Phylotranscriptomics Shed Light on Intrageneric Relationships and Historical Biogeography of Ceratozamia (Cycadales)
Abstract
:1. Introduction
2. Results
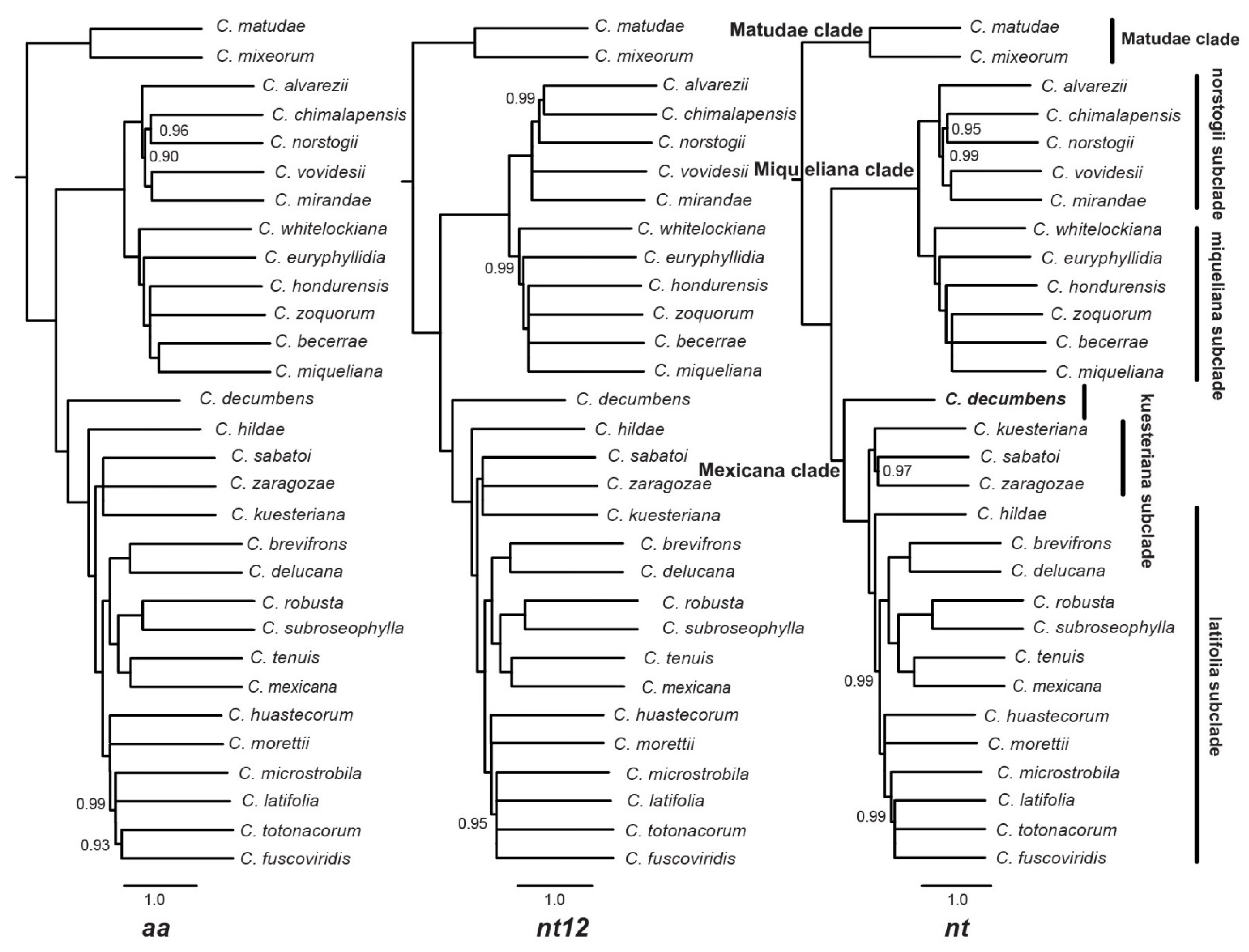
2.1. Phylogenetic Trees
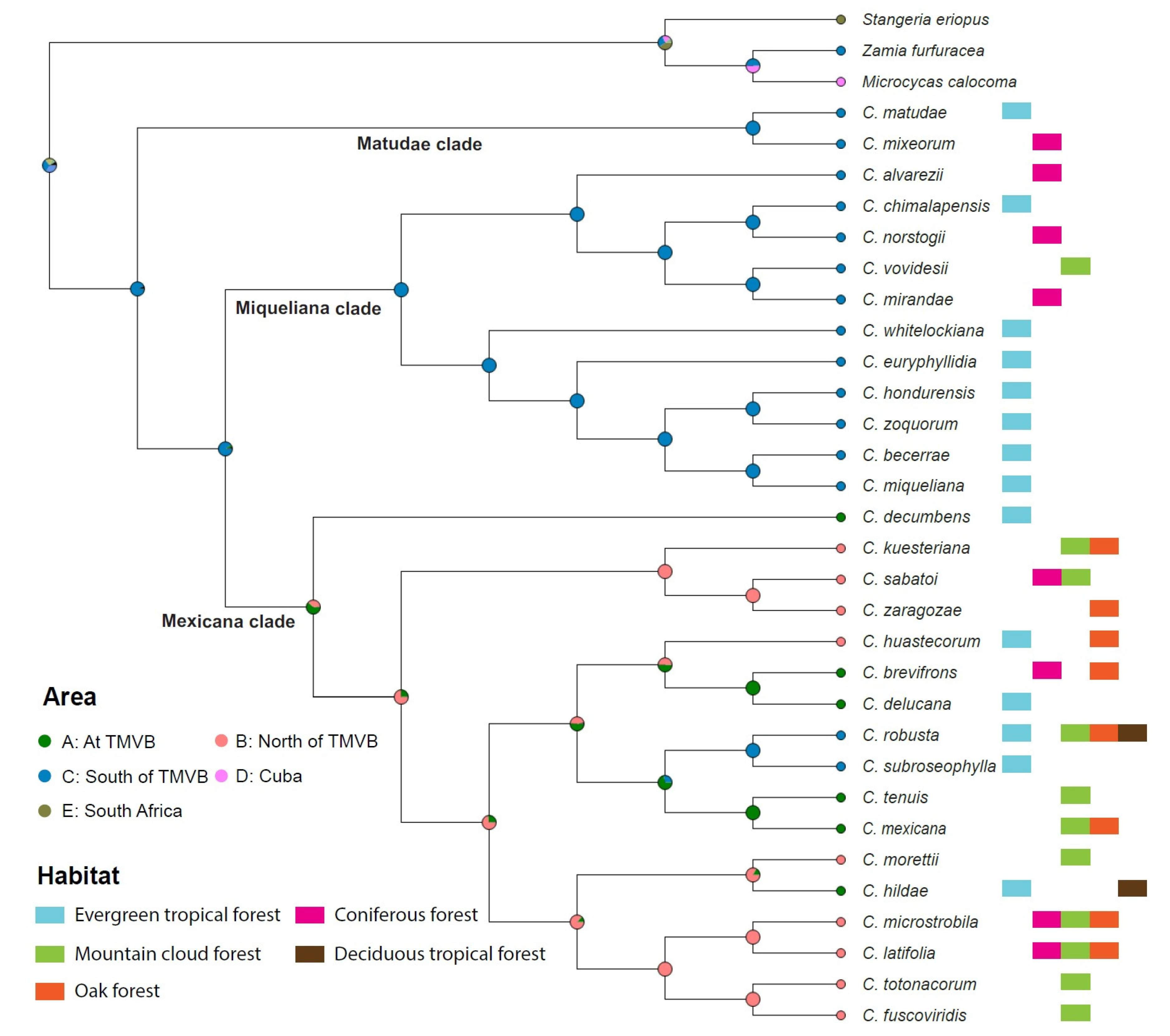
2.2. Biogeography and Habitat Description
2.3. Character Evolution
3. Discussion
3.1. Phylogeny Based on Extensive Transcriptome Data
3.2. Evolution of Species Complexes in Context of Phylogeny
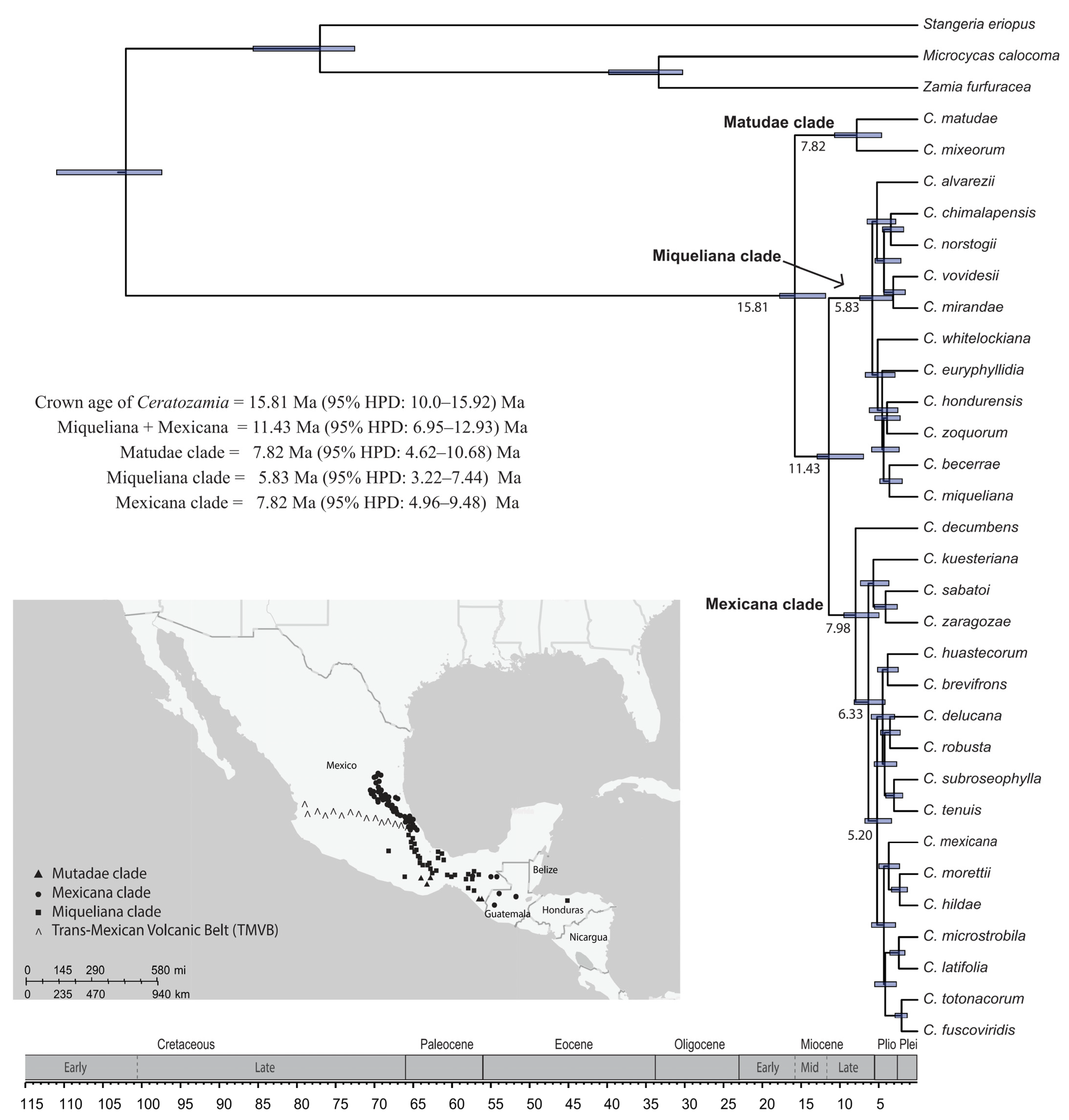
3.3. Age Estimations and Biogeographic Associations
3.4. Evolution of Morphological Characters
4. Materials and Methods
4.1. Sampling, Transcriptome Data Processing and Phylogenomic Analyses
4.2. Molecular Dating, Ancestral Area Reconstruction, and Habitat Assessment
4.3. Morphological Characters State Mapping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calonje, M.; Stevenson, D.W.; Stanberg, L. The World List of Cycads. Available online: http://www.cycadlist.org (accessed on 24 November 2022).
- Liu, Y.; Wang, S.; Li, L.; Yang, T.; Dong, S.; Wei, T.; Wu, S.; Liu, Y.; Gong, Y.; Feng, X.; et al. The Cycas genome and the early evolution of seed plants. Nat. Plants 2022, 8, 389–401. [Google Scholar] [CrossRef]
