Hydrangea arborescens ‘Annabelle’ Flower Formation and Flowering in the Current Year
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Observation in the Current-year Flower Development of H. arborescens ‘Annabelle’
2.2. Quality Analysis of Transcriptome Sequencing
2.3. Comparative Transcriptomic Analysis of Seven Flower Developmental Stages
2.4. Identification of Candidate DEGs Involved in Current-Year Flower Development
2.5. Construction of Gene Co-Expression Network (GCN) and Detection of Functional Modules
2.6. Analysis of Modules Highly Correlated with the Early Flower Developmental Phase
2.7. Analysis of Modules Highly Correlated with the Late Flower Developmental Phase
2.8. Real-time Quantitative PCR (qRT-PCR) Validation of RNA-seq Data of Flower Development-Related Genes
2.9. Changes in Phytohormone Content during Current-Year Flower Development
3. Discussion
3.1. Key Genes and Important Transcription Factors (TFs) Involved in Current-Year Flower Development
3.2. GA-, Aging-, and Photoperiod-Related Flowering Pathways Contribute to the Current-Year Flower Development of H. arborescens ‘Annabelle’
3.3. The Putative Roles of Phytohormone Crosswalk in the Current-Year Flower Development of H. arborescens ‘Annabelle’
4. Materials and Methods
4.1. Plant Material and Phenological Observation
4.2. Quantification of Phytohormones during Flower Development
4.3. RNA Extraction and RNA-seq Library Construction
4.4. Functional Annotation of Unigenes and Screening for Differentially Expressed Genes
4.5. Construction of the Genes Co-Expression Network (GCN) and Cluster Recognition
4.6. Module Identification, Functional Enrichment, Key Genes and transcription factors Analysis
4.7. Validation of RNA-seq Data via Quantitative Real-Time PCR (qRT-PCR)
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ke, Y.G.; Zhou, Y.W.; Lv, Y.Y.; Qi, Y.; Wei, H.Y.; Yu, L.; Huang, F.Y.; Abbas, F. Integrated Metabolome and Transcriptome Analysis Provides Insights on the Floral Scent Formation in Hydrangea arborescens. Physiol. Plant. 2023, 175, 114–139. [Google Scholar] [CrossRef]
- Cai, M.; Wang, K.; Luo, L.; Pan, H.T.; Zhang, Q.X.; Yang, Y.Y. Production of Interspecific Hybrids between Hydrangea Macrophylla and Hydrangea arborescens via Ovary Culture. HortScience 2015, 50, 1765–1769. [Google Scholar] [CrossRef]
- Dirr, M.A. Manual of Woody Landscape Plants: Their Identification, Ornamental Characteristics, Culture, Propagation and Uses; Stipes: Champaign, IL, USA, 2009; pp. 330–332. [Google Scholar]
- Adkins, J.A.; Dirr, M.A. Remontant Flowering Potential of Ten Hydrangea macrophylla (Thunb.) Ser. Cultivars. HortScience 2003, 38, 1337–1340. [Google Scholar] [CrossRef]
- Yu, S.L.; Hao, Z.D.; Ye, P.; Liu, S.Q.; Hu, L.F.; Shen, Y.B.; Shi, J.S.; Chen, J.H. Morphological, Phenological, and Transcriptional Analyses Provide Insight into the Diverse Flowering Traits of a Mutant of the Relic Woody Plant Liriodendron chinense. Hortic. Res. 2021, 8, 174–189. [Google Scholar]
