Host Tree and Geography Induce Metabolic Shifts in the Epiphytic Liverwort Radula complanata
Abstract
:1. Introduction
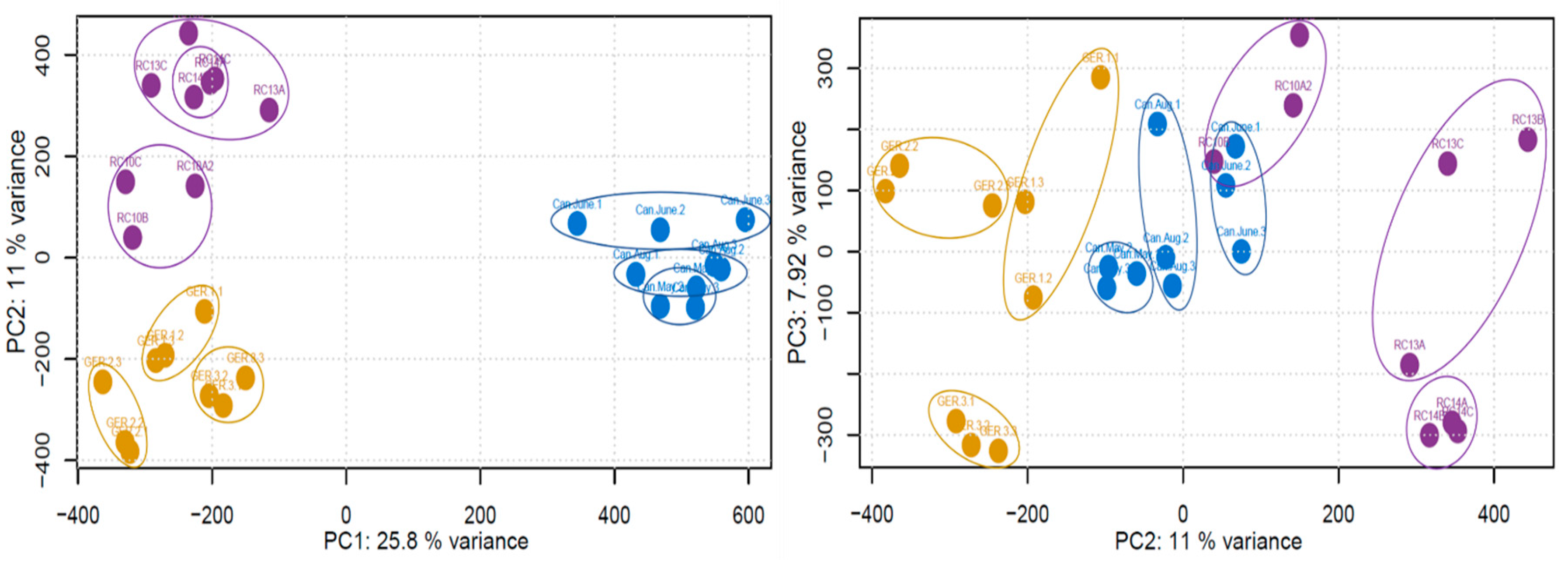
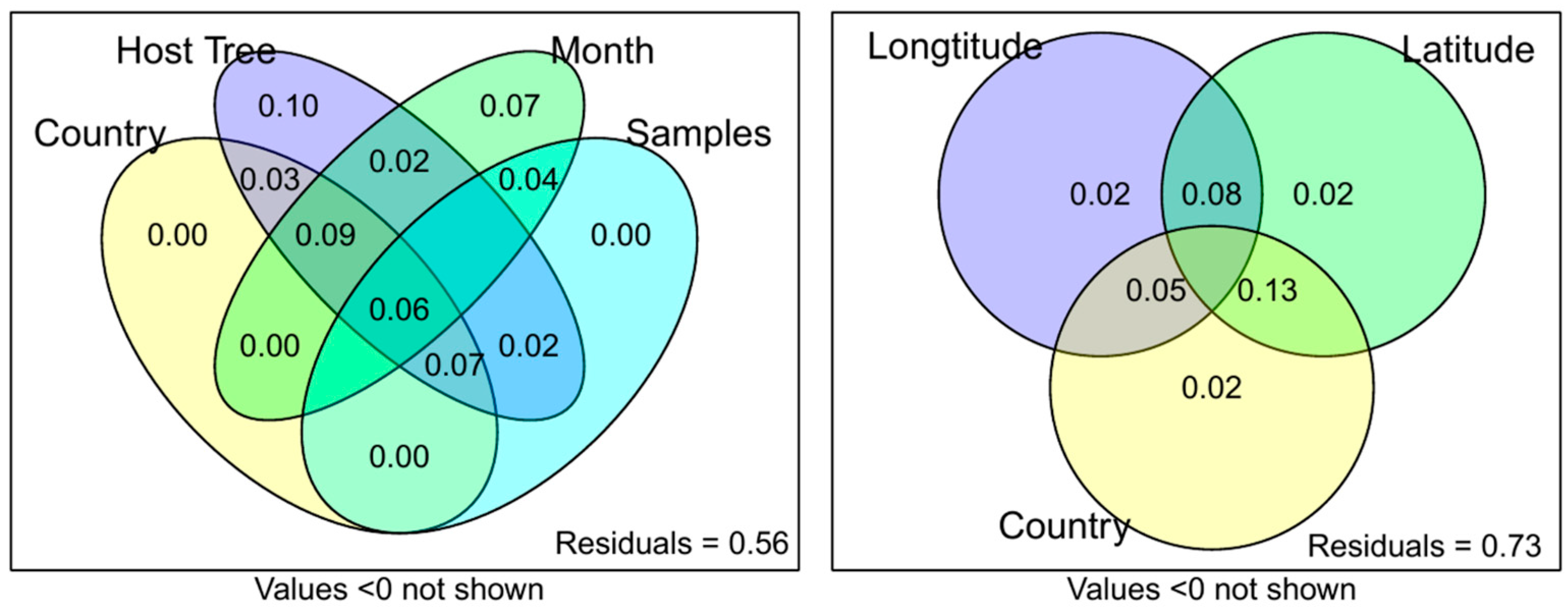
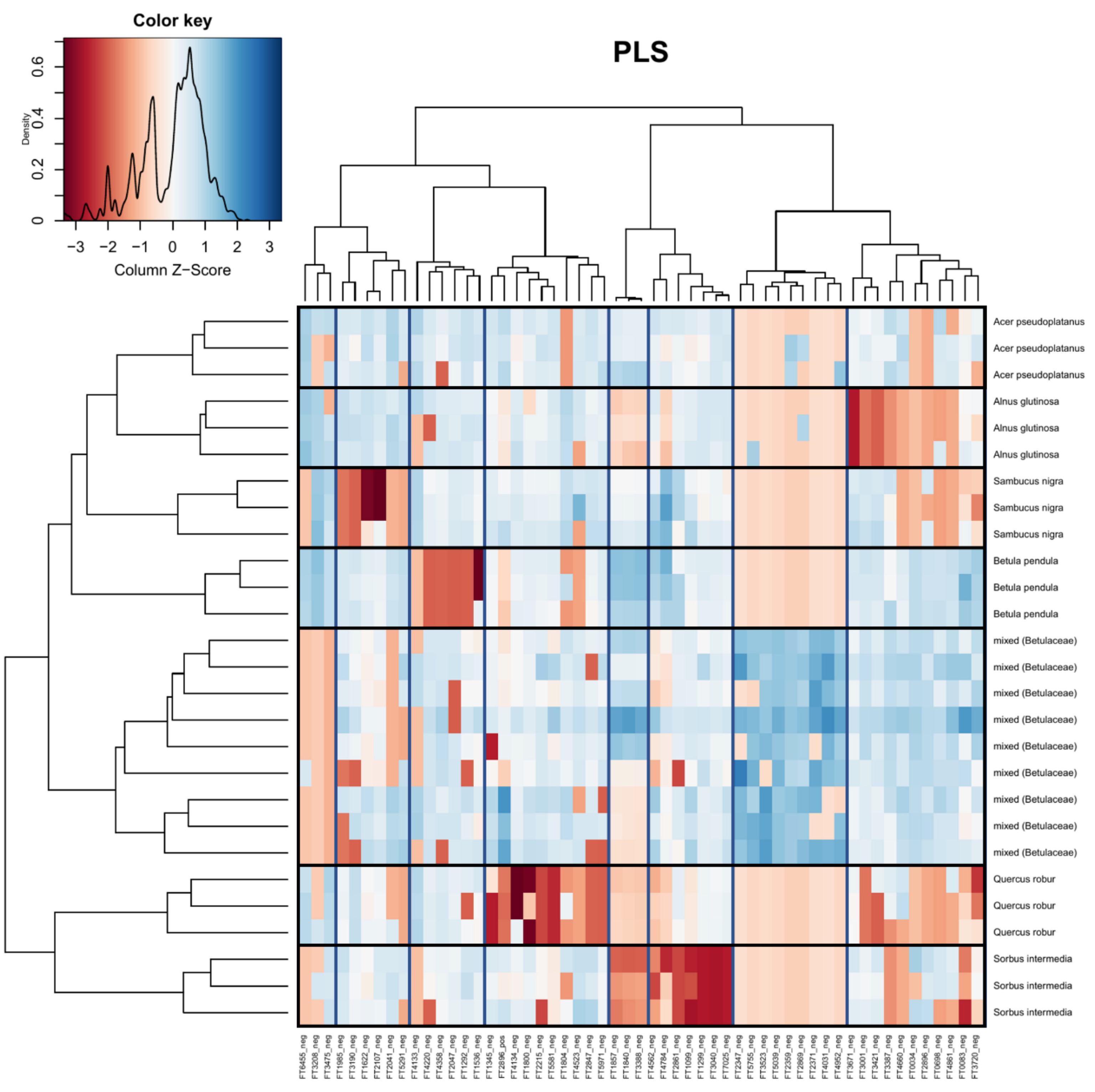
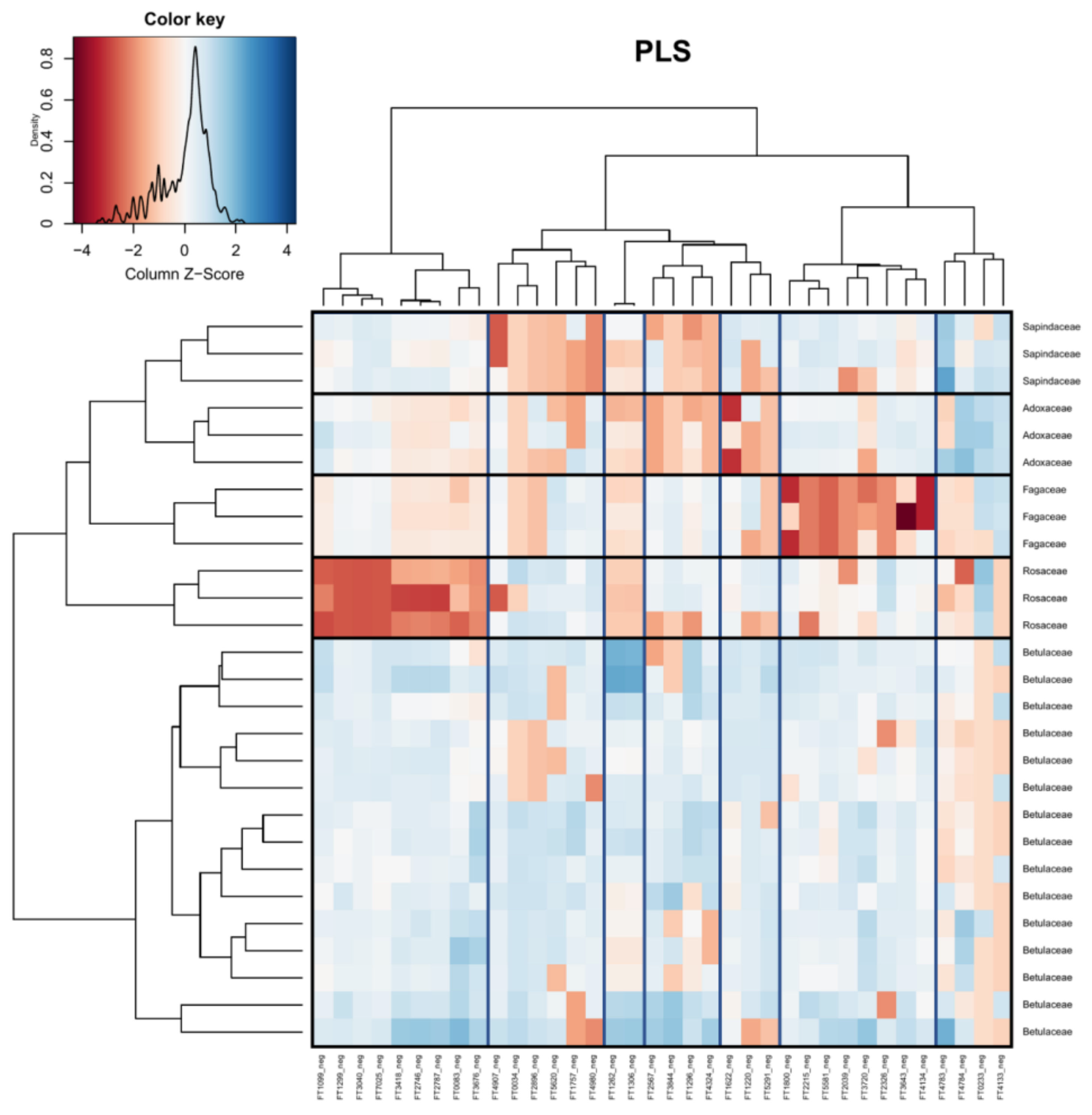
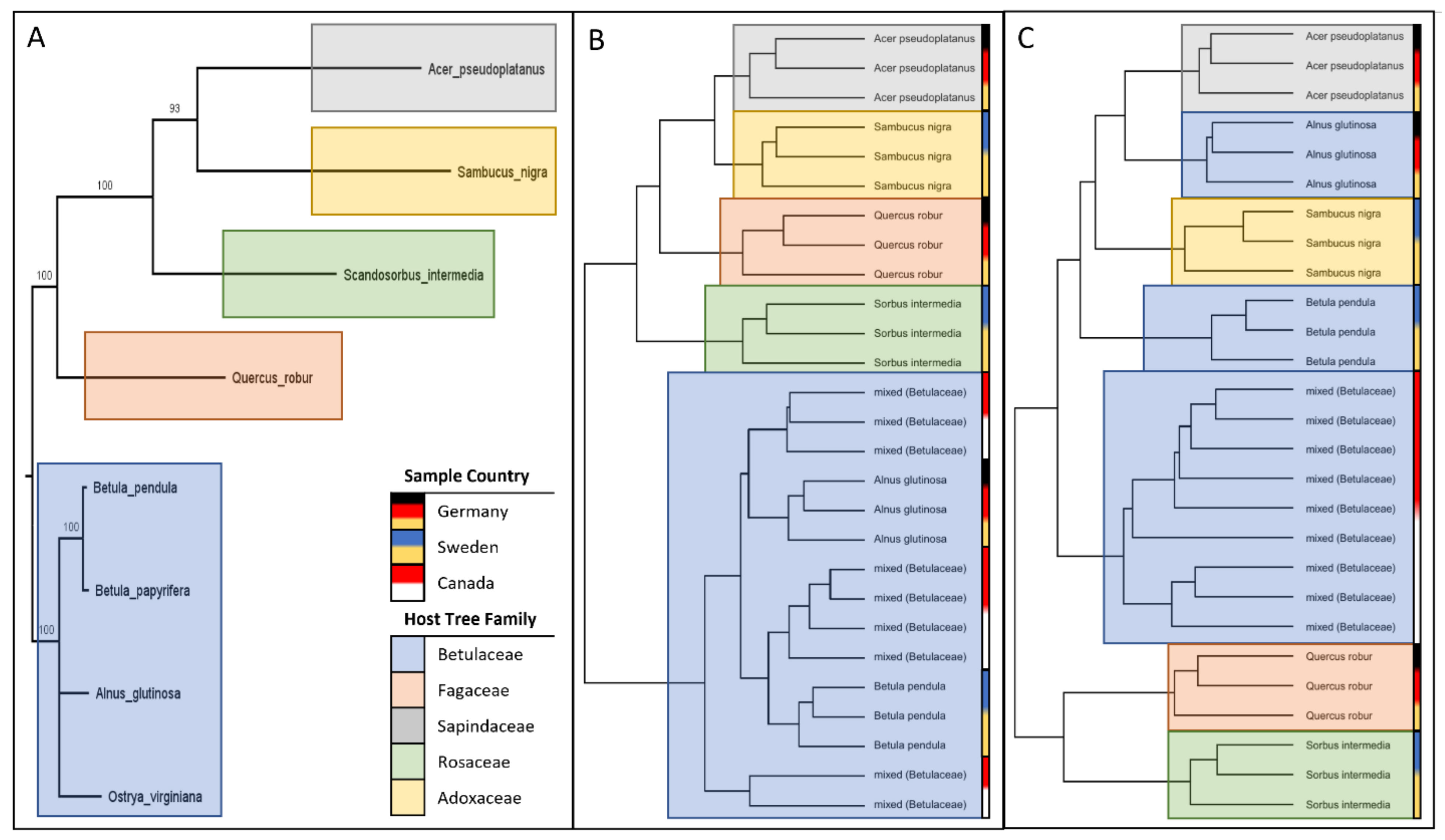
2. Results
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Collection
5.2. HPLC Analysis and Raw Data Collection
5.3. Peak Detection and Data Treatment
5.4. Statistical Analyses
5.5. Compound Annotation and Classification
5.6. Phylogenetic Analysis
5.7. Climate Data
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dey, A.; Mukherjee, A. Therapeutic potential of bryophytes and derived compounds against cancer. J. Acute Dis. 2015, 4, 236–248. [Google Scholar] [CrossRef] [Green Version]
- Nandy, S.; Dey, A. Bibenzyls and bisbybenzyls of bryophytic origin as promising source of novel therapeutics: Pharmacology, synthesis and structure-activity. DARU J. Pharm. Sci. 2020, 28, 701–734. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Espley, R.V.; Gertsch, J.; Whare, T.; Stehle, F.; Kayser, O. Demystifying the liverwort Radula marginata, a critical review on its taxonomy, genetics, cannabinoid phytochemistry and pharmacology. Phytochem. Rev. 2019, 8, 953–965. [Google Scholar] [CrossRef]
- He, X.; Sun, Y.; Zhu, R.L. The Oil Bodies of Liverworts: Unique and Important Organelles in Land Plants. Crit. Rev. Plant Sci. 2013, 32, 293–302. [Google Scholar] [CrossRef]
- Suire, C. A comparative, transmission-electron microscopic study on the formation of oil-bodies in liverworts. J. Hattori Bot. Lab. 2000, 232, 209–232. [Google Scholar] [CrossRef]