- Gutiérrez-Ortega, J.S.; Salinas-Rodríguez, M.M.; Martínez, J.F.; Molina-Freaner, F.; Pérez-Farrera, M.A.; Vovides, A.P.; Matsuki, Y.; Suyama, Y.; Ohsawa, T.A.; Watano, Y.; et al. The phylogeography of the cycad genus Dioon (Zamiaceae) clarifies its Cenozoic expansion and diversification in the Mexican transition zone. Ann. Bot. 2018, 121, 535–548. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Domínguez, L.; Nicolalde-Morejón, F.; Vergara-Silva, F.; Stevenson, D. Monograph of Ceratozamia (Zamiaceae, Cycadales): An endangered genus. PhytoKeys 2022, 208, 1–102. [Google Scholar] [CrossRef]
- Pérez-Farrera, M.A.; Gutiérrez-Ortega, J.S.; Gregory, T.; Chemnick, J.; Salas, S.; Calonje, M.; Díaz Jiménez, P. Ceratozamia schiblii (Zamiaceae): A new cycad species from the eastern mountains of Oaxaca, Mexico. Taxonomy 2022, 2, 25. [Google Scholar] [CrossRef]
- Martínez-Domínguez, L.; Nicolalde-Morejón, F.; Vergara-Silva, F.; Stevenson, D.W. Ceratozamia oliversacksii (Zamiaceae), a new species of gymnosperm from western Oaxaca, Mexico. Kew Bull. 2022, 77, 211–219. [Google Scholar] [CrossRef]