- Yu, Y.F.; Shan, C.L.; Li, C.H. Hydrangea arborescens “Annabelle” Introduction and Cultivation Techniques. Terr. Natrl. Resour. Study 2016, 163, 86–87. [Google Scholar]
- Ma, N.; Ma, C.; Liu, Y.; Shahid, M.O.; Wang, C.P.; Gao, J.P. Petal Senescence: A Hormone View. J. Exp. Bot. 2018, 69, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Zhang, D.; An, N.; Fan, S.; Zuo, X.Y.; Zhang, X.; Zhang, L.Z.; Gao, C.; Han, M.Y.; Xing, L.B. Transcriptomic Analysis Reveals the Regulatory Module of Apple (Malus × Domestica) Floral Transition in Response to 6-BA. BMC. Plant Biol. 2019, 19, 93–110. [Google Scholar] [CrossRef]
- Fornara, F.; Montaigu, A.; Coupland, G. SnapShot: Control of Flowering in Arabidopsis. Cell 2010, 141, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hao, X.Y.; Lu, Q.H.; Zhang, W.F.; Zhang, H.J.; Wang, L.; Yang, Y.J.; Xiao, B.; Wang, X.C. Genome-Wide Identification and Expression Analysis of Flowering-Related Genes Reveal Putative Floral Induction and Differentiation Mechanisms in Tea Plant (Camellia sinensis). Genomics 2020, 112, 2318–2326. [Google Scholar] [CrossRef]
- Shan, H.Y.; Cheng, J.; Zhang, R.; Yao, X.; Kong, H.Z. Developmental Mechanisms Involved in the Diversification of Flowers. Nat. Plants 2019, 5, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.F.; Li, X.M.; Liao, W.J. Advances in the Genetic Regulating Pathways of Plant Flowering Time. Biodivers. Sci. 2021, 29, 825–842. [Google Scholar] [CrossRef]
- Andrés, F.; Coupland, G. The Genetic Basis of Flowering Responses to Seasonal Cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, T.T.; Khan, W.; An, X.M. Regulatory Roles of MiRNAs Associated with Aging Pathway in Tree Vegetative Phase Change. J. For. Res. 2023, 3, 9–15. [Google Scholar] [CrossRef]
- Boss, P.K. Multiple Pathways in the Decision to Flower: Enabling, Promoting, and Resetting. Plant Cell 2004, 16, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Lee, H.; Kim, M.; Lee, I. Analysis of Flowering Pathway Integrators in Arabidopsis. Plant. Cell. Physiol. 2005, 46, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Torti, S.; Coupland, G. Genetic and Spatial Interactions Between FT, TSF and SVP during the Early Stages of Floral Induction in Arabidopsis. Plant. J. 2009, 60, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W. Regulation of Flowering Time by the MiR156-Mediated Age Pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, J.D.; Sedgley, M.; Olesen, T. Regulation of Floral Initiation in Horticultural Trees. J. Exp. Bot. 2008, 59, 3215–3228. [Google Scholar] [CrossRef]
- Zhang, S.X.; Dai, J.H.; Ge, Q.S. Progress of Responses of First Flowering Date to Climate Change and the Correlations of Plant Hormone Regulation. Prog. Geogr. 2019, 38, 1045–1055. [Google Scholar]
- Lin, G.Y.; Zheng, C.S.; Sun, X.Z.; Wang, W.L. Effects of Photoperiod on Floral Bud Differentiation and Contents of Endogenous Hormones in Chrysanthemum. Shandong. Agric. Sci. 2008, 1, 35–39. [Google Scholar]
