The Mining of Genetic Loci and the Analysis of Candidate Genes to Identify the Physical and Chemical Markers of Anti-Senescence in Rice
Abstract
:1. Introduction
2. Results
2.1. Phenotype Analysis of Anti-Senescence-Related Traits in Both Parents and RIL Populations
2.2. Analysis of QTL Mapping Results Related to Anti-Senescence Traits
2.3. Anti-Senescence Candidate Gene Expression Level Analysis
3. Discussion
3.1. QTL Mapping Showed Major and Minor QTLs Related to Anti-Senescence in Rice
3.2. Analysis of Anti-Senescence-Related Candidate Genes
4. Materials and Methods
4.1. Experimental Materials
4.2. Germination Cultivation and Management of Rice Seeds
4.3. Analysis of Data on Hormone Treatment and Anti-Senescence-Related Traits in Rice
4.4. Linkage Map Construction and QTL Mapping
4.5. Differential Analysis of Candidate Gene Expression
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lee, S.; Masclaux-Daubresse, C. Current understanding of leaf senescence in rice. Int. J. Mol. Sci. 2021, 22, 4515. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Ito, D.; Goto, M.; Suzuki, S.; Masuda, S.; Iba, K.; Kusumi, K. Regulation of ppGpp synthesis and its impact on chloroplast biogenesis during early leaf development in rice. Plant Cell Physiol. 2022, 63, 919–931. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, L.J.; Zhang, Z.H.; Wu, J.L. Identification and comparative analysis of premature senescence leaf mutants in rice (Oryza sativa L.). Int. J. Mol. Sci. 2018, 19, 140. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.J.; Lu, S.C.; Lv, B.; Zhang, B.; Shen, J.B.; He, J.M.; Luo, L.Q.; Xi, D.D.; Chen, X.; Ming, F. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef]
- Guo, P.R.; Li, Z.H.; Huang, P.X.; Li, B.S.; Fang, S.; Chu, J.F.; Guo, H.W. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 2017, 29, 2854–2870. [Google Scholar] [CrossRef]
- Guo, Y.F.; Ren, G.D.; Zhang, K.W.; Li, Z.H.; Miao, Y.; Guo, H.W. Leaf senescence: Progression, regulation, and application. Mol. Hortic. 2021, 1, 5. [Google Scholar] [CrossRef]
