An Interplay between Mitochondrial and ER Targeting of a Bacterial Signal Peptide in Plants
Abstract
:1. Introduction
1.1. Intracellular Protein Targeting in Plant Systems
1.2. Role of the ER in Unconventional Targeting Pathways
1.3. The Processing of Bacterial Signal Peptides in Plant Cells
1.4. Targeting of the Bacterial Enzyme PmoB
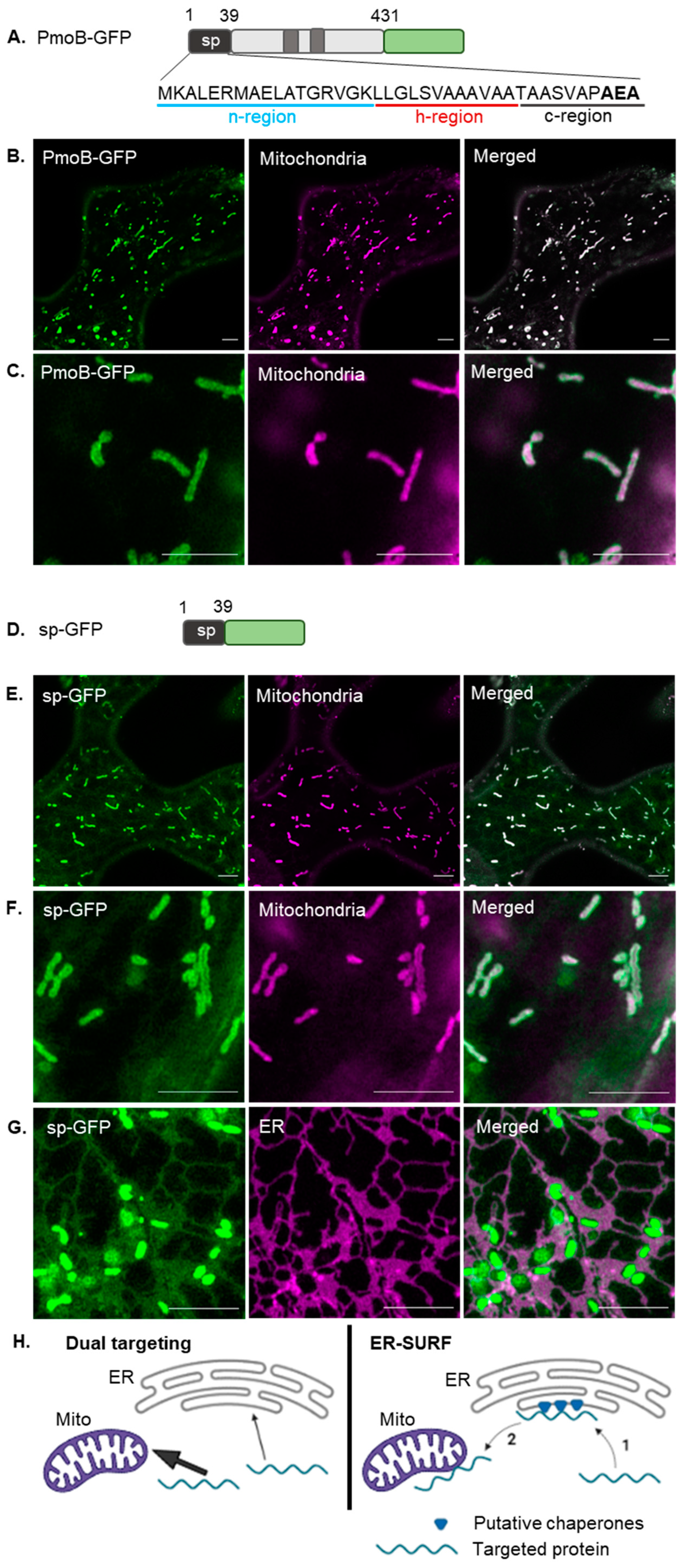
2. Results
2.1. Mitochondrial Targeting of a Bacterial Signal Peptide in Plants
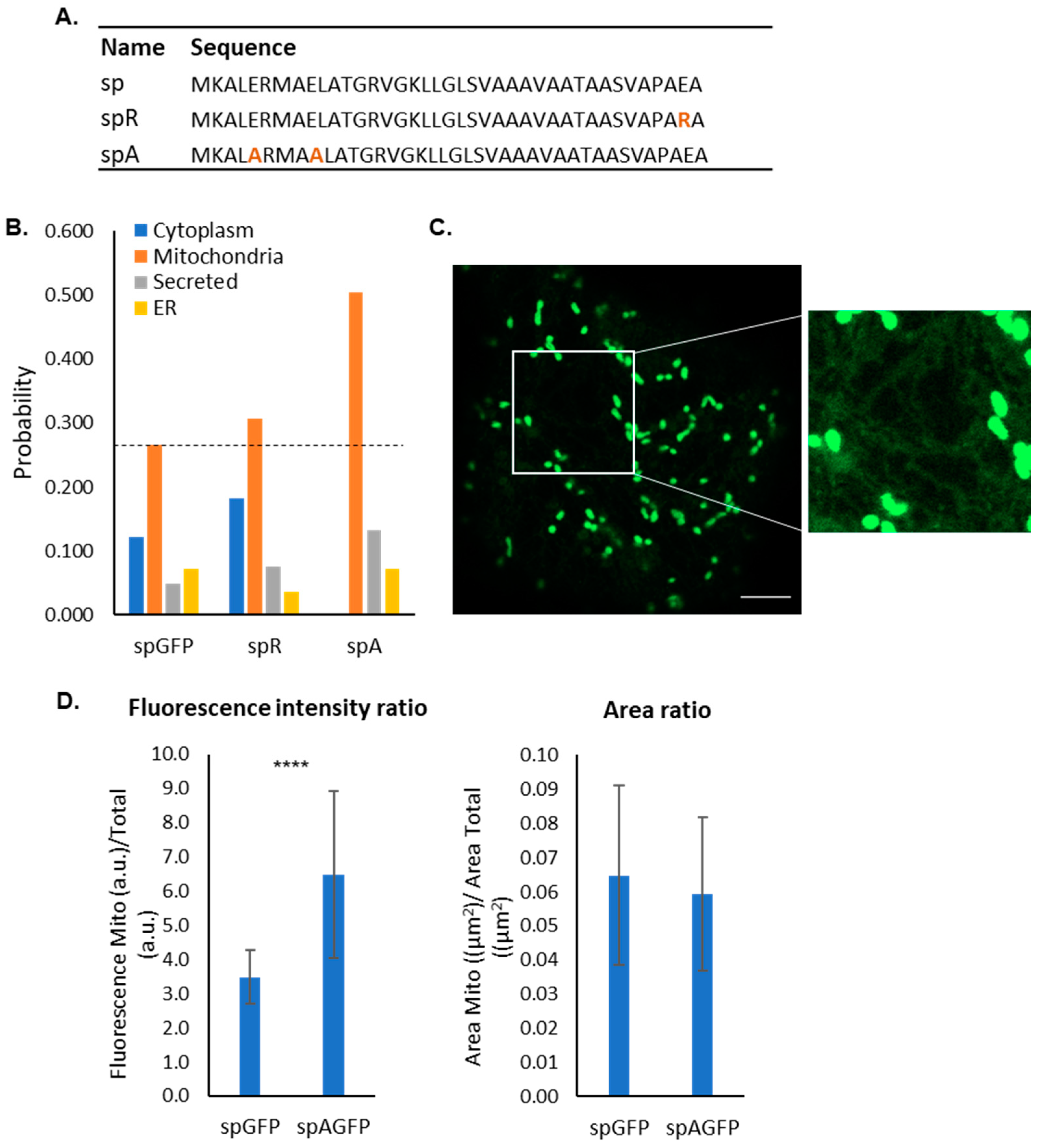
2.2. Sequence Analysis of the Bacterial Signal Peptide Expressed in Plants
2.3. Bioinformatic Analysis of Mutants
2.4. Impact of Negative Residues on the Mitochondrial Targeting Capacity of the PmoB Signal Peptide
3. Discussion
3.1. Targeting of Bacterial Signal Peptides in Plants
3.2. Dual Localisation between the Mitochondria and ER
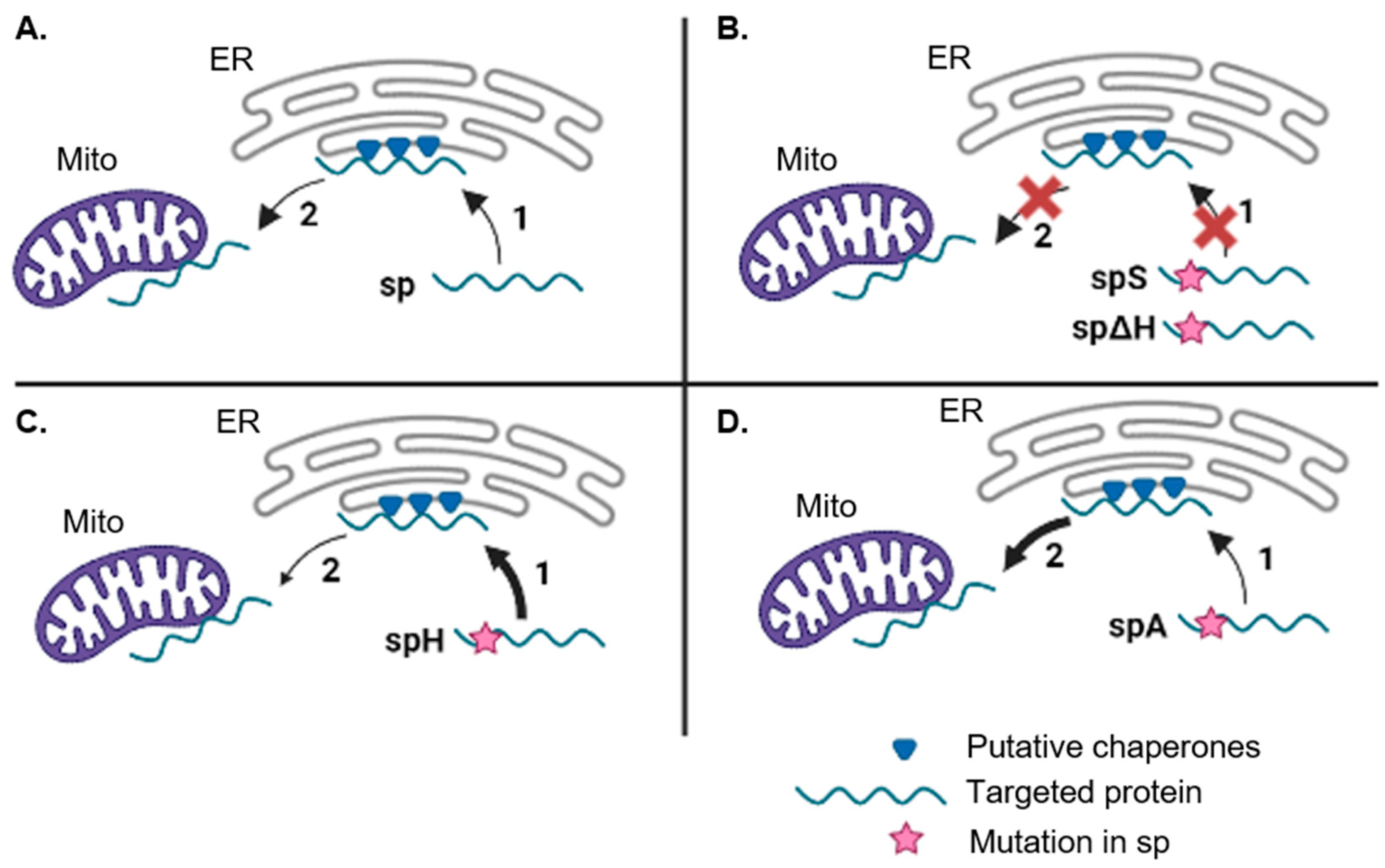
3.3. The PmoB Signal Peptide might Be Targeting Proteins via the ER-SURF Pathway
4. Materials and Methods
4.1. Vector Construction
4.2. Transient Transformation of Tobacco
4.3. Confocal Microscopy
4.4. Subcellular Localisation Prediction
4.5. Mitochondrial Targeting Efficiency Image Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shanmugaraj, B.; Bulaon, C.J.I.; Phoolcharoen, W. Plant Molecular Farming: A Viable Platform for Recombinant Biopharmaceutical Production. Plants 2020, 9, 842. [Google Scholar] [CrossRef]
- Rozov, S.M.; Deineko, E.V. Increasing the Efficiency of the Accumulation of Recombinant Proteins in Plant Cells: The Role of Transport Signal Peptides. Plants 2022, 11, 2561. [Google Scholar] [CrossRef] [PubMed]
- Vitale, A.; Pedrazzini, E. Recombinant Pharmaceuticals from Plants: The Plant Endomembrane System as Bioreactor. Mol. Interv. 