- Fan, L.C.; Gao, Y.; Jin, C.L.; Wen, X.H.; Geng, H.T.; Cheng, Y.; Qu, H.Y.; Liu, X.; Feng, S.; Zhang, F.; et al. The Chromosome-Level Genome of Gypsophila paniculata Reveals the Molecular Mechanism of Floral Development and Ethylene Insensitivity. Hort. Res. 2022, 9, 176–189. [Google Scholar]
- Domagalska, M.A.; Sarnowska, E.; Nagy, F.; Davis, S.J. Genetic Analyses of Interactions among Gibberellin, Abscisic Acid, and Brassinosteroids in the Control of Flowering Time in Arabidopsis thaliana. PLoS ONE 2010, 5, e14012. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.D.; Feng, S.X.; Pan, Y.L.; Zhong, J.D.; Chen, Y.; Yuan, C.C.; Li, H.L. Transcriptome Analysis and Identification of Genes Associated with Floral Transition and Flower Development in Sugar Apple (Annona squamosa L.). Front. Plant Sci. 2016, 7, 1695–1711. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.X.; Li, Q.Z.; Yang, L.Y.; Li, X.; Zhen, W.; Zhang, Y.C. Changes in Carbohydrate Metabolism and Endogenous Hormone Regulation during Bulblet Initiation and Development in Lycoris radiata. BMC Plant Biol. 2020, 20, 180. [Google Scholar] [CrossRef]
- Liu, Q.B.; Feng, Z.; Huang, C.J.; Wen, J.; Li, L.B.; Yu, S. Insights into the Genomic Regions and Candidate Genes of Senescence-Related Traits in Upland Cotton via GWAS. Int. J. Mol. Sci. 2022, 23, 8584. [Google Scholar] [CrossRef] [PubMed]
- Salleh, F.M.; Mariotti, L.; Spadafora, N.D.; Price, A.; Picciarelli, P.; Wagstaff, C.; Lombardi, L.; Rogers, H.J. Interaction of Plant Growth Regulators and Reactive Oxygen Species to Regulate Petal Senescence in Wallflowers (Erysimum linifolium). BMC Plant Biol. 2016, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.Z.; Zhang, X.; Wu, F.M.; Feng, H.L.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.M.; Li, C.Y. Transcriptional Mechanism of Jasmonate Receptor COI1-Mediated Delay of Flowering Time in Arabidopsis. Plant Cell 2015, 27, 2814–2828. [Google Scholar] [CrossRef]
- Wada, K.C.; Yamada, M.; Shiraya, T.; Takeno, K. Salicylic Acid and the Flowering Gene FLOWERING LOCUS T Homolog Are Involved in Poor-Nutrition Stress-Induced Flowering of Pharbitis nil. J. Plant Physiol. 2010, 167, 447–452. [Google Scholar] [CrossRef]
- Li, J.H. Studies on Influence of Endogenous Brassinosteroid on the Flowering-Time and the Mechanism in Arabidopsis. Ph.D. Thesis, Lanzhou University, Lanzhou, China, 2011. [Google Scholar]
- Galopin, G.; Codarin, S.; Viemont, J.D.; Morel, P. Architectural Development of Inflorescence in Hydrangea macrophylla Cv. Hermann Dienemann. HortScience 2008, 43, 361–365. [Google Scholar] [CrossRef]