- Zhang, C.; Li, N.; Hu, Z.X.; Liu, H.; Hu, Y.Y.; Tan, Y.N.; Sun, Q.N.; Liu, X.Q.; Xiao, L.T.; Wang, W.P.; et al. Mutation of Leaf Senescence 1 encoding a C2H2 zinc finger protein induces ROS accumulation and accelerates leaf senescence in rice. Int. J. Mol. Sci. 2022, 23, 14464. [Google Scholar] [CrossRef]
- Huang, Q.N.; Shi, Y.F.; Zhang, X.B.; Song, L.X.; Feng, B.H.; Wang, H.M.; Xu, X.; Li, X.H.; Guo, D.; Wu, J.L. Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J. Integr. Plant Biol. 2016, 58, 12–28. [Google Scholar] [CrossRef]
- Zhou, Q.Y.; Yu, Q.; Wang, Z.Q.; Pan, Y.F.; Lv, W.T.; Zhu, L.L.; Chen, R.Z.; He, G.C. Knockdown of GDCH gene reveals reactive oxygen species-induced leaf senescence in rice. Plant Cell Environ. 2013, 36, 1476–1489. [Google Scholar] [CrossRef]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef]
- Woo, H.R.; Masclaux-Daubresse, C.; Lim, P.O. Plant senescence: How plants know when and how to die. J. Exp. Bot. 2018, 69, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.C.; Jiao, R.; Wang, S.; Wu, X.M.; Ye, H.F.; Pan, C.Y.; Li, S.F.; Xin, D.D.; Zhou, W.Y.; Dai, G.X.; et al. SPL36 encodes a receptor-like protein kinase that regulates programmed cell death and defense responses in rice. Rice 2021, 14, 34. [Google Scholar]
- Abdelkhalik, A.F.; Shishido, R.; Nomura, K.; Ikehashi, H. QTL-based analysis of leaf senescence in an indica/japonica hybrid in rice (Oryza sativa L.). Theor. Appl. Genet. 2005, 110, 1226–1235. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.H.; He, Y.Q.; Xu, C.G.; Li, X.H.; Zhang, Q. The genetic basis of stay-greenin rice analyzed in a population of double haploid lines derivedfrom an indica by japonica cross. Theor. Appl. Genet. 2004, 108, 688–698. [Google Scholar] [CrossRef]
- Yue, B.; Xue, W.Y.; Luo, L.J.; Xing, Y.Z. QTL analysis for flag leaf characteristics and their relationships with yield and yield traits in rice. J. Genet. Genom. 2006, 33, 824–832. [Google Scholar] [CrossRef]
- Yoo, S.C.; Cho, S.H.; Zhang, H.T.; Paik, H.C.; Lee, C.H.; Li, J.J.; Yoo, J.H.; Lee, B.W.; Koh, H.J.; Seo, H.S.; et al. Quantitative trait loci associated with functional stay-green SNU-SG1 in rice. Mol. Cells 2007, 24, 83–94. [Google Scholar]