- Gutiérrez-Ortega, J.S.; Pérez-Farrera, M.A.; Vovides, A.; Chávez-Cortázar, A.; López, S.; Santos-Hernández, N.; Ruiz Roblero, S. Ceratozamia sanchezae (Zamiaceae): A new cycad species from Chiapas Highlands (Mexico). Phytotaxa 2021, 500, 201–216. [Google Scholar] [CrossRef]
- Pérez-Farrera, M.A.; Gutiérrez-Ortega, J.S.; Haynes, J.; Chemnick, J.; Salas, S.; Calonje, M.; Vovides, A. Ceratozamia aurantiaca (Zamiaceae): A new cycad species from the northern rainforests of Oaxaca, Mexico. Taxonomy 2021, 1, 18. [Google Scholar] [CrossRef]
- Martínez-Domínguez, L.; Nicolalde-Morejón, F.; Lorea-Hernández, F.; Vergara-Silva, F.; Stevenson, D. A novelty in Ceratozamia (Zamiaceae, Cycadales) from the Sierra Madre del Sur, Mexico: Biogeographic and morphological patterns, DNA barcoding and phenology. PhytoKeys 2020, 156, 1–25. [Google Scholar] [CrossRef]
- Gutiérrez-Ortega, J.S.; Pérez-Farrera, M.A.; Vovides, A.; Salas, S.; Chemnick, J. Dioon oaxacensis (Zamiaceae): A new cycad species from the arid central valleys of Oaxaca (Mexico). Phytotaxa 2020, 474, 51–61. [Google Scholar] [CrossRef]
- IUCN Red List of Threatened Species. Version 2022-1. Available online: https://www.iucnredlist.org (accessed on 12 November 2022).