2005, 5, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Barbante, A.; Irons, S.; Hawes, C.; Frigerio, L.; Vitale, A.; Pedrazzini, E. Anchorage to the Cytosolic Face of the Endoplasmic Reticulum Membrane: A New Strategy to Stabilize a Cytosolic Recombinant Antigen in Plants. Plant Biotechnol. J. 2008, 6, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.S.; Tilbrook, K.; Warden, A.C.; Campbell, P.C.; Rolland, V.; Singh, S.P.; Wood, C.C. Expression of 16 Nitrogenase Proteins within the Plant Mitochondrial Matrix. Front. Plant Sci. 2017, 8, 287. [Google Scholar] [CrossRef] [Green Version]
- Strand, S.E.; Zhang, L.; Flury, M. Theoretical Analysis of Engineered Plants for Control of Atmospheric Nitrous Oxide and Methane by Modification of the Mitochondrial Proteome. ACS Sustain. Chem. Eng. 2022, 10, 5441–5452. [Google Scholar] [CrossRef]
- von Heijne, G. Signal Sequences. The Limits of Variation. J. Mol. Biol. 1985, 184, 99–105. [Google Scholar] [CrossRef]
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A Comprehensive Review of Signal Peptides: Structure, Roles, and Applications. Eur. J. Cell Biol. 2018, 97, 422–441. [Google Scholar] [CrossRef]
- Rapoport, T.A.; Li, L.; Park, E. Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell Dev. Biol. 2017, 33, 369–390. [Google Scholar] [CrossRef]
- Foresti, O.; Denecke, J. Intermediate Organelles of the Plant Secretory Pathway: Identity and Function. Traffic 2008, 9, 1599–1612. [Google Scholar] [CrossRef]
- Wojcik, S.; Kriechbaumer, V. Go Your Own Way: Membrane-Targeting Sequences. Plant Physiol. 2021, 185, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Shodai, T.; Muto, T.; Mihara, K.; Torii, H.; Nishikawa, S.I.; Endo, T.; Kohda, D. Structural Basis of Presequence Recognition by the Mitochondrial Protein Import Receptor Tom20. Cell 2000, 100, 551–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.W.; Lee, S.; Lee, J.; Woo, S.; Razzak, M.A.; Vitale, A.; Hwang, I. Molecular Mechanism of the Specificity of Protein Import into Chloroplasts and Mitochondria in Plant Cells. Mol. Plant 2019, 12, 951–966. [Google Scholar] [CrossRef] [PubMed]