- Wu, H.B.; Wang, B.; Chen, Y.L.; Liu, Y.G.; Chen, L.T. Characterization and fine mapping of the rice premature senescence mutant ospse1. Theor. Appl. Genet. 2013, 126, 1897–1907. [Google Scholar] [CrossRef]
- Lee, R.H.; Wang, C.H.; Huang, L.T.; Chen, S.C. Leaf senescence in riceplants: Cloning and characterization of scenescence up-regulatedgenes. J. Exp. Bot. 2001, 52, 1117–1121. [Google Scholar] [CrossRef]
- Lee, R.H.; Lin, M.C.; Chen, S.C.G. A novel alkaline α-galactosidase gene is involved in rice leaf senescence. Plant Mol. Biol. 2004, 55, 281–295. [Google Scholar] [CrossRef]
- Ansari, M.I.; Lee, R.H.; Chen, S.C.G. A novel senescence-associated gene encoding γ-aminobutyric acid (GABA): Pyruvate transaminase is upregulated during rice leaf senescence. Physiol. Plant. 2005, 123, 1–8. [Google Scholar] [CrossRef]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef]
- Xu, J.M.; Pan, C.Y.; Lin, H.; Ye, H.F.; Wang, S.; Lu, T.; Chen, Q.Y.; Yang, K.R.; Lu, M.; Qian, Q.; et al. A rice XANTHINE DEHYDROGENASE gene regulates leaf senescence and response to abiotic stresses. Crop J. 2022, 10, 310–322. [Google Scholar] [CrossRef]
- Xu, F.F.; Sang, X.C.; Ren, D.Y.; Tang, Y.Q.; Hu, H.W.; Yang, Z.L.; Zhao, F.M.; He, G.H. Genetic analysis and gene mapping of early senescence leaf mutant esl2 in rice. Acta Agron. Sin. 2012, 38, 1347–1353. [Google Scholar] [CrossRef]
- Wang, Y.T.; Wang, X.W.; Xie, J.; Yin, W.Z.; Zhang, T.; Zhu, X.Y.; Yu, P.; Huang, J.Y.; Yang, Z.L.; He, G.H.; et al. Identification and gene mapping of an early senescent leaf mutant esl11 of rice. Crop Sci. 2018, 58, 1932–1941. [Google Scholar] [CrossRef]
- Xiong, E.H.; Li, Z.Y.; Zhang, C.; Zhang, J.; Liu, Y.; Peng, T.; Chen, Z.; Zhao, Q.Z. A study of leaf-senescence genes in rice based on a combination of genomics, proteomics and bioinformatics. Brief. Bioinform. 2021, 22, bbaa305. [Google Scholar] [CrossRef] [PubMed]