- Nagalingum, N.S.; Marshall, C.R.; Quental, T.B.; Rai, H.S.; Little, D.P.; Mathews, S. Recent synchronous radiation of a living fossil. Science 2011, 334, 796–799. [Google Scholar] [CrossRef]
- Salas-Leiva, D.E.; Meerow, A.W.; Calonje, M.; Griffith, M.P.; Francisco-Ortega, J.; Nakamura, K.; Stevenson, D.W.; Lewis, C.E.; Namoff, S. Phylogeny of the cycads based on multiple single-copy nuclear genes: Congruence of concatenated parsimony, likelihood and species tree inference methods. Ann. Bot. 2013, 112, 1263–1278. [Google Scholar] [CrossRef] [Green Version]
- Condamine, F.L.; Nagalingum, N.S.; Marshall, C.R.; Morlon, H. Origin and diversification of living cycads: A cautionary tale on the impact of the branching process prior in Bayesian molecular dating. BMC Evol. Biol. 2015, 15, 65. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, D.W. Morphology and Systematics of the Cycadales. Mem. N. Y. Bot. Gard. 1990, 57, 8–55. [Google Scholar]
- Stevenson, D.W. A formal classification of the extant cycads. Brittonia 1992, 44, 220–223. [Google Scholar] [CrossRef]
- González, D.; Vovides, A.P. A modification to the SCAR (Sequence Characterized Amplified Region) method provides phylogenetic insights within Ceratozamia (Zamiaceae). Rev. Mex. Biodivers. 2012, 83, 929–938. [Google Scholar]
- Medina-Villarreal, A.; González-Astorga, J.; Espinosa de los Monteros, A. Evolution of Ceratozamia cycads: A proximate-ultimate approach. Mol. Phylogenet. Evol. 2019, 139, 106530. [Google Scholar] [CrossRef]
- Habib, S.; Gong, Y.; Dong, S.; Lindstrom, A.; Stevenson, D.W.; Liu, Y.; Wu, H.; Zhang, S. Phylotranscriptomics reveal the spatio-temporal distribution and morphological evolution of Macrozamia, an Australian endemic genus of Cycadales. Ann. Bot. 2022, 130, 671–685. [Google Scholar] [CrossRef]
- Little, D.P.; Stevenson, D.W. A comparison of algorithms for the identification of specimens using DNA barcodes: Examples from gymnosperms. Cladistics 2007, 22, 1–21. [Google Scholar] [CrossRef]
- Sass, C.; Little, D.P.; Stevenson, D.W.; Specht, C.D. DNA Barcoding in the Cycadales: Testing the potential of proposed barcoding markers for species identification of Cycads. PLoS ONE 2007, 2, e1154. [Google Scholar] [CrossRef]
- Zhang, N.; Zeng, L.; Shan, H.; Ma, H. Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol. 2012, 195, 923–937. [Google Scholar] [CrossRef]
- Zeng, L.; Zhang, Q.; Sun, R.; Kong, H.; Zhang, N.; Ma, H. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 2014, 5, 4956. [Google Scholar] [CrossRef] [Green Version]
- Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Carpenter, E.; Matasci, N.; Ayyampalayam, S.; Barker, M.S.; Burleigh, J.G.; Gitzendanner, M.A.; et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 2014, 111, E4859–E4868. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Sun, R.; Hu, Y.; Zeng, L.; Zhang, N.; Cai, L.; Zhang, Q.; Koch, M.A.; Al-Shehbaz, I.; Edger, P.P.; et al. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 2015, 33, 394–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, J.; Yu, Y.; Xie, D.-F.; Peng, C.; Liu, Q.; Zhou, S.-D.; He, X.-J. A transcriptome-based study on the phylogeny and evolution of the taxonomically controversial subfamily Apioideae (Apiaceae). Ann. Bot. 2020, 125, 937–953. [Google Scholar] [CrossRef]
- Dong, S.; Yu, J.; Zhang, L.; Goffinet, B.; Liu, Y. Phylotranscriptomics of liverworts: Revisiting the backbone phylogeny and ancestral gene duplications. Ann. Bot. 2022, mcac113. [Google Scholar] [CrossRef]
- Brongniart, A. Note sur un nouveau genre de Cycadées du Mexique. Ann. Des Sci. Nat. Sér 1846, 35, 5–10. [Google Scholar]
- Vovides, A.; Pérez-Farrera, M.A.; González, D.; Avendaño, S. Relationships and phytogeography in Ceratozamia (Zamiaceae). In Cycad Classification: Concepts and Recommendations; Walters, T., Osborne, R., Eds.; CABI publishing: Wallingford, UK, 2004; pp. 109–125. [Google Scholar]