- Jing, D.L.; Chen, W.W.; Hu, R.Q.; Zhang, Y.C.; Xia, Y.; Wang, S.M.; He, Q.; Guo, Q.G.; Liang, G.L. An Integrative Analysis of Transcriptome, Proteome and Hormones Reveals Key Differentially Expressed Genes and Metabolic Pathways Involved in Flower Development in Loquat. Int. J. Mol. Sci. 2020, 21, 5107. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.H.; Muino, J.M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.D.; Liu, S.; Westphal, A.H.; Boeren, S.; et al. Characterization of MADS-Domain Transcription Factor Complexes in Arabidopsis Flower Development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Zhao, Z.Q.; Wang, J.S.; Yu, H.F.; Shen, Y.S.; Zeng, X.Y.; Gu, H.H. Genome Wide Analysis of MADS-Box Gene Family in Brassica oleracea Reveals Conservation and Variation in Flower Development. BMC Plant Biol. 2019, 19, 106. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Xian, D.Y.; Song, M.; Tang, Q.L. Research Progress of MIKC-Type MADS-Box Protein Regulation on Flowering. Biotechnol. Bull. 2014, 0, 8–15. [Google Scholar]
- Alter, P.; Bircheneder, S.; Zhou, L.Z.; Schlüter, U.; Gahrtz, M.; Sonnewald, U.; Dresselhaus, T. Flowering Time-Regulated Genes in Maize Include the Transcription Factor ZmMADS1. Plant Physiol. 2016, 172, 389–404. [Google Scholar] [CrossRef]
- Immink, R.G.H.; Posé, D.; Ferrario, S.; Ott, F.; Kaufmann, K.; Valentim, F.L.; Folter, S.; Wal, F.; Dijk, A.D.J.; Schmid, M.; et al. Characterization of SOC1’S Central Role in Flowering by the Identification of its Upstream and Downstream Regulators. Plant Physiol. 2012, 160, 433–449. [Google Scholar] [CrossRef]
- Li, C.C.; Gu, H.Y.; Jiang, W.; Zou, C.H.; Wei, D.Y.; Wang, Z.M.; Tang, Q.L. Protein Interactions of SOC1 with SVP Are Regulated by a Few Crucial Amino Acids in Flowering Pathways of Brassica juncea. Acta Physiol. Plant. 2019, 41, 43–50. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Wu, M.F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The MicroRNA-Regulated SBP-Box Transcription Factor SPL3 Is a Direct Upstream Activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Wellmer, F.; Muiño, J.M.; Ferrier, T.; Wuest, S.E.; Kumar, V.; Serrano-Mislata, A.; Madueño, F.; Krajewski, P.; Meyerowitz, E.M.; et al. Orchestration of Floral Initiation by APETALA1. Science 2010, 328, 85–89. [Google Scholar] [CrossRef]
- Hong, Y.G.; Jackson, S. Floral Induction and Flower Formation-the Role and Potential Applications of MiRNAs. Plant Biotechnol. J. 2015, 13, 282–292. [Google Scholar] [CrossRef]
- Xu, M.L.; Hu, T.Q.; Zhao, J.F.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of MiR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-like (SPL) Genes in Arabidopsis Thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chen, X.H.; Yu, H.; Tian, Q.F.; Lu, L.M. Identification and Characterization of CONSTANS-like Genes from Curcuma alismatifolia. Hortic. Environ. Biotechnol. 2021, 62, 279–286. [Google Scholar] [CrossRef]
- Zhang, J.X.; Zhao, X.L.; Tian, R.X.; Zeng, S.J.; Wu, K.L.; Teixeira, J.A.; Duan, J. Molecular Cloning and Functional Analysis of Three CONSTANS-like Genes from Chinese Cymbidium. J. Plant Growth Regul. 2019, 39, 1061–1074. [Google Scholar] [CrossRef]