- Pabuayon, I.C.M.; Kitazumi, A.; Cushman, K.R.; Singh, R.K.; Gregorio, G.B.; Dhatt, B.; Zabet-Moghaddam, M.; Walia, H.; de Los Reyes, B.G. Novel and transgressive salinity tolerance in recombinant inbred lines of rice created by physiological coupling-uncoupling and network rewiring effects. Front. Plant Sci. 2021, 12, 615277. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.Y.; Wan, J.M.; Su, C.C.; Wang, C.M.; Shen, W.B.; Li, J.M.; Wang, H.L.; Jiang, L.; Liu, S.J.; Chen, L.M.; et al. QTL detection for eating quality of cooked rice in a population of chromosome segment substitution lines. Theor. Appl. Genet. 2004, 110, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Koide, Y.; Sakaguchi, S.; Uchiyama, T.; Ota, Y.; Tezuka, A.; Nagano, A.J.; Ishiguro, S.; Takamure, I.; Kishima, Y. Genetic properties responsible for the transgressive segregation of days to heading in rice. G3 Genes Genomes Genet. 2019, 9, 1655–1662. [Google Scholar] [CrossRef]
- Wang, L. Analysis and Gene Mapping of Leaf Early Senescence and Late Growth Stage Leaf Shape of Rice; Chinese Academy of Agricultural Sciences: Beijing, China, 2014. [Google Scholar]
- Ishimaru, K.; Yano, M.; Aoki, N.; Ono, K.; Hirose, T.; Lin, S.Y.; Monna, L.; Sasaki, T.; Ohsugi, R. Toward the mapping of physiological and agronomic characters on a rice function map: QTL analysis and comparison between QTLs and expressed sequence tags. Theor. Appl. Genet. 2001, 102, 793–800. [Google Scholar] [CrossRef]
- You, Q.Y.; Zhai, K.R.; Yang, D.L.; Yang, W.B.; Wu, J.N.; Liu, J.Z.; Pan, W.B.; Wang, J.J.; Zhu, X.D.; Jian, Y.K.; et al. An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 2016, 20, 758–769. [Google Scholar] [CrossRef]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008, 279, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Ito, Y.; Serizawa, A.; Kurata, N. OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 2003, 36, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Cançado, G.M.A.; Nogueir, F.T.S.; Camargo, S.R.; Drummond, R.D.; Jorge, R.A.; Menossi, M. Gene expression profiling in maize roots under aluminum stress. Biol. Plant. 2008, 52, 475–485. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Moreno-Alvarado, M.; García-Morales, S.; Trejo-Téllez, L.I.; Hidalgo-Contreras, J.V.; Gómez-Merino, F.C. Aluminum enhances growth and sugar concentration, alters macronutrient status and regulates the expression of NAC transcription factors in rice. Front. Plant Sci. 2017, 8, 73. [Google Scholar] [CrossRef]