- Carrie, C.; Whelan, J. Widespread Dual Targeting of Proteins in Land Plants: When, Where, How and Why. Plant Signal. Behav. 2013, 8, 23257241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berglund, A.K.; Spånning, E.; Biverståhl, H.; Maddalo, G.; Tellgren-Roth, C.; Mäler, L.; Glaser, E. Dual Targeting to Mitochondria and Chloroplasts: Characterization of Thr–TRNA Synthetase Targeting Peptide. Mol. Plant 2009, 2, 1298–1309. [Google Scholar] [CrossRef]
- Dinur-Mills, M.; Tal, M.; Pines, O. Dual Targeted Mitochondrial Proteins Are Characterized by Lower MTS Parameters and Total Net Charge. PLoS ONE 2008, 3, e2161. [Google Scholar] [CrossRef] [Green Version]
- Regev-Rudzki, N.; Yogev, O.; Pines, O. The Mitochondrial Targeting Sequence Tilts the Balance between Mitochondrial and Cytosolic Dual Localization. J. Cell Sci. 2008, 121, 2423–2431. [Google Scholar] [CrossRef] [Green Version]
- Jarvis, P. Targeting of Nucleus-Encoded Proteins to Chloroplasts in Plants. New Phytol. 2008, 179, 257–285. [Google Scholar] [CrossRef]
- Bykov, Y.S.; Rapaport, D.; Herrmann, J.M.; Schuldiner, M. Cytosolic Events in the Biogenesis of Mitochondrial Proteins. Trends Biochem. Sci. 2020, 45, 650–667. [Google Scholar] [CrossRef]
- Hansen, K.G.; Aviram, N.; Laborenz, J.; Bibi, C.; Meyer, M.; Spang, A.; Schuldiner, M.; Herrmann, J.M. An ER Surface Retrieval Pathway Safeguards the Import of Mitochondrial Membrane Proteins in Yeast. Science 2018, 361, 1118–1122. [Google Scholar] [CrossRef]
- Schuldiner, M.; Metz, J.; Schmid, V.; Denic, V.; Rakwalska, M.; Schmitt, H.D.; Schwappach, B.; Weissman, J.S. The GET Complex Mediates Insertion of Tail-Anchored Proteins into the ER Membrane. Cell 2008, 134, 634. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Schuldiner, M.; Herrmann, J.M. ER-SURF: Riding the Endoplasmic Reticulum Surface to Mitochondria. Int. J. Mol. Sci. 2021, 22, 9655. [Google Scholar] [CrossRef] [PubMed]
- Kudva, R.; Denks, K.; Kuhn, P.; Vogt, A.; Müller, M.; Koch, H.G. Protein Translocation across the Inner Membrane of Gram-Negative Bacteria: The Sec and Tat Dependent Protein Transport Pathways. Res. Microbiol. 2013, 164, 505–534. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, T.A. Protein Translocation across the Eukaryotic Endoplasmic Reticulum and Bacterial Plasma Membranes. Nature 2007, 450, 663–669. [Google Scholar] [CrossRef]
