Canola with Stacked Genes Shows Moderate Resistance and Resilience against a Field Population of Plasmodiophora brassicae (Clubroot) Pathotype X
Abstract
:1. Introduction
2. Materials and Methods
2.1. Resistance to Pathotype X
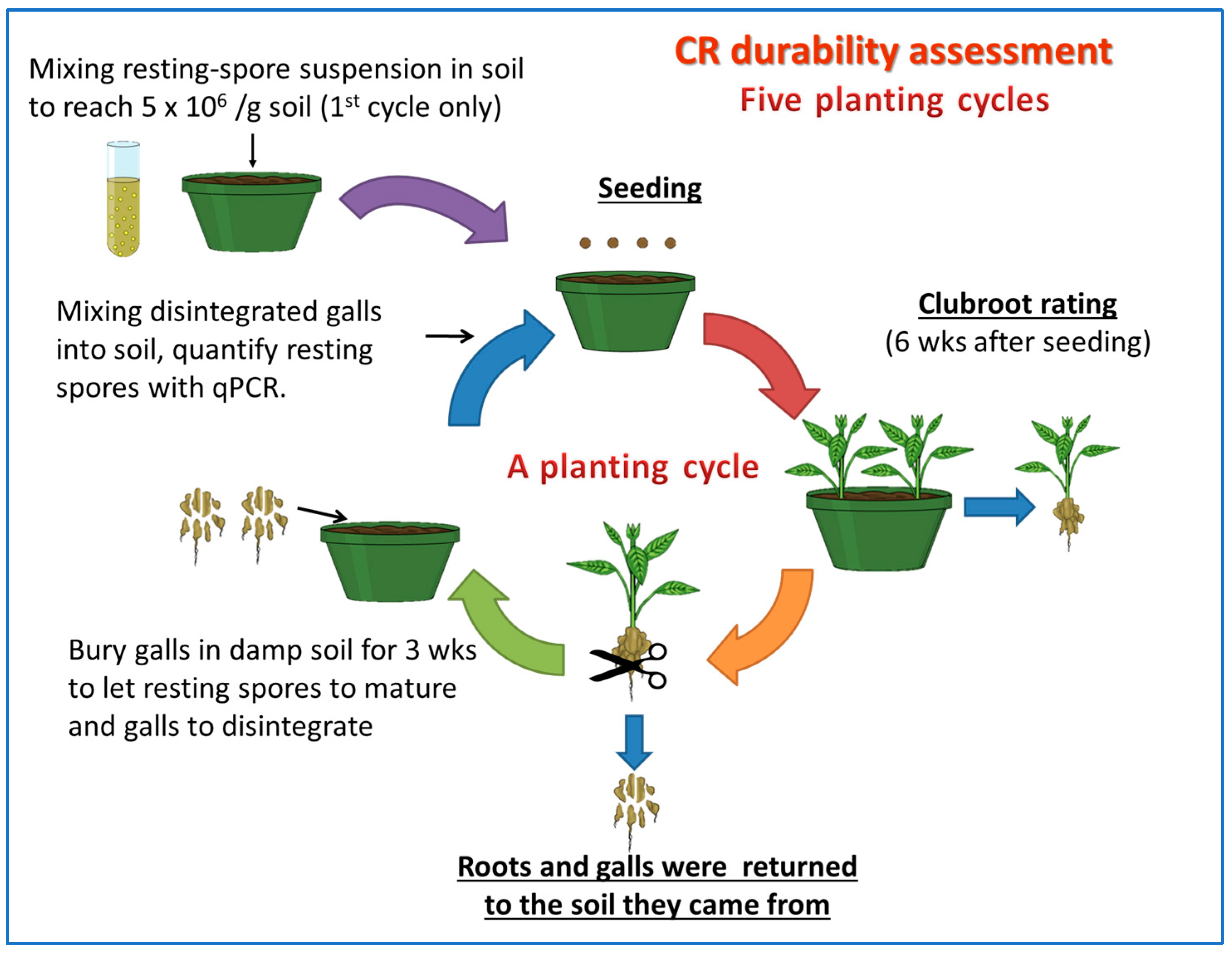
2.2. Durability of Resistance
2.3. Quantification of Resting Spores