- Escobar-Sepúlveda, H.F.; Trejo-Téllez, L.L.; García-Morales, S.; Gómez-Merino, F.C. Expression patterns and promoter analyses of aluminum-responsive NAC genes suggest a possible growth regulation of rice mediated by aluminum, hormones and NAC transcription factors. PLoS ONE 2017, 12, e0186084. [Google Scholar] [CrossRef]
- Bourdais, G.; Burdiak, P.; Gauthier, A.; Nitsch, L.; Salojärvi, J.; Rayapuram, C.; Idänheimo, N.; Hunter, K.; Kimura, S.; Merilo, E.; et al. Large-Scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 2015, 11, e1005373. [Google Scholar] [CrossRef]
- Chen, K.G.; Du, L.Q.; Chen, Z.X. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 2003, 53, 61–74. [Google Scholar] [CrossRef]
- Chen, K.G.; Fan, B.F.; Du, L.Q.; Chen, Z.X. Activation of hypersensitive cell death by pathogeninduced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 2004, 56, 271–283. [Google Scholar] [CrossRef]
- Burdiak, P.; Rusaczonek, A.; Witoń, D.; Głów, D.; Karpiński, S. Cysteine-rich receptor-like kinase CRK5 as a regulator of growth, development, and ultraviolet radiation responses in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 3325–3337. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Khalid, N.; Islam, W.; Sanaullah, T.; Anwar, M.; Khan, S.; Ye, W.F.; Lou, Y.G. Zinc finger protein transcription factors: Integrated line of action for plant antimicrobial activity. Microb. Pathog. 2019, 132, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yang, J.; Tong, H.; Li, T.; Wang, L.; Chen, H. Genome-wide analysis of fatty acid desaturase genes in rice (Oryza sativa L.). Sci. Rep. 2019, 9, 19445. [Google Scholar]
- Vaid, N.; Macovei, A.; Tuteja, N. Knights in action: Lectin receptor-like kinases in plant development and stress responses. Mol. Plant 2013, 6, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Passricha, N.; Saifi, S.K.; Kharb, P.; Tuteja, N. Rice lectin receptor-like kinase provides salinity tolerance by ion homeostasis. Biotechnol. Bioeng. 2020, 117, 498–510. [Google Scholar] [CrossRef]