- Hall, J.; Hazlewood, G.P.; Surani, M.A.; Hirst, B.H.; Gilbert, H.J. Eukaryotic and Prokaryotic Signal Peptides Direct Secretion of a Bacterial Endoglucanase by Mammalian Cells. J. Biol. Chem. 1990, 265, 19996–19999. [Google Scholar] [CrossRef]
- Nagamine, T.; Kawasaki, Y.; Iizuka, T.; Okano, K.; Matsumoto, S.; Choudary, P.V. Functional Characterization of Bacterial Signal Peptide OmpA in a Baculovirus-Mediated Expression System. Cell Struct. Funct. 2003, 28, 131–142. [Google Scholar] [CrossRef] [Green Version]
- Moeller, L.; Gan, Q.; Wang, K. A Bacterial Signal Peptide Is Functional in Plants and Directs Proteins to the Secretory Pathway. J. Exp. Bot. 2009, 60, 3337–3352. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Al-Babili, S.; Klöti, A.; Zhang, J.; Lucca, P.; Beyer, P.; Potrykus, I. Engineering the Provitamin A (Beta-Carotene) Biosynthetic Pathway into (Carotenoid-Free) Rice Endosperm. Science 2000, 287, 303–305. [Google Scholar] [CrossRef] [Green Version]
- Paine, J.A.; Shipton, C.A.; Chaggar, S.; Howells, R.M.; Kennedy, M.J.; Vernon, G.; Wright, S.Y.; Hinchliffe, E.; Adams, J.L.; Silverstone, A.L.; et al. Improving the Nutritional Value of Golden Rice through Increased Pro-Vitamin A Content. Nat. Biotechnol. 2005, 23, 482–487. [Google Scholar] [CrossRef]
- Jani, D.; Singh Meena, L.; Rizwan-Ul-Haq, Q.M.; Singh, Y.; Sharma, A.K.; Tyagi, A.K. Expression of Cholera Toxin B Subunit in Transgenic Tomato Plants. Transgenic Res. 2002, 11, 447–454. [Google Scholar] [CrossRef]
- Sahoo, A.; Mandal, A.K.; Dwivedi, K.; Kumar, V. A Cross Talk between the Immunization and Edible Vaccine: Current Challenges and Future Prospects. Life Sci. 2020, 261, 118343. [Google Scholar] [CrossRef] [PubMed]