- Wu, W.X.; Zheng, X.M.; Chen, D.B.; Zhang, Y.X.; Ma, W.W.; Zhang, H.; Sun, L.P.; Yang, Z.F.; Zhao, C.D.; Zhan, X.D.; et al. OsCOL16, Encoding a CONSTANS-like Protein, Represses Flowering by Up-Regulating Ghd7 Expression in Rice. Plant Sci. 2017, 260, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W. Regulation of Arabidopsis tapetum Development and Function by DYSFUNCTIONAL TAPETUM1 (DYT1) Encoding a Putative BHLH Transcription Factor. Development 2006, 133, 3085–3095. [Google Scholar] [CrossRef]
- Niu, Y.J.; Figueroa, P.; Browse, J. Characterization of JAZ-Interacting BHLH Transcription Factors That Regulate Jasmonate Responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef] [PubMed]
- Ori, N. Dissecting the Biological Functions of ARF and Aux/IAA Genes. Plant Cell 2019, 31, 1210–1211. [Google Scholar] [CrossRef]
- Bao, S.J.; Hua, C.M.; Shen, L.S.; Yu, H. New Insights into Gibberellin Signaling in Regulating Flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef]
- Sang, Q.; Vayssières, A.; Ó’Maoiléidigh, D.S.; Yang, X.L.; Vincent, C.; Bertran, E.; Cerise, M.; Franzen, R.; Coupland, G. MicroRNA172 Controls Inflorescence Meristem Size through Regulation of APETALA2 in Arabidopsis. New Phytol. 2022, 235, 356–371. [Google Scholar] [CrossRef]
- Xu, S.D.; Geng, X.M.; Wang, L.L. A Review of the Structure, Function and Expression Regulation of Ethylene Response Factors (ERF) in Plant. J. Zhejiang AF Univ. 2021, 38, 624–633. [Google Scholar]
- Park, M.J.; Kwon, Y.J.; Gil, K.E.; Park, C.M. Late elongated hypocotyl Regulates Photoperiodic Flowering via the Circadian Clock in Arabidopsis. BMC Plant Biol. 2016, 16, 114. [Google Scholar] [CrossRef]
- Shen, Y.P.; Feng, S.H.; Ma, L.G.; Lin, R.C.; Qu, L.J.; Chen, Z.L.; Wang, H.Y.; Deng, X.W. Arabidopsis FHY1 Protein Stability Is Regulated by Light via Phytochrome a and 26S Proteasome. Plant Physiol. 2005, 139, 1234–1243. [Google Scholar] [CrossRef]
- Tian, S.B.; Guo, C.X.; Zheng, C.S. Molecular Mechanism of Controlling Flower Formation by Photoperiod Inducement in Plants. Yuan Yi Xue Bao 2010, 37, 325–330. [Google Scholar]
- Inoue, K.; Araki, T.; Endo, M. Correction To: Circadian Clock during Plant Development. J. Plant Res. 2018, 131, 59–66. [Google Scholar] [CrossRef]
- Ponnu, J.; Hoecker, U. Illuminating the COP1/SPA Ubiquitin Ligase: Fresh Insights into Its Structure and Functions during Plant Photomorphogenesis. Front. Plant Sci. 2021, 12, 662793. [Google Scholar] [CrossRef]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Figueroa, J.D.; Begoña, R.M.; Pollmann, S.; Granell, A.; Molina, R.V.; Jesús, V.C.; et al. Multifaceted Role of Cycling DOF Factor 3 (CDF3) in the Regulation of Flowering Time and Abiotic Stress Responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef]
- Fu, J.X.; Yang, L.W.; Dai, S.L. Identification and Characterization of the CONSTANS-like Gene Family in the Short-Day Plant Chrysanthemum lavandulifolium. Mol. Genet. Genom. 2014, 290, 1039–1054. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xiao, W.F.; Chen, H.M.; Zhu, G.F.; Lv, F.B. Transcriptome Analysis Reveals Endogenous Hormone Changes during Spike Development in Phalaenopsis. Int. J. Mol. Sci. 2022, 23, 10461. [Google Scholar] [CrossRef]