- Tobias, C.M.; Chow, E.K. Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 2005, 220, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.J.; Pujar, A.; Youens-Clark, K.; Yap, I.; Jaiswal, P.; Tecle, I.; Tung, C.W.; Ren, L.Y.; Spooner, W.; Wei, X.H.; et al. Gramene QTL database: Development, content and applications. Database 2009, 2009, bap005. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Garcia, R.S.; Coronejo, S.; Tapia, R. Comparative transcriptomics of rice genotypes with contrasting responses to nitrogen stress reveals genes influencing nitrogen uptake through the regulation of root architecture. Int. J. Mol. Sci. 2020, 21, 5759. [Google Scholar] [CrossRef]
- Mondal, S.; Kumar, V.; Singh, S.P. Phylogenetic distribution and structural analyses of cyanobacterial glutaredoxins (Grxs). Comput. Biol. Chem. 2020, 84, 107141. [Google Scholar] [CrossRef]
- Malik, W.A.; Wang, X.G.; Wang, X.L.; Shu, N.; Cui, R.F.; Chen, X.G.; Wang, D.L.; Lu, X.K.; Yin, Z.J.; Wang, J.J.; et al. Genome-wide expression analysis suggests glutaredoxin genes response to various stresses in cotton. Int. J. Biol. Macromol. 2020, 153, 470–491. [Google Scholar] [CrossRef]
- Asis, S.; Ambrose, K.D.; Yoshiaki, U.; Wu, L.B.; Boby, M.; Michael, F. Genome-wide association study to identify candidate loci and genes for Mn toxicity tolerance in rice. PLoS ONE 2018, 13, e0192116. [Google Scholar]
- Garg, R.; Jhanwar, S.; Tyagi, A.K.; Jain, M. Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 2010, 17, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Shang, L.G.; Yu, H.; Zeng, L.J.; Hu, J.; Ni, S.; Rao, Y.C.; Li, S.F.; Chu, J.F.; Meng, X.B.; et al. A strigolactone biosynthesis gene contributed to the green revolution in rice. Mol. Plant 2020, 13, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2019, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Xie, W.B.; Feng, Q.; Yu, H.H.; Huang, X.H.; Zhao, Q.; Xing, Y.Z.; Yu, S.B.; Han, B.; Zhang, Q.F. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 10578–10583. [Google Scholar] [CrossRef]