- Ludwiczuk, A.; Asakawa, Y. Bryophytes as a source of bioactive volatile terpenoids—A review. Food Chem. Toxicol. 2019, 132, 110649. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wei, G.; Chen, X.; Guo, H.; Crandall-Stotler, B.; Köllner, T.G.; Chen, F. Sesquiterpene biosynthesis in a leafy liverwort Radula lindenbergiana Gottsche ex C. Hartm. Phytochemistry 2021, 190, 112847. [Google Scholar] [CrossRef]
- Tanaka, M.; Esaki, T.; Kenmoku, H.; Koeduka, T.; Kiyoyama, Y.; Masujima, T.; Asakawa, Y.; Matsui, K. Direct evidence of specific localization of sesquiterpenes and marchantin A in oil body cells of Marchantia polymorpha L. Phytochemistry 2016, 130, 77–84. [Google Scholar] [CrossRef]
- Kanazawa, T.; Morinaka, H.; Ebine, K.; Shimada, T.L.; Ishida, S.; Minamino, N.; Yamaguchi, K.; Shigenobu, S.; Kohchi, T.; Nakano, A.; et al. The liverwort oil body is formed by redirection of the secretory pathway. Nat. Commun. 2020, 11, 6152. [Google Scholar] [CrossRef]
- Romani, F.; Banić, E.; Florent, S.N.; Kanazawa, T.; Goodger, J.Q.D.; Mentink, R.A.; Dierschke, T.; Zachgo, S.; Ueda, T.; Bowman, J.L.; et al. Oil Body Formation in Marchantia polymorpha Is Controlled by MpC1HDZ and Serves as a Defense against Arthropod Herbivores. Curr. Biol. 2020, 30, 2815–2828. [Google Scholar] [CrossRef]
- Davies, K.M.; Jibran, R.; Zhou, Y.; Albert, N.W.; Brummell, D.A.; Jordan, B.R.; Bowman, J.L.; Schwinn, K.E. The Evolution of Flavonoid Biosynthesis: A Bryophyte Perspective. Front. Plant Sci. 2020, 11, 1–21. [Google Scholar] [CrossRef] [Green Version]
- GILBERT, O.L. BRYOPHYTES AS INDICATORS OF AIR POLLUTION IN THE TYNE VALLEY. New Phytol. 1968, 67, 15–30. [Google Scholar] [CrossRef]