- Vovides, A.P.; González, D.; Pérez-Farrera, M.A.; Avendaño, S.; Bárcenas, C. A review of research on the cycad genus Ceratozamia Brongn. (Zamiaceae) in Mexico. Taxon 2004, 53, 291–297. [Google Scholar] [CrossRef]
- González, D.; Vovides, A.P. Low intralineage divergence in Ceratozamia (Zamiaceae) detected with nuclear ribosomal DNA ITS and chloroplast DNA trnL-F non-coding Region. Syst. Bot. 2002, 27, 654–661. [Google Scholar]
- Medina-Villarreal, A.; González-Astorga, J. Morphometric and geographical variation in the Ceratozamia mexicana Brongn. (Zamiaceae) complex: Evolutionary and taxonomic implications. Biol. J. Linn. Soc. Lond. 2016, 119, 213–233. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Domínguez, L.; Nicolalde-Morejón, F.; Vergara-Silva, F.; Stevenson, D.W. Integrative taxonomy of Mexican cycads: Biogeography, morphology and DNA barcoding corroborate a new sympatric species in Ceratozamia (Zamiaceae). Phytotaxa 2016, 268, 25–45. [Google Scholar] [CrossRef]
- Pérez-Farrera, M.A.; Vovides, A.P.; Martínez-Camilo, R.; Martínez-Meléndez, N.; Iglesias, C. A reassessment of the Ceratozamia miqueliana species complex (Zamiaceae) of southeastern Mexico, with comments on species relationships. Syst. Biodivers. 2009, 7, 433–443. [Google Scholar] [CrossRef]
- Martínez-Domínguez, L.; Nicolalde-Morejón, F.; Stevenson, D.W.; Vergara-Silva, F. A new species of Ceratozamia (Zamiaceae) from the Sierra Norte of Puebla, Mexico. Brittonia 2017, 69, 516–524. [Google Scholar] [CrossRef]
- Pérez-Farrera, M.A.; Vovides, A.P.; Avendaño, S. Morphology and leaflet anatomy of the Ceratozamia norstogii (Zamiaceae, Cycadales) species complex in Mexico with comments on relationships and speciation. Int. J. Plant Sci. 2014, 175, 110–121. [Google Scholar] [CrossRef]
- Vovides, A.P.; Stevenson, D.W.; Pérez-Farrera, M.Á.; López, S.; Avendaño, S. What is Ceratozamia mexicana (Zamiaceae)? Bol. Soc. Bot. Mex. 2016, 94, 419–429. [Google Scholar] [CrossRef]
- Haynes, J.; Whitelock, L.; Schutzman, B.; Adams, R. A new endemic Ceratozamia from Honduras (Cycadales: Zamiaceae). Cycad Newsl. 2008, 31, 16–21. [Google Scholar]
- Vovides, A.P.; Pérez-Farrera, M.Á.; Schutzman, B.; Iglesias, C.; Hernández-Sandoval, L.; Martínez, M. A new species of Ceratozamia (Zamiaceae) from Tabasco and Chiapas, Mexico. Bot. J. Linn. Soc. 2004, 146, 123–128. [Google Scholar] [CrossRef]
- Martínez-Domínguez, L.; Nicolalde-Morejón, F.; Vergara-Silva, F.; Stevenson, D.W.; del Callejo, E. Cryptic diversity, sympatry, and other integrative taxonomy scenarios in the Mexican Ceratozamia miqueliana complex (Zamiaceae). Org. Divers. Evol. 2017, 17, 727–752. [Google Scholar] [CrossRef]
- Chemnick, J.; Gregory, T. A new species of Ceratozamia (Zamiaceae) from Oaxaca, Mexico with comments on distribution, habitat, and relationships. Phytologia 1995, 79, 51–57. [Google Scholar]
- Pérez-Farrera, M.A.; González-Astorga, J.; Avendaño, S.; Iglesias, C.G. A new species of Ceratozamia (Zamiaceae) from the Sierra Madre of Chiapas, Mexico, with comments on species relationships. Bot. J. Linn. Soc. 2007, 153, 393–400. [Google Scholar] [CrossRef] [Green Version]
- Vovides, A.; Pérez-Farrera, M.A.; Arturo, G.A.; Iglesias, C. A new species of Ceratozamia (Zamiaceae) from Oaxaca, Mexico with comments on habitat and relationships. Bot. J. Linn. Soc. 2008, 157, 169–175. [Google Scholar] [CrossRef] [Green Version]
- Vovides, A.P.; Pérez-Farrera, M.A.; Gutiérrez-Ortega, J.S.; Avendaño, S.; Medina-Villarreal, A.; González-Astorga, J.; Galicia, S. A revision of the Ceratozamia miqueliana (Zamiaceae) species complex based on analyses of leaflet anatomical characters. Flora 2002, 270, e151649. [Google Scholar] [CrossRef]
- Vovides, A.P.; Avendaño, S.; Pérez-Farrera, M.A.; Stevenson, D.W. What is Ceratozamia brevifrons (Zamiaceae)? Brittonia 2012, 64, 35–42. [Google Scholar] [CrossRef]
- Lundell, C.L. The vegetation and natural resources of British Honduras. Chron. Bot. 1942, 7, 169–171. [Google Scholar]