- Arends, D.; Prins, P.; Jansen, R.C.; Broman, K.W. R/qtl: High-throughput multiple QTL mapping. Bioinformatics 2010, 26, 2990–2992. [Google Scholar] [CrossRef]
- Ren, D.Y.; Rao, Y.C.; Huang, L.C.; Leng, Y.J.; Hu, J.; Lu, M.; Zhang, G.H.; Zhu, L.; Gao, Z.Y.; Dong, G.J.; et al. Fine mapping identifies new QTL for brown rice rate in rice (Oryza sativa L.). Rice 2016, 9, 4. [Google Scholar] [CrossRef]
- Mccouch, S.; Cho, Y.G.; Yano, M.; Paul, E.; Blinstrub, M.; Morishima, H.; Kinoshita, T. Report on QTL nomenclature. Rice Genet. Newsl. 1997, 14, 11–13. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
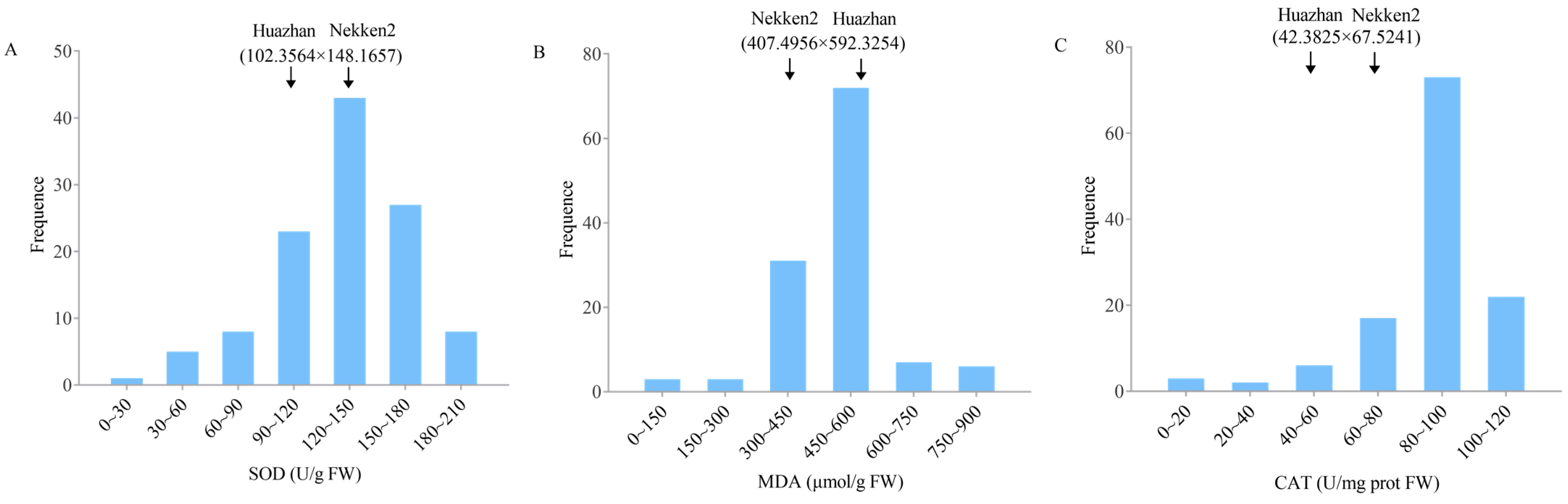
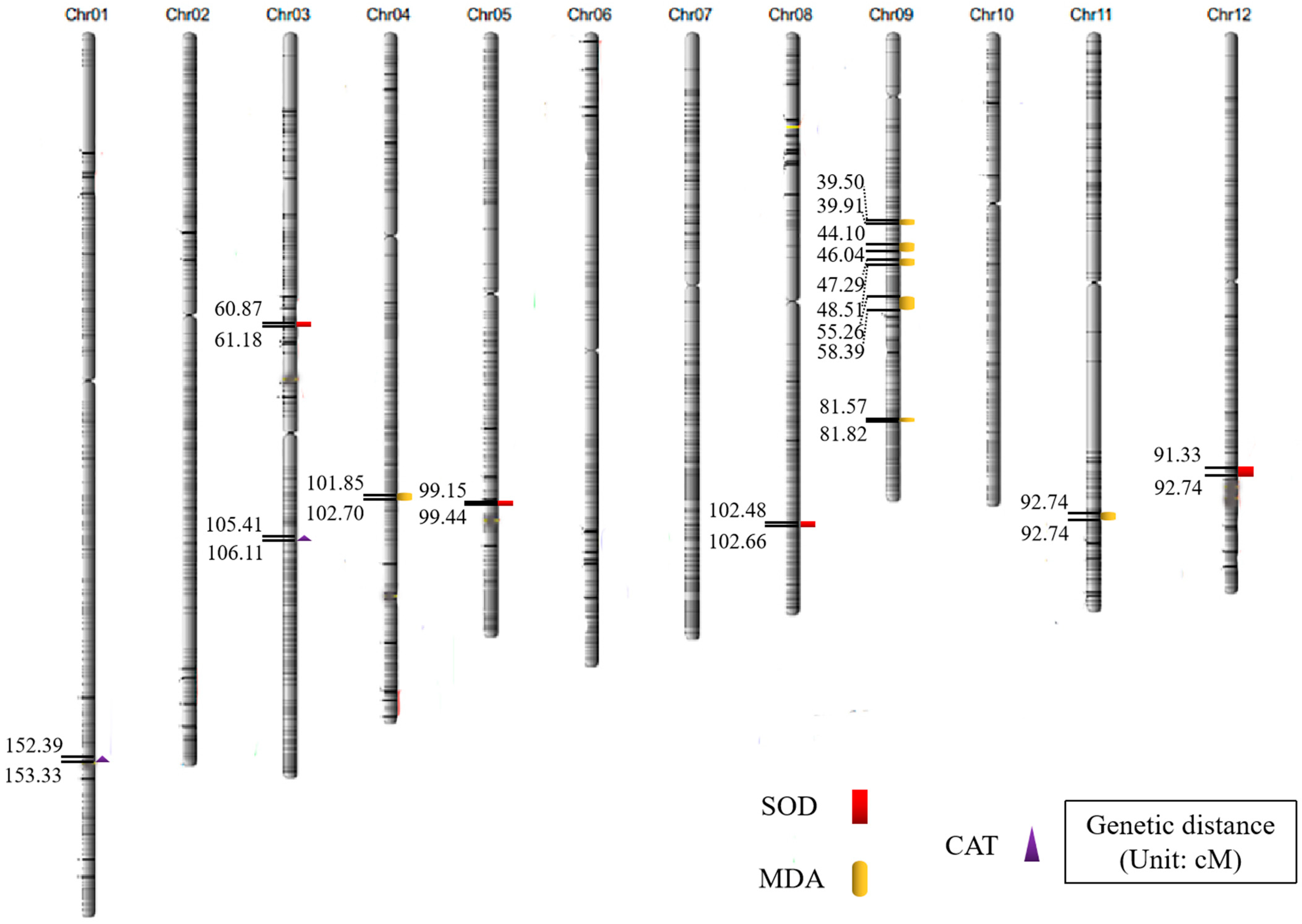
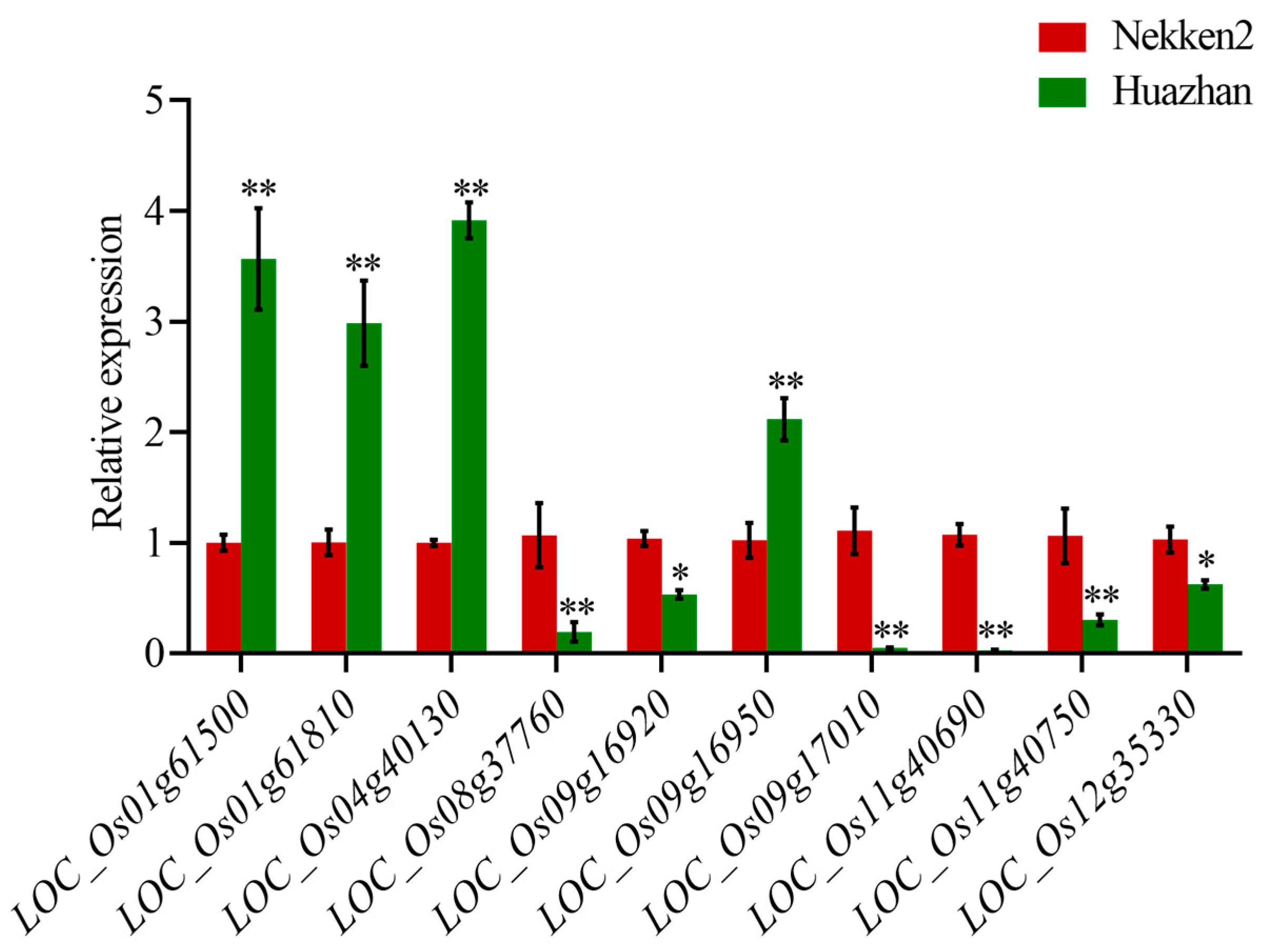
| Traits | QTL | Chromosome | Physical Distance (bp) | Position of Support (cM) | LOD |
|---|---|---|---|---|---|
| SOD activity | qSOD3 | 3 | 14,200,134~14,271,458 | 60.87~61.18 | 2.92 |
| qSOD5 | 5 | 23,130,097~23,196,156 | 99.15~99.44 | 2.50 | |
| qSOD8 | 8 | 23,905,607~23,948,650 | 102.48~102.66 | 3.14 | |
| qSOD12 | 12 | 21,305,692~21,634,437 | 91.33~92.74 | 2.55 | |
| MDA content | qMDA4 | 4 | 23,759,183~23,911,317 | 101.85~102.70 | 3.56 |
| qMDA9.1 | 9 | 9,232,289~9,309,844 | 39.50~39.91 | 2.59 | |
| qMDA9.2 | 9 | 10,288,160~10,740,128 | 44.10~46.04 | 3.58 | |
| qMDA9.3 | 9 | 11,031,216~11,316,376 | 47.29~48.51 | 2.86 | |
| qMDA9.4 | 9 | 12,891,121~13,620,974 | 55.26~58.39 | 2.77 | |
| qMDA9.5 | 9 | 19,029,258~19,086,513 | 81.57~81.82 | 2.62 | |
| qMDA11 | 11 | 24,097,660~24,422,854 | 103.30~104.69 | 3.33 | |
| CAT activity | qCAT1 | 1 | 35,548,884~35,768,322 | 152.39~153.33 | 5.70 |
| qCAT3 | 3 | 24,589,489~24,754,215 | 105.41~106.11 | 2.95 |
| Gene ID | SOD | Gene ID | MDA | Gene ID | CAT |
|---|---|---|---|---|---|
| Putative Function | Putative Function | Putative Function | |||
| LOC_Os03g24930 | tyrosine protein kinase domain containing protein | LOC_Os04g40130 | Rf1, mitochondrial precursor | LOC_Os01g61500 | BAG protein |