- Wang, X.; Cao, J.; Dai, X.; Xiao, J.; Wu, Y.; Wang, Q. Total flavonoid concentrations of bryophytes from Tianmu Mountain, Zhejiang Province (China): Phylogeny and ecological factors. PLoS ONE 2017, 12, 2–11. [Google Scholar] [CrossRef] [Green Version]
- Markert, B.A.; Breure, A.M.; Zechmeister, H.G. Bioindicators & Biomonitors: Principles, Concepts and Applications, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Vitt, D.H.; House, M. Bryophytes as key indicators of ecosystem function and structure of northern peatlands. Bryophyt. Divers. Evol. 2021, 43, 253–264. [Google Scholar] [CrossRef]
- Patiño, J.; González-Mancebo, J.M. Exploring the effect of host tree identity on epiphyte bryophyte communities in different Canarian subtropical cloud forests. Plant Ecol. 2011, 212, 433–449. [Google Scholar] [CrossRef]
- Maul, K.; Wei, Y.M.; Nebel, M.; Luebert, F.; Ho, B.C.; Quandt, D.; Kessler, M. Different Predictors Shape the Diversity Patterns of Epiphytic and Non-epiphytic Liverworts in Montane Forests of Uganda. Front. Plant Sci. 2020, 11, 765. [Google Scholar] [CrossRef]
- Bates, J.W. Influence of Chemical and Physical Factors on Quercus and Fraxinus Epiphytes at Loch Sunart, Western Scotland: A Multivariate Analysis. J. Ecol. 1992, 80, 163. [Google Scholar] [CrossRef]
- Mukhia, S.; Mandal, P.; Singh, D.K.; Singh, D. The abundance of epiphytic liverworts on the bark of Cryptomeria japonica in relation to different physical and biochemical attributes, found in Senchal Wildlife Sanctuary, Darjeeling, Eastern Himalaya. BMC Ecol. 2019, 19, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hauck, M.; Jürgens, S.R.; Willenbruch, K.; Huneck, S.; Leuschner, C. Dissociation