- Miranda, F. Vegetación de la Vertiente del Pacífico de la Sierra Madre de Chiapas (México) y sus Relaciones Florísticas; National Research Council of the Philippines, University of the Philippines: Kalakhang Maynila, Philipines, 1957. [Google Scholar]
- Miranda, F. Estudios Acerca de la Vegetación. Ediciones del Instituto Mexicano de Recursos Naturales Renovables, AC: Mexico City, Mexico, 1958; pp. 215–271. [Google Scholar]
- Toledo, V.M. Pleistocene changes of vegetation in tropical Mexico. In Biological Diversification in the Tropics; Prance, G.T., Ed.; Columbia University Press: New York, NY, USA, 1982; pp. 93–111. [Google Scholar]
- Brown, K.S. Centros de evolução, refúgios quaternários e conservação de patrimônios genéticos na região neotropical: Padrões de diferenciação em Ithomiinae (Lepidoptera: Nymphalidae). Acta Amaz. 1977, 7, 75–137. [Google Scholar] [CrossRef] [Green Version]
- Flower, B.P.; Kennett, J.P. The middle Miocene climatic transition: East Antarctic ice sheet development, deep ocean circulation and global carbon cycling. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1994, 108, 537–555. [Google Scholar] [CrossRef]
- Ramírez, E.; Martinez-Hernandez, E.; Flores, H.; Ochoterena, H.; Prámparo, M.B. Correlation of the Late Eocene–Early Oligocene izúcar de matamoros evaporites (Cuayuca formation) in Mexico based on parsimony analysis of endemicity. Palynology 2008, 32, 231–252. [Google Scholar]
- Ramírez-Arriaga, E.; Prámparo, M.B.; Martínez-Hernández, E.; Helenes-Escamilla, J. Palaeoevironmental reconstruction based on palynomorphs from the upper Oligocene San Gregorio Formation (core LB1), in a semiarid coastal marine setting, Baja California Sur, Mexico. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2021, 575, 110476. [Google Scholar] [CrossRef]
- Sosa, V.; De-Nova, J.A.; Vásquez-Cruz, M. Evolutionary history of the flora of Mexico: Dry forests cradles and museums of endemism. J. Syst. Evol. 2018, 56, 523–536. [Google Scholar] [CrossRef]
- Ramírez-Arriaga, E.; Prámparo, M.B.; Nieto-Samaniego, A.F.; Martínez-Hernández, E.; Valiente-Banuet, A.; Macías-Romo, C.; Dávalos-Álvarez, O.G. Palynological evidence for Middle Miocene vegetation in the Tehuacán Formation of Puebla, Mexico. Palynology 2014, 38, 1–27. [Google Scholar] [CrossRef]
- Caldwell, O.W.; Baker, C. The identity of Microcycas calocoma. Bot. Gaz. 1907, 43, 330–335. [Google Scholar] [CrossRef] [Green Version]
- Hill, K.; Chase, M.; Stevenson, D.; Hills, H.; Schutzman, B. The families and genera of cycads: A molecular phylogenetic analysis of Cycadophyta based on nuclear and plastid DNA sequences. Int. J. Plant Sci. 2003, 164, 933–948. [Google Scholar] [CrossRef]
- Singh, R.; Radha, P. A new species of Cycas from the Malabar coast, western Ghats, India. Brittonia 2006, 58, 119–123. [Google Scholar] [CrossRef]
- González-Astorga, J.; Vovides, A.P.; Iglesias, C. Morphological and geographic variation of the cycad Dioon edule Lindl. (Zamiaceae): Ecological and evolutionary implications. Bot. J. Linn. Soc. 2003, 141, 465–470. [Google Scholar] [CrossRef] [Green Version]
- Chaiprasongsuk, M.; Mingmuang, M.; Thongpan, A.; Namwongprom, K. Molecular identification of Encephalartos (Zamiaceae) species and their relationships to morphological characters. Witthayasan Kasetsat 2007, 41, 43–60. [Google Scholar]
- Calonje, M.; Meerow, A.; Griffith, M.; Salas-Leiva, D.; Vovides, A.; Coiro, M.; Francisco-Ortega, J. A time-calibrated species tree phylogeny of the New World cycad genus Zamia L. (Zamiaceae, Cycadales). Int. J. Plant Sci. 2019, 180, 286–314. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laetsch, D.R.; Blaxter, M.L. KinFin: Software for taxon-aware analysis of clustered protein sequences. G3 Genes Genom. Genet. 2017, 7, 3349–3357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010, 38, W7–W13. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Hodcroft, E. TreeCollapserCL4. Available online: http://emmahodcroft.com/TreeCollapseCL.html (accessed on 13 June 2022).