| LOC_Os05g39450 | glutaredoxin | LOC_Os09g16920 | cytochrome b5 | LOC_Os01g61810 | B subunit of nuclear factor Y |
| LOC_Os05g39500 | DUF640 domain containing protein | LOC_Os09g16950 | cysteine-rich receptor-like protein kinase 25 precursor | LOC_Os01g61780 | vacuolar ATP synthase 98 kDA subunit |
| LOC_Os05g39520 | methyltransferase | LOC_Os09g17010 | lectin-like receptor kinase 1 | LOC_Os01g61690 | OsSCP5-putative serine carboxypeptidase homologue |
| LOC_Os08g37760 | zinc finger, C3HC4 type domain containing protein | LOC_Os11g40690 | dehydrogenase | LOC_Os03g43860 | endosomal sorting complex required for transport; ESCRT-III component |
| LOC_Os12g35330 | glutaredoxin | LOC_Os11g40750 | citrate synthase | LOC_Os03g44000 | LTPL15-Protease inhibitor/seed storage/LTP family protein precursor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, W.; Huang, Z.; Zhong, Q.; Tang, L.; Wu, R.; Li, S.; Mao, Y.; Zhu, X.; Wang, C.; Rao, Y.; et al. The Mining of Genetic Loci and the Analysis of Candidate Genes to Identify the Physical and Chemical Markers of Anti-Senescence in Rice. Plants 2023, 12, 3812. https://doi.org/10.3390/plants12223812
Yin W, Huang Z, Zhong Q, Tang L, Wu R, Li S, Mao Y, Zhu X, Wang C, Rao Y, et al. The Mining of Genetic Loci and the Analysis of Candidate Genes to Identify the Physical and Chemical Markers of Anti-Senescence in Rice. Plants. 2023; 12(22):3812. https://doi.org/10.3390/plants12223812
Chicago/Turabian StyleYin, Wenjing, Zhao Huang, Qianqian Zhong, Luyao Tang, Richeng Wu, Sanfeng Li, Yijian Mao, Xudong Zhu, Changchun Wang, Yuchun Rao, and et al. 2023. "The Mining of Genetic Loci and the Analysis of Candidate Genes to Identify the Physical and Chemical Markers of Anti-Senescence in Rice" Plants 12, no. 22: 3812. https://doi.org/10.3390/plants12223812
APA StyleYin, W., Huang, Z., Zhong, Q., Tang, L., Wu, R., Li, S., Mao, Y., Zhu, X., Wang, C., Rao, Y., & Wang, Y. (2023). The Mining of Genetic Loci and the Analysis of Candidate Genes to Identify the Physical and Chemical Markers of Anti-Senescence in Rice. Plants, 12(22), 3812. https://doi.org/10.3390/plants12223812