and metal-binding characteristics of yellow lichen substances suggest a relationship with site preferences of lichens. Ann. Bot. 2009, 103, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Govindapyari, H.; Leleeka, M.; Nivedita, M.; Uniyal, P. Bryophytes: Indicators and monitoring agents of pollution. NeBIO 2010, 1, 35–41. [Google Scholar]
- Shi, X.-M.; Song, L.; Liu, W.-Y.; Lu, H.-Z.; Qi, J.-H.; Li, S.; Chen, X.; Wu, J.-F.; Liu, S.; Wu, C.-S. Epiphytic bryophytes as bio-indicators of atmospheric nitrogen deposition in a subtropical montane cloud forest: {Response} patterns, mechanism, and critical load. Environ. Pollut. 2017, 229, 932–941. [Google Scholar] [CrossRef]
- Glime, J.M. Chapter 1-10 Aquatic and Wet Marchantiophyta, Class Jungermanniopsida: Radulaceae and Ptilidiaceae. In Bryophyte Ecology; Glime, J.M., Ed.; Volume 4, Habitat and Role. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. Last Updated 11 April 2021; Available online: http://digitalcommons.mtu.edu/bryophyte-ecology/ (accessed on 24 October 2022).
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Li, J.; Xu, H.; Song, Y.; Tang, L.; Gong, Y.; Yu, R.; Shen, L.; Wu, X.; Liu, Y.; Zeng, W. Geography plays a more important role than soil composition on structuring genetic variation of Pseudometallophyte Commelina communis. Front. Plant Sci. 2016, 7, 1085. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.W.; Fridman, E.; Mascher, M.; Himmelbach, A.; Schmid, K. Physical geography, isolation by distance and environmental variables shape genomic variation of wild barley (Hordeum vulgare L. ssp. spontaneum) in the Southern Levant. Heredity 2022, 128, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Lasky, J.R.; Des Marais, D.L.; Lowry, D.B.; Povolotskaya, I.; McKay, J.K.; Richards, J.H.; Keitt, T.H.; Juenger, T.E. Natural variation in abiotic stress responsive gene expression and local adaptation to climate in Arabidopsis thaliana. Mol. Biol. Evol. 2014, 31, 2283–2296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghirardo, A.; Nosenko, T.; Kreuzwieser, J.; Winkler, J.B.; Kruse, J.; Albert, A.; Merl-Pham, J.; Lux, T.; Ache, P.; Zimmer, I.; et al. Protein expression plasticity contributes to heat and drought tolerance of date palm. Oecologia 2021, 197, 903–919. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Sun, X.; Feng, Q.; Luo, H.; Wassie, M.; Amee, M.; Amombo, E.; Chen, L. Comparative physiological and metabolomic analyses reveal mechanisms of Aspergillus aculeatus-mediated abiotic stress tolerance in tall fescue. Plant Physiol. Biochem. 2019, 142, 342–350. [Google Scholar] [CrossRef]
- Ravi, S.; D’Odorico, P. A field-scale analysis of the dependence of wind erosion threshold velocity on air humidity. Geophys. Res. Lett. 2005, 32, 1–4. [Google Scholar] [CrossRef]
- WHO. Review of Evidence on Health Aspects of Air Pollution—REVIHAAP Project; Technical Report; WHO: Geneva, Switzerland, 2013. [Google Scholar]
- Ilek, A.; Siegert, C.M.; Wade, A. Hygroscopic contributions to bark water storage and controls exerted by internal bark structure over water vapor absorption. Trees 2021, 35, 831–843. [Google Scholar] [CrossRef]
- Alonso-Serra, J.; Safronov, O.; Lim, K.-J.; Fraser-Miller, S.J.; Blokhina, O.B.; Campilho, A.; Chong, S.-L.; Fagerstedt, K.; Haavikko, R.; Helariutta, Y.; et al. Tissue-specific study across the stem reveals the chemistry and transcriptome dynamics of birch bark. New Phytol. 2019, 222, 1816–1831. [Google Scholar] [CrossRef]