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.A.; O’Meara, B.C. treePL: Divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 2012, 28, 2689–2690. [Google Scholar] [CrossRef] [Green Version]
- Helfrich, P.; Rieb, E.; Abrami, G.; Lücking, A.; Mehler, A. TreeAnnotator: Versatile Visual Annotation of Hierarchical Text Relations. In Proceedings of the 11th edition of the Language Resources and Evaluation Conference, Miyazaki, Japan, 7–12 May 2018. [Google Scholar]
- Matzke, N. BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis with R Scripts; Version 1.1.1; GitHub: San Francisco, CA, USA, 2018; Available online: https://github.com/nmatzke/BioGeoBEARS (accessed on 15 September 2022).
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.70. Available online: http://www.mesquiteproject.org (accessed on 27 August 2022).
- Lewis, P.O. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 2001, 50, 913–925. [Google Scholar] [CrossRef] [Green Version]
- Yessoufou, K.; Daru, B.H.; Tafirei, R.; Elansary, H.O.; Rampedi, I. Integrating biogeography, threat and evolutionary data to explore extinction crisis in the taxonomic group of cycads. Ecol. Evol. 2017, 7, 2735–2746. [Google Scholar] [CrossRef]



| No. | Sample ID * | Taxon Names | Raw Data (Gb) | Clean Reads (Gb) * | Accession No. * | Herbarium Code |
|---|---|---|---|---|---|---|
| 1 | SAMN32670532 | Ceratozamia alvarezii | 6 | 3.32 | SRR23044200 | SING |
| 2 | SAMN32670533 | Ceratozamia becerrae | 6 | 3.32 | SRR23044199 | SING |
| 3 | SAMN32670534 | Ceratozamia brevifrons | 6 | 3.35 | SRR23044188 | W |
| 4 | SAMN32670535 | Ceratozamia chimalapensis | 6 | 3.26 | SRR23044177 | SING |
| 5 | SAMN32670536 | Ceratozamia decumbens | 6 | 3.29 | SRR23044173 | W |
| 6 | SAMN32670537 | Ceratozamia delucana | 6 | 3.47 | SRR23044172 | SING |
| 7 | SAMN32670538 | Ceratozamia euryphyllidia | 6 | 3.21 | SRR23044171 | SING |
| 8 | SAMN32670539 | Ceratozamia fuscoviridis | 6 | 2.94 | SRR23044170 | W |
| 9 | SAMN32670540 | Ceratozamia hildae | 12 | 6.49 | SRR23044169 | W |
| 10 | SAMN32670541 | Ceratozamia hondurensis | 6 | 3.40 | SRR23044168 | SING |
| 11 | SAMN32670542 | Ceratozamia huastecorum | 6 | 3.38 | SRR23044198 | SING |
| 12 | SAMN32670543 | Ceratozamia kuesteriana | 6 | 3.09 | SRR23044197 | W |
| 13 | SAMN32670544 | Ceratozamia latifolia | 6 | 3.36 | SRR23044196 | W |
| 14 | SAMN32670545 | Ceratozamia matudae | 6 | 3.31 | SRR23044195 | SING |
| 15 | SAMN32670546 | Ceratozamia mexicana | 6 | 3.35 | SRR23044194 | SING |
| 16 | SAMN32670547 | Ceratozamia microstrobila | 6 | 3.38 | SRR23044193 | W |
| 17 | SAMN32670548 | Ceratozamia miqueliana | 6 | 3.27 | SRR23044192 | W |
| 18 | SAMN32670549 | Ceratozamia mirandae | 6 | 3.35 | SRR23044191 | W |
| 19 | SAMN32670550 | Ceratozamia mixeorum | 6 | 3.46 | SRR23044190 | W |
| 20 | SAMN32670551 | Ceratozamia morettii | 6 | 3.44 | SRR23044189 | SING |
| 21 | SAMN32670552 | Ceratozamia norstogii | 6 | 3.41 | SRR23044187 | W |
| 22 | SAMN32670553 | Ceratozamia robusta | 6 | 3.31 | SRR23044186 | SING |
| 23 | SAMN32670554 | Ceratozamia sabatoi | 6 | 3.33 | SRR23044185 | W |
| 24 | SAMN32670555 | Ceratozamia subroseophylla | 6 | 3.38 | SRR23044184 | W |
