 * | BB | 351.79 ± 19.70 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Li, G.; Qi, X.; Yu, Z.; Abdallah, Y.; Ogunyemi, S.O.; Zhang, S.; Ren, H.; Mohany, M.; S. Al-Rejaie, S.; et al. The Effects of Accompanying Ryegrass on Bayberry Trees by Change of Soil Property, Rhizosphere Microbial Community Structure, and Metabolites. Plants 2023, 12, 3669. https://doi.org/10.3390/plants12213669
Li C, Li G, Qi X, Yu Z, Abdallah Y, Ogunyemi SO, Zhang S, Ren H, Mohany M, S. Al-Rejaie S, et al. The Effects of Accompanying Ryegrass on Bayberry Trees by Change of Soil Property, Rhizosphere Microbial Community Structure, and Metabolites. Plants. 2023; 12(21):3669. https://doi.org/10.3390/plants12213669
Chicago/Turabian StyleLi, Changxin, Gang Li, Xingjiang Qi, Zheping Yu, Yasmine Abdallah, Solabomi Olaitan Ogunyemi, Shuwen Zhang, Haiying Ren, Mohamed Mohany, Salim S. Al-Rejaie, and et al. 2023. "The Effects of Accompanying Ryegrass on Bayberry Trees by Change of Soil Property, Rhizosphere Microbial Community Structure, and Metabolites" Plants 12, no. 21: 3669. https://doi.org/10.3390/plants12213669
APA StyleLi, C., Li, G., Qi, X., Yu, Z., Abdallah, Y., Ogunyemi, S. O., Zhang, S., Ren, H., Mohany, M., S. Al-Rejaie, S., Li, B., & Liu, E. (2023). The Effects of Accompanying Ryegrass on Bayberry Trees by Change of Soil Property, Rhizosphere Microbial Community Structure, and Metabolites. Plants, 12(21), 3669. https://doi.org/10.3390/plants12213669