- Elias, J.P.C.; Mortara, S.R.; Nunes-Freitas, A.F.; van den Berg, E.; Ramos, F.N. Host tree traits in pasture areas affect forest and pasture specialist epiphyte species differently. Am. J. Bot. 2021, 108, 598–606. [Google Scholar] [CrossRef]
- Kovářová, M.; Pyszko, P.; Plášek, V. How Does the pH of Tree Bark Change with the Presence of the Epiphytic Bryophytes from the Family Orthotrichaceae in the Interaction with Trunk Inclination? Plants 2021, 11, 63. [Google Scholar] [CrossRef]
- Ebeling, A.; Pompe, S.; Baade, J.; Eisenhauer, N.; Hillebrand, H.; Proulx, R.; Roscher, C.; Schmid, B.; Wirth, C.; Weisser, W.W. A trait-based experimental approach to understand the mechanisms underlying biodiversity–ecosystem functioning relationships. Basic Appl. Ecol. 2014, 15, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Blatt-Janmaat, K.; Neumann, S.; Schmidt, F.; Ziegler, J.; Peters, K.; Qu, Y. Impact of in vitro hormone treatments on the bibenzyl production of Radula complanata. Botany 2022. In Press. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Libiseller, G.; Dvorzak, M.; Kleb, U.; Gander, E.; Eisenberg, T.; Madeo, F.; Neumann, S.; Trausinger, G.; Sinner, F.; Pieber, T.; et al. IPO: A tool for automated optimization of XCMS parameters. BMC Bioinform. 2015, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Tautenhahn, R.; Bottcher, C.; Neumann, S. Highly sensitive feature detection for high resolution {LC}/{MS}. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef] [Green Version]