- Murrell, J.C.; Gilbert, B.; McDonald, I.R. Molecular Biology and Regulation of Methane Monooxygenase. Arch. Microbiol. 2000, 173, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, R.L.; Rosenzweig, A.C. Crystal Structure of a Membrane-Bound Metalloenzyme That Catalyses the Biological Oxidation of Methane. Nature 2005, 43, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.V.; Walsh, M.L.; Chen, L.B. Localization of Mitochondria in Living Cells with Rhodamine 123. Cell Biol. 1980, 77, 990–994. [Google Scholar] [CrossRef] [Green Version]
- Hawes, C.; Wang, P.; Kriechbaumer, V. Labeling the ER for Light and Fluorescence Microscopy. Methods Mol. Biol. 2018, 1691, 1–14. [Google Scholar]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing Mitochondrial Proteins: Machineries and Mechanisms. Cell 2009, 138, 628. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Taylor, N.L.; Whelan, J.; Millar, A.H. Refining the Definition of Plant Mitochondrial Presequences through Analysis of Sorting Signals, N-Terminal Modifications, and Cleavage Motifs. Plant Physiol. 2009, 150, 1272. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Sparkes, I.; Gattolin, S.; Dzimitrowicz, N.; Roberts, L.M.; Hawes, C.; Frigerio, L. An Arabidopsis Reticulon and the Atlastin Homologue RHD3-Like2 Act Together in Shaping the Tubular Endoplasmic Reticulum. New Phytol. 2013, 197, 481–489. [Google Scholar] [CrossRef] [Green Version]
- von Heijne, G. Mitochondrial Targeting Sequences May Form Amphiphilic Helices. EMBO J. 1986, 5, 1335–1342. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A Protein Secondary Structure Prediction Server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 Years On. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A Web Server to Screen Sequences with Specific α-Helical Properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Lee, D.W.; Yoo, Y.J.; Duncan, O.; Oh, Y.J.; Lee, Y.J.; Lee, G.; Whelan, J.; Hwang, I. Mitochondrial Targeting of the Arabidopsis F1-ATPase γ-Subunit via Multiple Compensatory and Synergistic Presequence Motifs. Plant Cell 2012, 24, 5037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terada, K.; Gimenez-Dejoz, J.; Kurita, T.; Oikawa, K.; Uji, H.; Tsuchiya, K.; Numata, K. Synthetic Mitochondria-Targeting Peptides Incorporating α-Aminoisobutyric Acid with a Stable Amphiphilic Helix Conformation in Plant Cells. ACS Biomater. Sci. Eng. 2021, 7, 1475–1484. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, Y.; Tsuji, J.; Fu, S.C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved Prediction of Mitochondrial Targeting Sequences and Their Cleavage Sites. Mol. Cell. Proteomics 2015, 14, 1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Wang, D.; Yao, Y.; Eubel, H.; Künzler, P.; Møller, I.M.; Xu, D. MULocDeep: A Deep-Learning Framework for Protein Subcellular and Suborganellar Localization Prediction with Residue-Level Interpretation. Comput. Struct. Biotechnol. J. 2021, 19, 4825–4839. [Google Scholar] [CrossRef]
- Murcha, M.W.; Kmiec, B.; Kubiszewski-Jakubiak, S.; Teixeira, P.F.; Glaser, E.; Whelan, J. Protein Import into Plant Mitochondria: Signals, Machinery, Processing, and Regulation. J. Exp. Bot. 2014, 65, 6301–6335. [Google Scholar] [CrossRef] [Green Version]
- Koo, C.W.; Rosenzweig, A.C. Biochemistry of Aerobic Biological Methane Oxidation. Chem. Soc. Rev. 2021, 50, 3424–3436. [Google Scholar] [CrossRef]
- Stern, B.; Olsen, L.C.; Tröße, C.; Ravneberg, H.; Pryme, I.F. Improving Mammalian Cell Factories: The Selection of Signal Peptide Has a Major Impact on Recombinant Protein Synthesis and Secretion in Mammalian Cells. Trends Cell Mol. Biol. 2007, 2, 1–17. [Google Scholar]
- Lee, D.W.; Lee, S.; Min, C.K.; Park, C.; Kim, J.M.; Hwang, C.S.; Park, S.K.; Cho, N.H.; Hwang, I. Cross-Species Functional Conservation and Possible Origin of the N-Terminal Specificity Domain of Mitochondrial Presequences. Front. Plant Sci. 2020, 11, 64. [Google Scholar] [CrossRef]