- Wei, Q.; Ma, C.; Xu, Y.J.; Wang, T.L.; Chen, Y.Y.; Lü, J.; Zhang, L.L.; Jiang, C.Z.; Hong, B.; Gao, J.P. Control of Chrysanthemum Flowering through Integration with an Aging Pathway. Nat. Commun. 2017, 8, 829–839. [Google Scholar] [CrossRef]
- Fukazawa, J.; Ohashi, Y.; Takahashi, R.; Nakai, K.; Takahashi, Y. DELLA Degradation by Gibberellin Promotes Flowering via GAF1-TPR-Dependent Repression of Floral Repressors in Arabidopsis. Plant Cell 2021, 33, 2258–2272. [Google Scholar] [CrossRef]
- Li, M.Z.; An, F.Y.; Li, W.Y.; Ma, M.D.; Feng, Y.; Zhang, X.; Guo, H.W. DELLA Proteins Interact with FLC to Repress Flowering Transition. J. Integr. Plant Biol. 2016, 58, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration. Plant Physiol. 2016, 173, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.X.; Huang, W.D. Immunohistochemical Localization of IAA and ABP1 in Strawberry Shoot Apexes during Floral Induction. Planta 2005, 222, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Willige, B.C.; Isono, E.; Richter, R.; Zourelidou, M.; Schwechheimer, C. Gibberellin Regulates PIN-FORMED Abundance and Is Required for Auxin Transport–Dependent Growth and Development in Arabidopsis thaliana. Plant Cell 2011, 23, 2184–2195. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.D.; Harberd, N.P. Auxin Promotes Arabidopsis Root Growth by Modulating Gibberellin Response. Nature 2003, 421, 740–743. [Google Scholar] [CrossRef]
- Zou, J.J.; Cai, X.; Yang, J.; Zeng, X.L.; Liu, D.X.; Huang, S.M.; Chen, X.; Yang, Q.Y.; Wang, C.Y.; Chen, H.G. DNA Hypomethylation Mediates Flower Opening and Senescence in Sweet Osmanthus through Auxin and Ethylene Responsive Pathways. Postharvest. Biol. Technol. 2023, 198, 112250. [Google Scholar] [CrossRef]
- Martignago, D.; Siemiatkowska, B.; Lombardi, A.; Conti, L. Abscisic Acid and Flowering Regulation: Many Targets, Different Places. Int. J. Mol. Sci. 2020, 21, 9700. [Google Scholar] [CrossRef]
- Li, Z.C.; He, Y. hui Roles of Brassinosteroids in Plant Reproduction. Int. J. Mol. Sci. 2020, 21, 872. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Liu, L.Y.; Song, S.S. Jasmonate Action in Plant Growth and Development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef]
- Cheng, H.; Zha, S.X.; Luo, Y.Y.; Li, L.; Wang, S.Y.; Wu, S.; Cheng, S.Y.; Li, L.L. JAZ1-3 and MYC2-1 Synergistically Regulate the Transformation from Completely Mixed Flower Buds to Female Flower Buds in Castanea mollisima. Int. J. Mol. Sci. 2022, 23, 6452. [Google Scholar] [CrossRef]
- Wada, K.C.; Mizuuchi, K.; Koshio, A.; Kaneko, K.; Mitsui, T.; Takeno, K. Stress Enhances the Gene Expression and Enzyme Activity of Phenylalanine Ammonia-Lyase and the Endogenous Content of Salicylic Acid to Induce Flowering in Pharbitis. J. Plant Physiol. 2014, 171, 895–902. [Google Scholar] [CrossRef]