- Peters, K.; Balcke, G.; Kleinenkuhnen, N.; Treutler, H.; Neumann, S. Untargeted in silico compound classification—A novel metabolomics method to assess the chemodiversity in bryophytes. Int. J. Mol. Sci. 2021, 22, 3251. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef]
- Ludwig, M.; Nothias, L.-F.; Dührkop, K.; Koester, I.; Fleischauer, M.; Hoffmann, M.A.; Petras, D.; Vargas, F.; Morsy, M.; Aluwihare, L.; et al. ZODIAC: Database-independent molecular formula annotation using Gibbs sampling reveals unknown small molecules. Nat. Mach. Intell. 2020, 2, 629–641. [Google Scholar] [CrossRef]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using {CSI}:{FingerID}. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.A.; Nothias, L.-F.; Ludwig, M.; Fleischauer, M.; Gentry, E.C.; Witting, M.; Dorrestein, P.C.; Dührkop, K.; Böcker, S. Assigning confidence to structural annotations from mass spectra with COSMIC. Biorxiv 2021. [Google Scholar] [CrossRef]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Dührkop, K.; Nothias, L.F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2021, 39, 462–471. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. Standley MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Hijmans, R.J. raster: Geographic Data Analysis and Modeling. R Package Version 3.6-3. 2022. Available online: https://CRAN.R-project.org/package=raster (accessed on 16 November 2022).
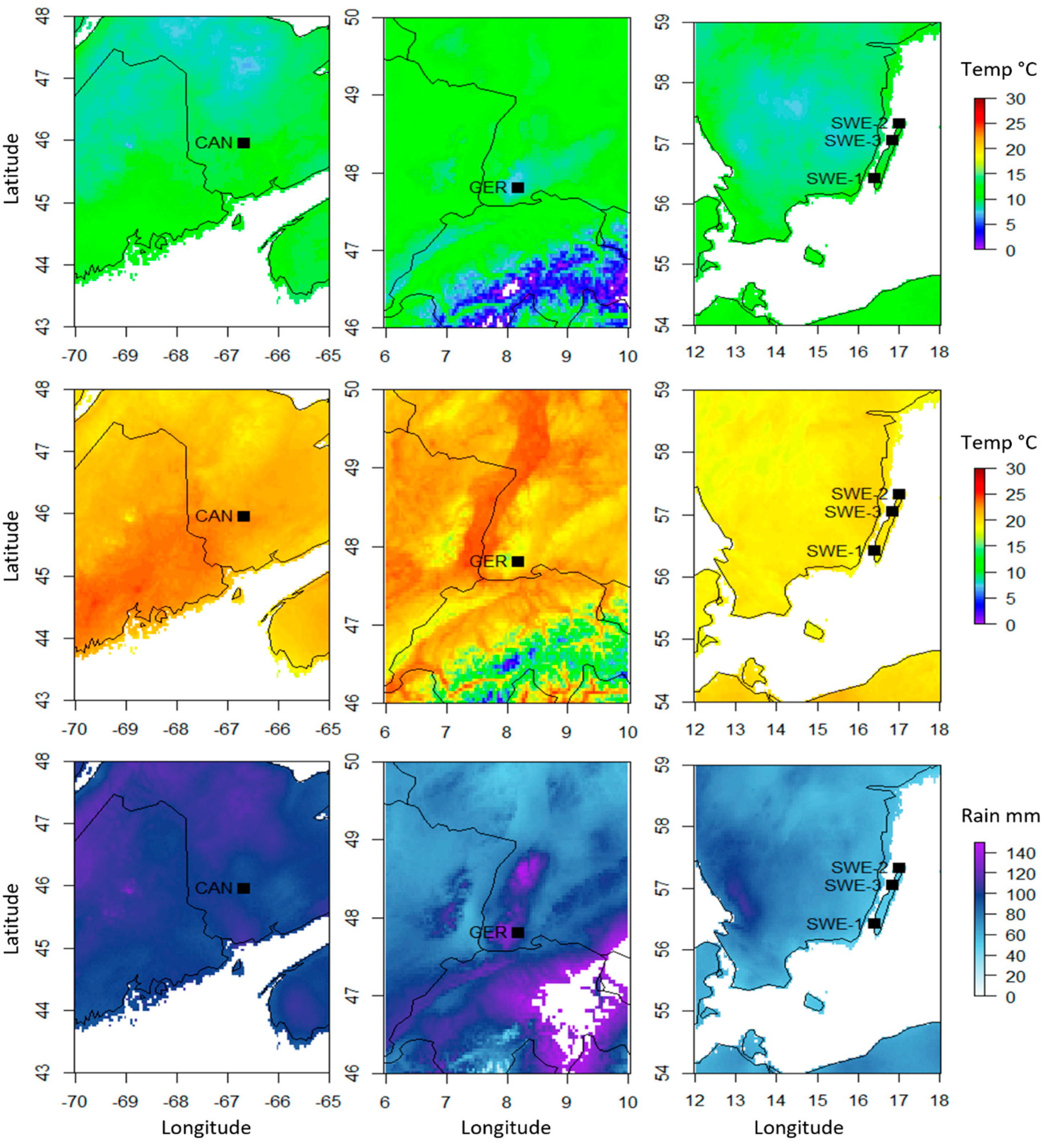

| Sample | Min Temperature (°C) | Max Temperature (°C) | Precipitation (mm) |
|---|---|---|---|
| GER-1 | 7.693678 | 18.45839 | 115.9697 |
| GER-2 | 7.693678 | 18.45839 | 115.9697 |
| GER-3 | 7.693678 | 18.45839 | 115.9697 |
| SWE-1 | 10.89388 | 19.29325 | 46.78941 |