- Yogev, O.; Pines, O. Dual Targeting of Mitochondrial Proteins: Mechanism, Regulation and Function. Biochim. Biophys. Acta-Biomembr. 2011, 1808, 1012–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bader, G.; Enkler, L.; Araiso, Y.; Hemmerle, M.; Binko, K.; Baranowska, E.; De Craene, J.O.; Ruer-Laventie, J.; Pieters, J.; Tribouillard-Tanvier, D.; et al. Assigning Mitochondrial Localization of Dual Localized Proteins Using a Yeast Bi-Genomic Mitochondrial-Split-GFP. Elife 2020, 9, e56649. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.H.; Lee, J.H.; Chandrasekar, S.; Shan, S.-O. A Ribosome-Associated Chaperone Enables Substrate Triage in a Cotranslational Protein Targeting Complex. Nat. Commun. 2020, 11, 5840. [Google Scholar] [CrossRef]
- Brandizzi, F.; Hanton, S.; DaSilva, L.L.P.; Boevink, P.; Evans, D.; Oparka, K.; Denecke, J.; Hawes, C. ER Quality Control Can Lead to Retrograde Transport from the ER Lumen to the Cytosol and the Nucleoplasm in Plants. Plant J. 2003, 34, 269–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gateway ® Technology A Universal Technology to Clone DNA Sequences for Functional Analysis and Expression in Multiple Systems; Thermo Fisher Scientific: Waltham, MA, USA, 2003; Volume 70.
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, Transient Expression of Fluorescent Fusion Proteins in Tobacco Plants and Generation of Stably Transformed Plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spatola Rossi, T.; Kriechbaumer, V. An Interplay between Mitochondrial and ER Targeting of a Bacterial Signal Peptide in Plants. Plants 2023, 12, 617. https://doi.org/10.3390/plants12030617
Spatola Rossi T, Kriechbaumer V. An Interplay between Mitochondrial and ER Targeting of a Bacterial Signal Peptide in Plants. Plants. 2023; 12(3):617. https://doi.org/10.3390/plants12030617
Chicago/Turabian StyleSpatola Rossi, Tatiana, and Verena Kriechbaumer. 2023. "An Interplay between Mitochondrial and ER Targeting of a Bacterial Signal Peptide in Plants" Plants 12, no. 3: 617. https://doi.org/10.3390/plants12030617
APA StyleSpatola Rossi, T., & Kriechbaumer, V. (2023). An Interplay between Mitochondrial and ER Targeting of a Bacterial Signal Peptide in Plants. Plants, 12(3), 617. https://doi.org/10.3390/plants12030617