- Nisar, S.; Ahmad Dar, R.; Tahir, I. Salicylic Acid Retards Senescence and Makes Flowers Last Longer in Nicotiana plumbaginifolia (Viv). Plant Physiol. Rep. 2021, 26, 128–136. [Google Scholar] [CrossRef]
- JACQMARD, A. Cell Division and Morphological Changes in the Shoot Apex of Arabidopsis thaliana during Floral Transition. Ann. Bot. 2003, 91, 571–576. [Google Scholar] [CrossRef]
- Zou, L.P.; Pan, C.; Wang, M.X.; Chen, L.; Han, B.Y. Progress on the Mechanism of Hormones Regulating Plant Flower Formation. Hereditas 2020, 42, 739–751. [Google Scholar] [PubMed]
- Wu, L.; Ma, N.; Jia, Y.C.; Zhang, Y.; Feng, M.; Jiang, C.Z.; Ma, C.; Gao, J.P. An Ethylene-Induced Regulatory Module Delays Flower Senescence by Regulating Cytokinin Content. Plant Physiol. 2016, 173, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Chen, S.; An, L. Involvement of Brassinosteroid Signals in the Floral-Induction Network of Arabidopsis. J. Exp. Bot. 2010, 61, 4221–4230. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.Y.; Gao, Z.; Chen, J.Q.; Qiu, Y.T.; Zhang, L.S.; Chen, X. Auxin Controls Circadian Flower Opening and Closure in the Waterlily. BMC Plant Biol. 2018, 18, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Arana, M.V.; Marin-de la Rosa, N.; Maloof, J.N.; Blazquez, M.A.; Alabadi, D. Circadian Oscillation of Gibberellin Signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 9292–9297. [Google Scholar] [CrossRef] [PubMed]
- He, Z. (Ed.) Guidance to Experiment on Chemical Control in Crop Plants. In Guidance to Experiment on Chemical Control in Crop Plants; Beijing Agricultural University Publishers: Beijing, China, 1993; pp. 60–68. [Google Scholar]
- Yang, J.C.; Zhang, J.H.; Wang, Z.Q.; Zhu, Q.S.; Wang, W. Hormonal Changes in the Grains of Rice Subjected to Water Stress during Grain Filling. Plant Physiol. 2001, 127, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Weiler, E.W.; Jourdan, P.; Conrad, W.E. Levels of Indole-3-Acetic Acid in Intact and Decapitated Coleoptiles as Determined by a Specific and Highly Sensitive Solid-Phase Enzyme Immunoassay. Planta 1981, 153, 561–571. [Google Scholar] [CrossRef]
- Wang, J.M.; Wang, Q.; Yang, Y.; Liu, X.H.; Gu, J.H.; Li, W.Q.; Ma, S.L.Y.; Lu, Y.Z. De Novo Assembly and Characterization of Stress Transcriptome and Regulatory Networks under Temperature, Salt and Hormone Stresses in Lilium lancifolium. Mol. Biol. Rep. 2014, 41, 8231–8245. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome. Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.Y.; Li, W.; Wang, J.; Zhang, Y.; Lu, Y. Identification of Gene Co-Expression Networks Involved in Cold Resistance of Lilium lancifolium. Biol. Plantarum. 2018, 62, 287–298. [Google Scholar] [CrossRef]
- Wong, D.C.; Sweetman, C.; Ford, C.M. Annotation of Gene Function in Citrus Using Gene Expression Information and Co-Expression Networks. BMC Plant Biol. 2014, 14, 186. [Google Scholar] [CrossRef] [PubMed]