| SWE-2 | 10.5323 | 19.41664 | 45.9303 |
| SWE-3 | 10.11017 | 19.82523 | 49.68862 |
| CAN-May | 10.66182 | 23.49243 | 97.7621 |
| CAN-June | 10.35292 | 23.09298 | 97.39983 |
| CAN-Aug | 10.35292 | 23.09298 | 97.39983 |
| Accession Number | Species | Voucher |
|---|---|---|
| KX676526.1 | Betula pendula Ehrh. Roth | HERB0072 |
| KX230018.1 | Scandosorbus intermedia (Ehrh.) Sennikov | OMHD19M |
| JQ412285.1 | Sambucus nigra L. | BS0146 |
| KX677621.1 | Quercus robur L. | HERB0346 |
| KX229854.1 | Alnus glutinosa (L.) Gaertn. | OMHD05M |
| KX229847.1 | Acer pseudoplatanus L. | OMHD21M |
| EU749296.1 | Betula papyrifera Marshall | OAC:JAG175 |
| HQ593376.1 | Ostrya virginiana (Mill.) K.Koch | AP243 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blatt-Janmaat, K.L.; Neumann, S.; Ziegler, J.; Peters, K. Host Tree and Geography Induce Metabolic Shifts in the Epiphytic Liverwort Radula complanata. Plants 2023, 12, 571. https://doi.org/10.3390/plants12030571
Blatt-Janmaat KL, Neumann S, Ziegler J, Peters K. Host Tree and Geography Induce Metabolic Shifts in the Epiphytic Liverwort Radula complanata. Plants. 2023; 12(3):571. https://doi.org/10.3390/plants12030571
Chicago/Turabian StyleBlatt-Janmaat, Kaitlyn L., Steffen Neumann, Jörg Ziegler, and Kristian Peters. 2023. "Host Tree and Geography Induce Metabolic Shifts in the Epiphytic Liverwort Radula complanata" Plants 12, no. 3: 571. https://doi.org/10.3390/plants12030571
APA StyleBlatt-Janmaat, K. L., Neumann, S., Ziegler, J., & Peters, K. (2023). Host Tree and Geography Induce Metabolic Shifts in the Epiphytic Liverwort Radula complanata. Plants, 12(3), 571. https://doi.org/10.3390/plants12030571