| 25 | SAMN32670556 | Ceratozamia tenuis | 6 | 3.91 | SRR23044183 | SING |
| 26 | SAMN32670557 | Ceratozamia totonacorum | 6 | 3.34 | SRR23044182 | W |
| 27 | SAMN32670558 | Ceratozamia vovidesii | 6 | 3.23 | SRR23044181 | W |
| 28 | SAMN32670559 | Ceratozamia whitelockiana | 6 | 3.4 | SRR23044180 | SING |
| 29 | SAMN32670560 | Ceratozamia zaragozae | 6 | 3.35 | SRR23044179 | W |
| 30 | SAMN32670561 | Ceratozamia zoquorum | 6 | 3.43 | SRR23044178 | W |
| 31 | SAMN32670562 | Microcycas calocoma | 12 | 6.6 | SRR23044176 | W |
| 32 | SAMN32670563 | Stangeria eriopus | 12 | 6.85 | SRR23044175 | W |
| 33 | SAMN32670564 | Zamia furfuracea | 12 | 7.79 | SRR23044174 | W |
| Area A: At TMVB | Area B: North of TMVB | Area C: South of TMVB | |
|---|---|---|---|
| Whole genus | 2.67 | 1.61 | 94.22 |
| Matudae clade | 0 | 0 | 100 |
| Miqueliana + Mexicana | 5.94 | 3.5 | 90.23 |
| Miqueliana clade | 0 | 0 | 100 |
| norstogii subclade | 0 | 0 | 100 |
| miqueliana subclade | 0 | 0 | 100 |
| Mexicana clade | 61.57 | 37.24 | >1 |
| kuesteriana subclade | 100 | 0 | 0 |
| latifolia subclade | 72.44 | 26.45 | 1.09 |
| No. | Characters | States | |||
|---|---|---|---|---|---|
| 0 | 1 | 2 | |||
| Stem | 1 | Habit | Epigeous | semi-hypogeous | |
| Leaves | 2 | Leaflet shape | elliptic-lanceolate | oblong to oblanceolate | obovate |
| 3 | Leaflet direction | Descending | ascending | ||
| 4 | Leaflet texture | Coriaceous | papyraceous | membranaceous | |
| 5 | Formation of prickles (Petiole) | Robust | thin | ||
| Microsporangiate strobili | 6 | Microsporophyll shape | Discoid | obconic | elliptic |
| 7 | Angle between horns of microsporophyll | Obtuse | acute | right | |
| 8 | Infertile portion shape | Orbicular | rounded | linear | |
| 9 | Fertile portion shape | Lobate | deeply lobate | ||
| Megasporangiate strobili | 10 | Angle between horns of megasporophylls | Obtuse | acute | right |
| 11 | Distal face of megasporophylls shape | Truncate | prominent | ||
| Seed | 12 | Sarcotesta color | light-brown | whitish-yellow | whitish-red |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habib, S.; Gong, Y.; Dong, S.; Lindstrom, A.; Stevenson, D.W.; Wu, H.; Zhang, S. Phylotranscriptomics Shed Light on Intrageneric Relationships and Historical Biogeography of Ceratozamia (Cycadales). Plants 2023, 12, 478. https://doi.org/10.3390/plants12030478
Habib S, Gong Y, Dong S, Lindstrom A, Stevenson DW, Wu H, Zhang S. Phylotranscriptomics Shed Light on Intrageneric Relationships and Historical Biogeography of Ceratozamia (Cycadales). Plants. 2023; 12(3):478. https://doi.org/10.3390/plants12030478
Chicago/Turabian StyleHabib, Sadaf, Yiqing Gong, Shanshan Dong, Anders Lindstrom, Dennis William Stevenson, Hong Wu, and Shouzhou Zhang. 2023. "Phylotranscriptomics Shed Light on Intrageneric Relationships and Historical Biogeography of Ceratozamia (Cycadales)" Plants 12, no. 3: 478. https://doi.org/10.3390/plants12030478
APA StyleHabib, S., Gong, Y., Dong, S., Lindstrom, A., Stevenson, D. W., Wu, H., & Zhang, S. (2023). Phylotranscriptomics Shed Light on Intrageneric Relationships and Historical Biogeography of Ceratozamia (Cycadales). Plants, 12(3), 478. https://doi.org/10.3390/plants12030478