- Kuruş, M.; Akbari, S.; Eskier, D.; Bursalı, A.; Ergin, K.; Erdal, E.; Karakülah, G. Transcriptome Dynamics of Human Neuronal Differentiation from IPSC. Front. Cell Dev. Biol. 2021, 9, 727–747. [Google Scholar] [CrossRef]
- Dai, Y.; Sun, X.; Wang, C.G.; Li, F.; Zhang, S.F.; Zhang, H.; Li, G.L.; Yuan, L.Y.; Chen, G.H.; Sun, R.F.; et al. Gene Co-Expression Network Analysis Reveals Key Pathways and Hub Genes in Chinese Cabbage (Brassica Rapa L.) during Vernalization. BMC Genom. 2021, 22, 236. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
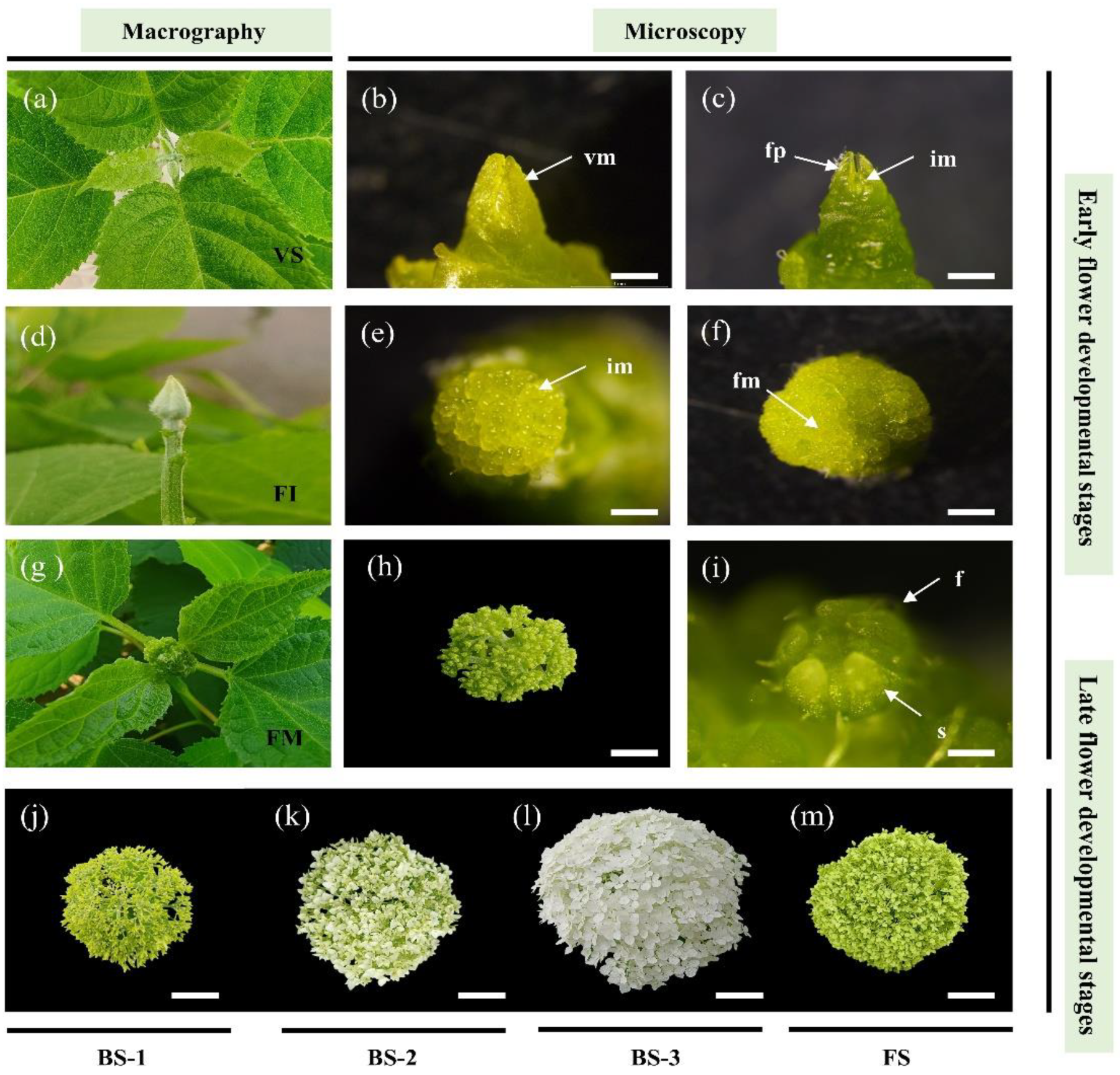
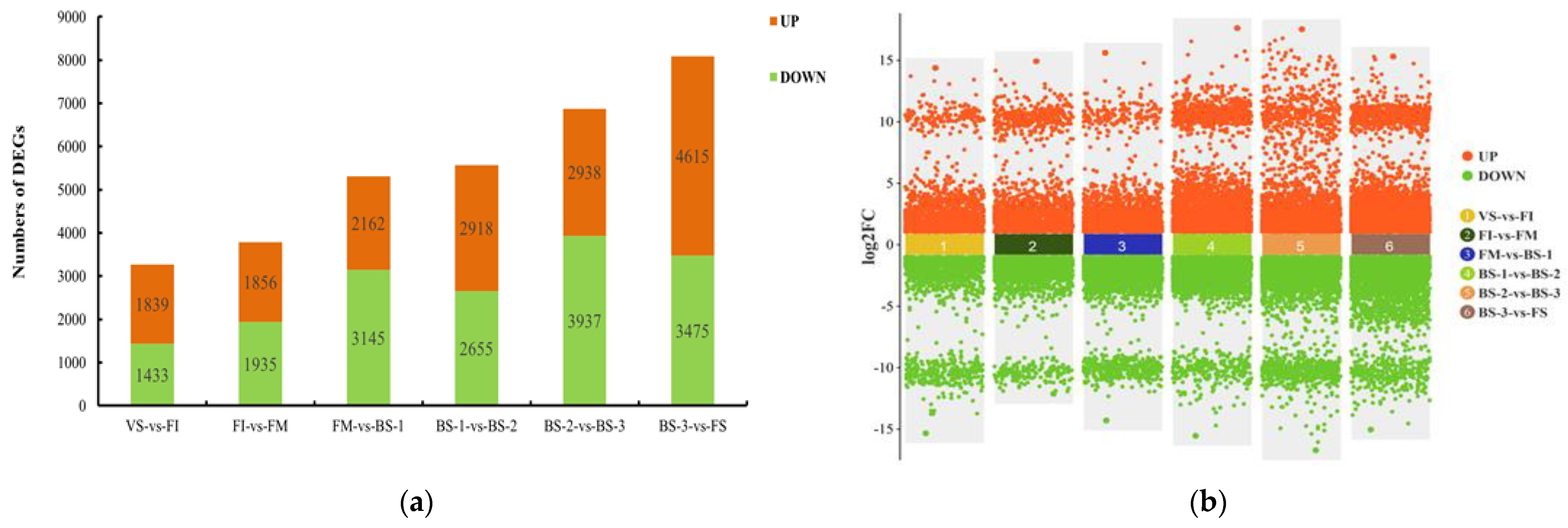

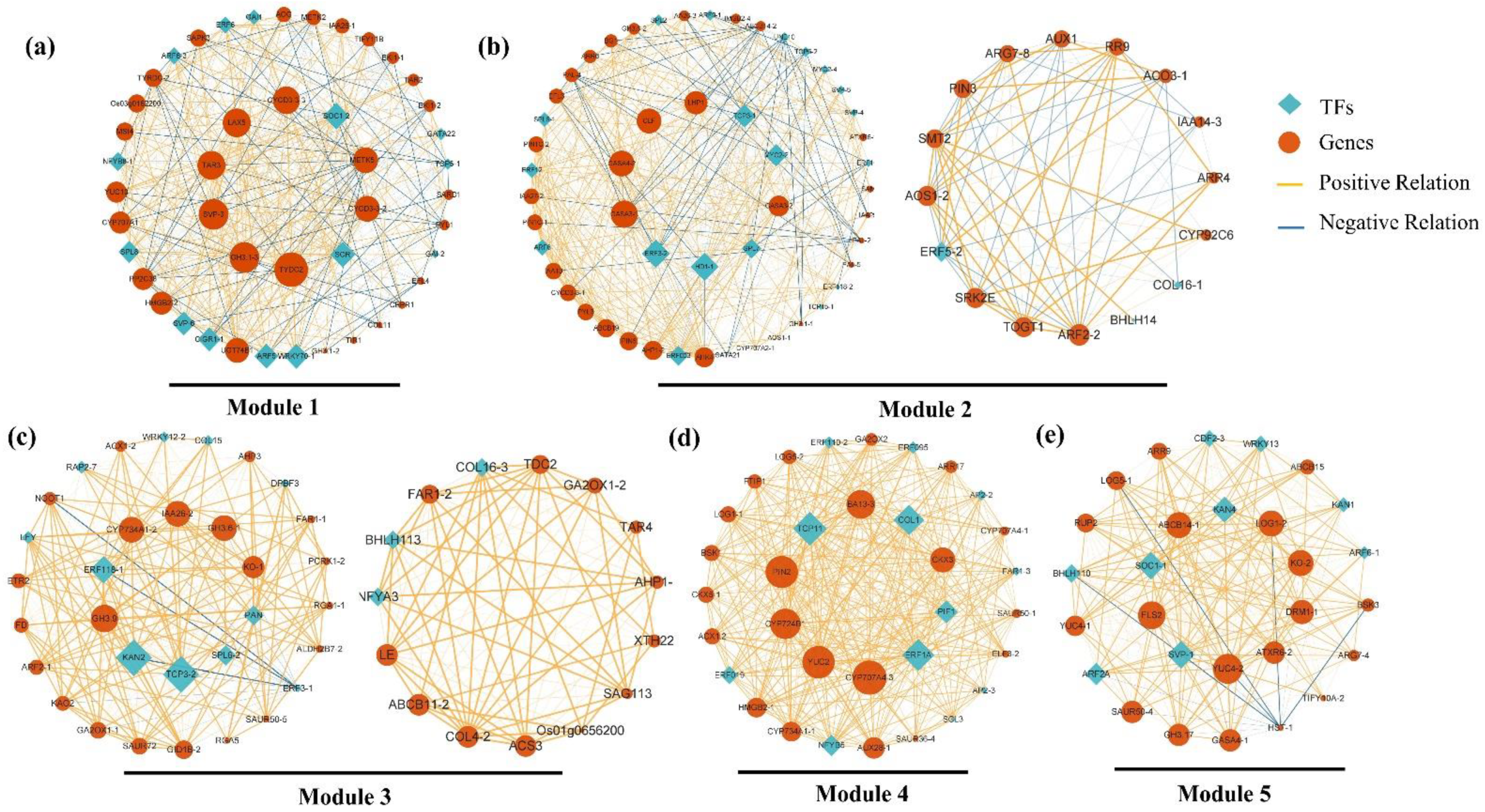
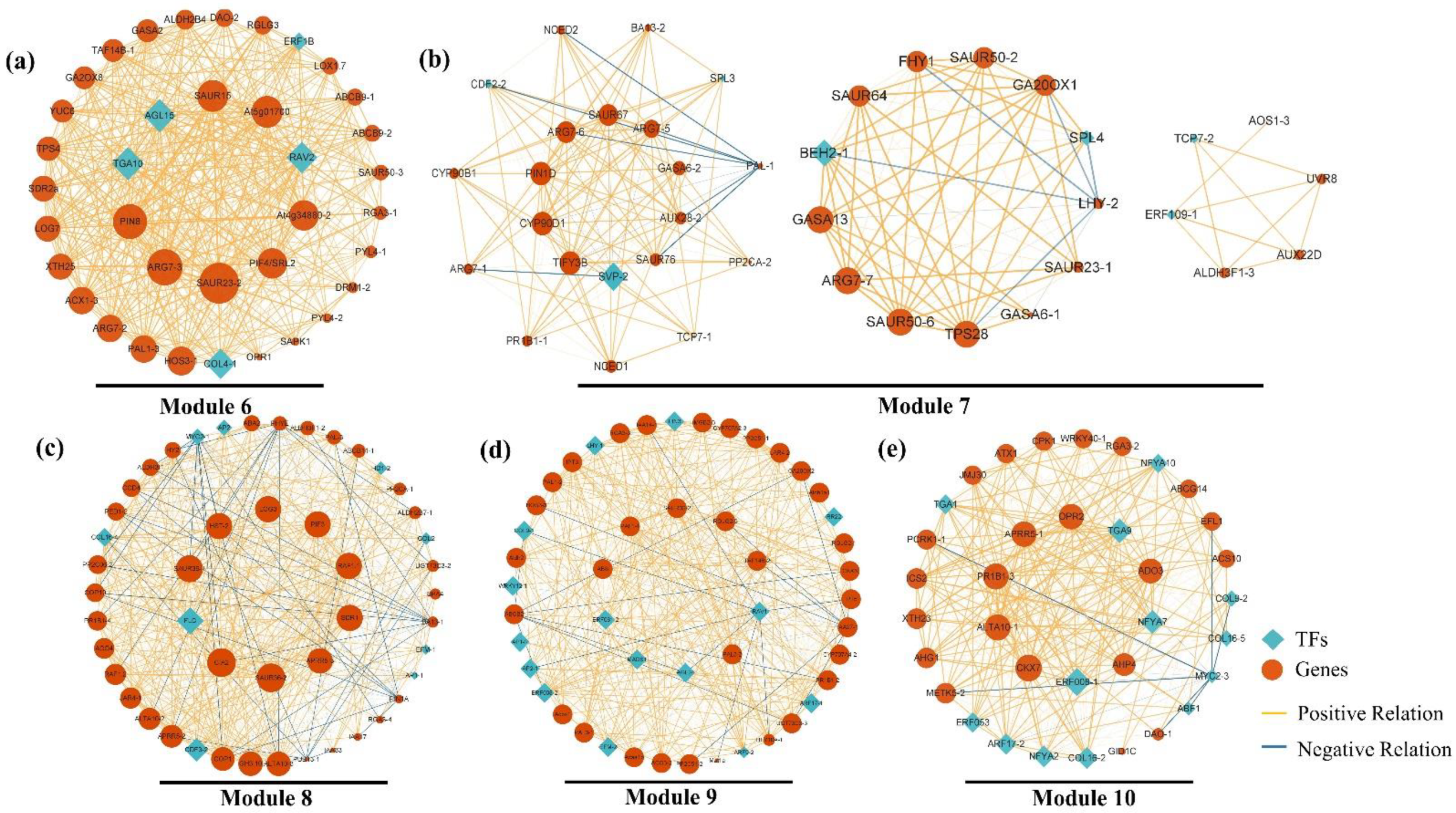
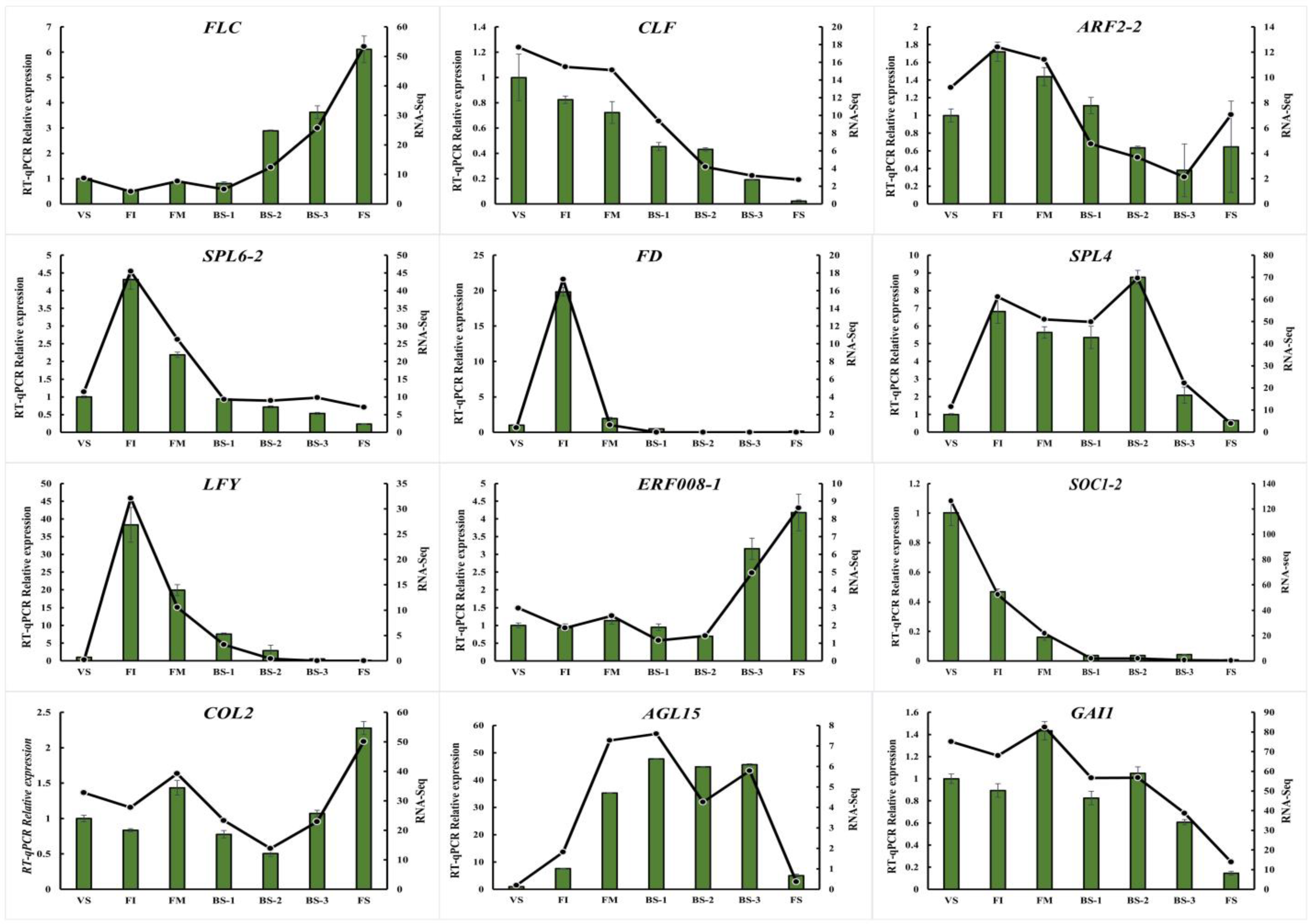

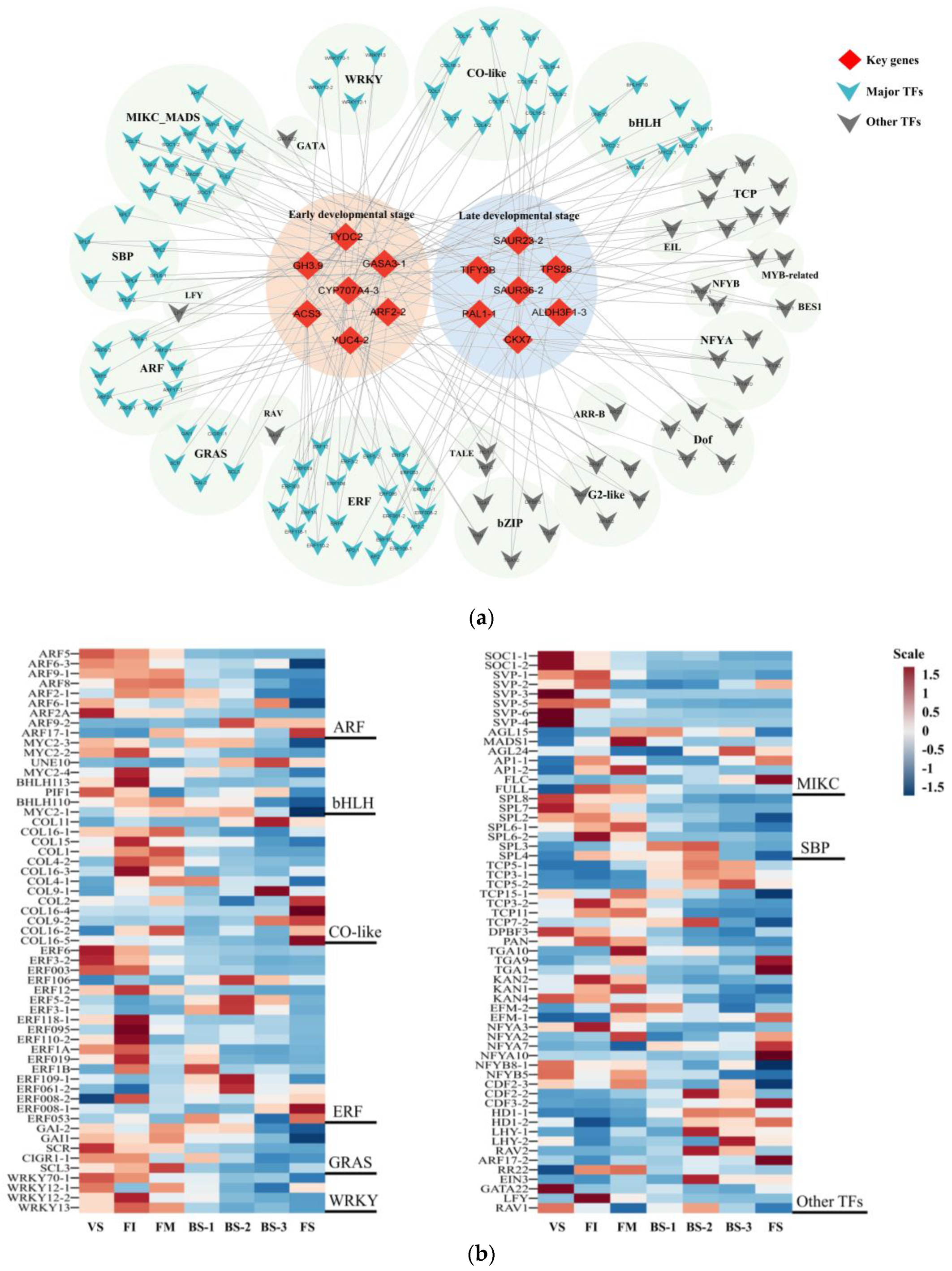

| Stage | Description | Figure |
|---|---|---|
| Vegetative Stage (VS) | Vegetative shoot apical meristem. | Figure 1a |
| Floral Initiation (FI) | Flower bud expansion: many flower primordia clearly distinguishable and forming a spherical shape. | Figure 1d |
| Floral Morphogenesis (FM) | Secondary branching elongation of spherical inflorescences and floral organ differentiation. | Figure 1g |
| Blooming Stage 1 (BS-1) | Early blooming stage: fifth branching of inflorescences formed; decorative floret displayed tender green. | Figure 1j |
| Blooming Stage 2 (BS-2) | Middle blooming stage: sepals of decorative florets were not fully open; the color of sepals gradually changed from green to white. | Figure 1k |
| Blooming Stage 3 (BS-3) | Full blooming stage: more than 80% of decorative florets were snowy-colored in inflorescence. | Figure 1l |
| Flower Senescence (FS) | The senescent inflorescence displayed a deep green. | Figure 1m |
| Simple Parameters | |||
|---|---|---|---|
| Number of nodes | 428 | Clustering coefficient | 0.739 |
| Number of edges | 4838 | Network density | 0.536 |
| Avg. number of neighbors | 26.784 | Network heterogeneity | 0.318 |
| Network of diameter | 3 | Network centralization | 0.358 |
| Characteristic path length | 1.468 | Connected components | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Lyu, T.; Li, Z.; Lyu, Y. Hydrangea arborescens ‘Annabelle’ Flower Formation and Flowering in the Current Year. Plants 2023, 12, 4103. https://doi.org/10.3390/plants12244103
Huang X, Lyu T, Li Z, Lyu Y. Hydrangea arborescens ‘Annabelle’ Flower Formation and Flowering in the Current Year. Plants. 2023; 12(24):4103. https://doi.org/10.3390/plants12244103
Chicago/Turabian StyleHuang, Xiaoxu, Tong Lyu, Zheng Li, and Yingmin Lyu. 2023. "Hydrangea arborescens ‘Annabelle’ Flower Formation and Flowering in the Current Year" Plants 12, no. 24: 4103. https://doi.org/10.3390/plants12244103
APA StyleHuang, X., Lyu, T., Li, Z., & Lyu, Y. (2023). Hydrangea arborescens ‘Annabelle’ Flower Formation and Flowering in the Current Year. Plants, 12(24), 4103. https://doi.org/10.3390/plants12244103