2.4. Data Analysis
3. Results
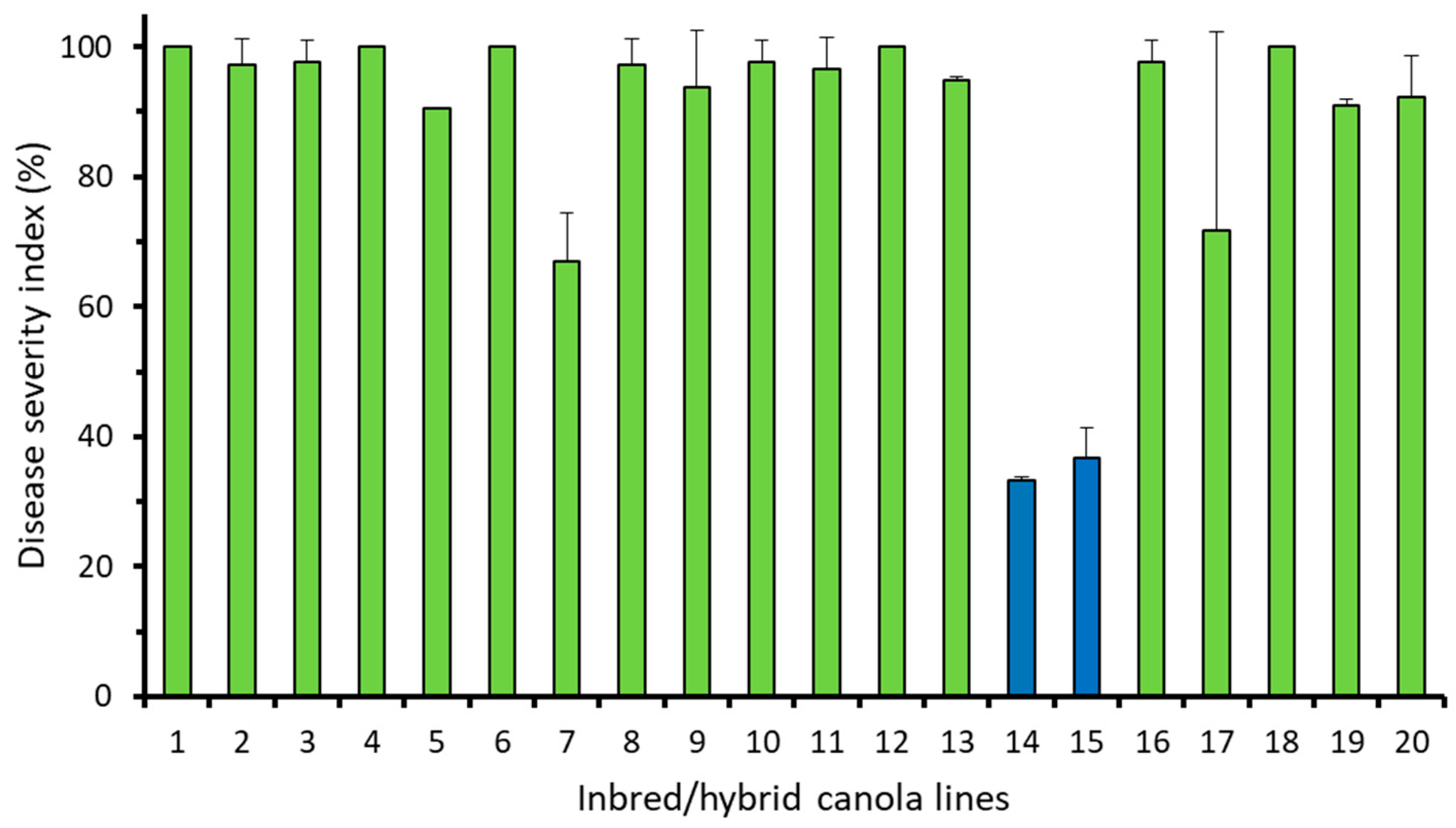
3.1. Resistance to Pathotype X
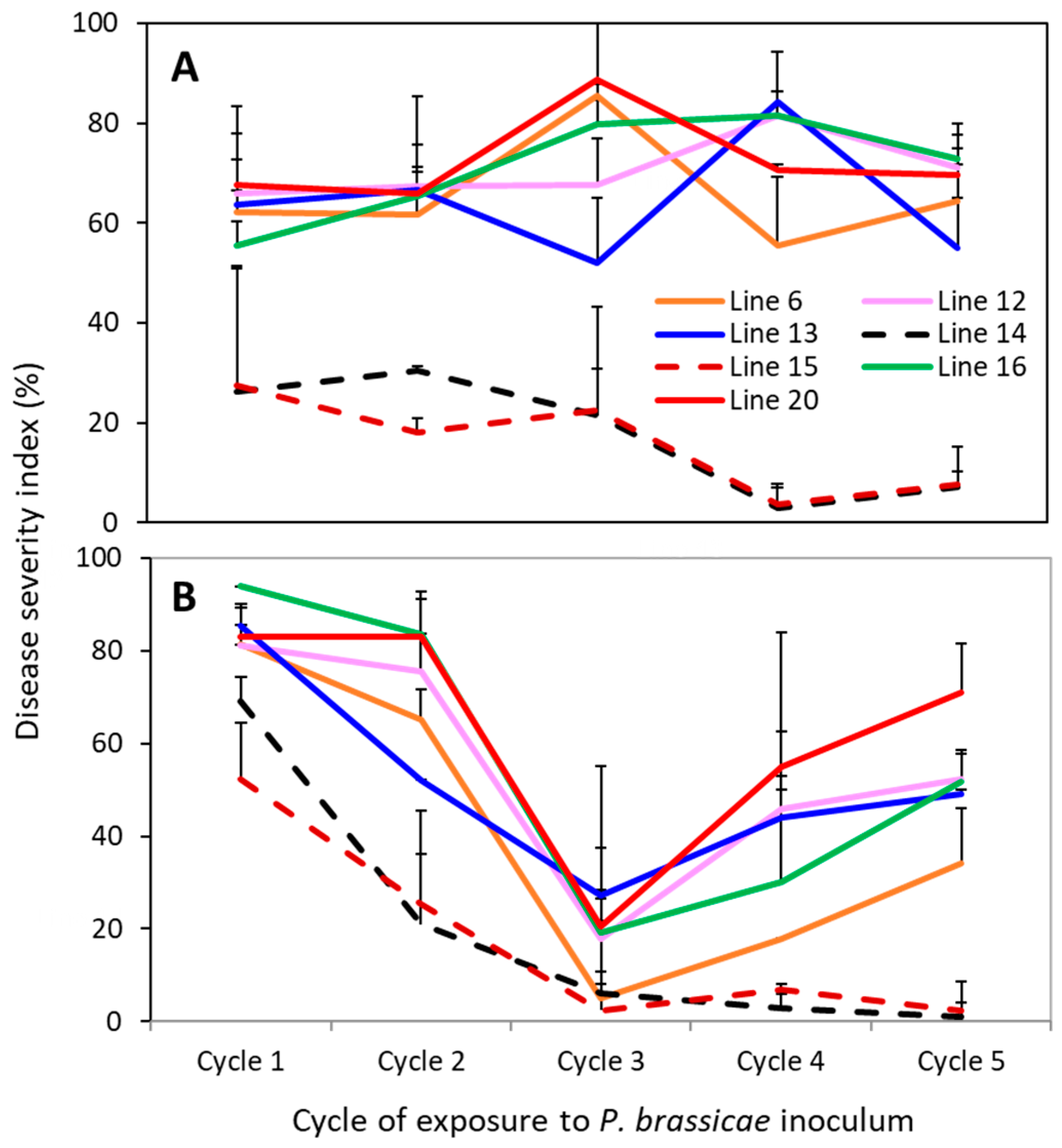
3.2. Durability of Clubroot Resistance
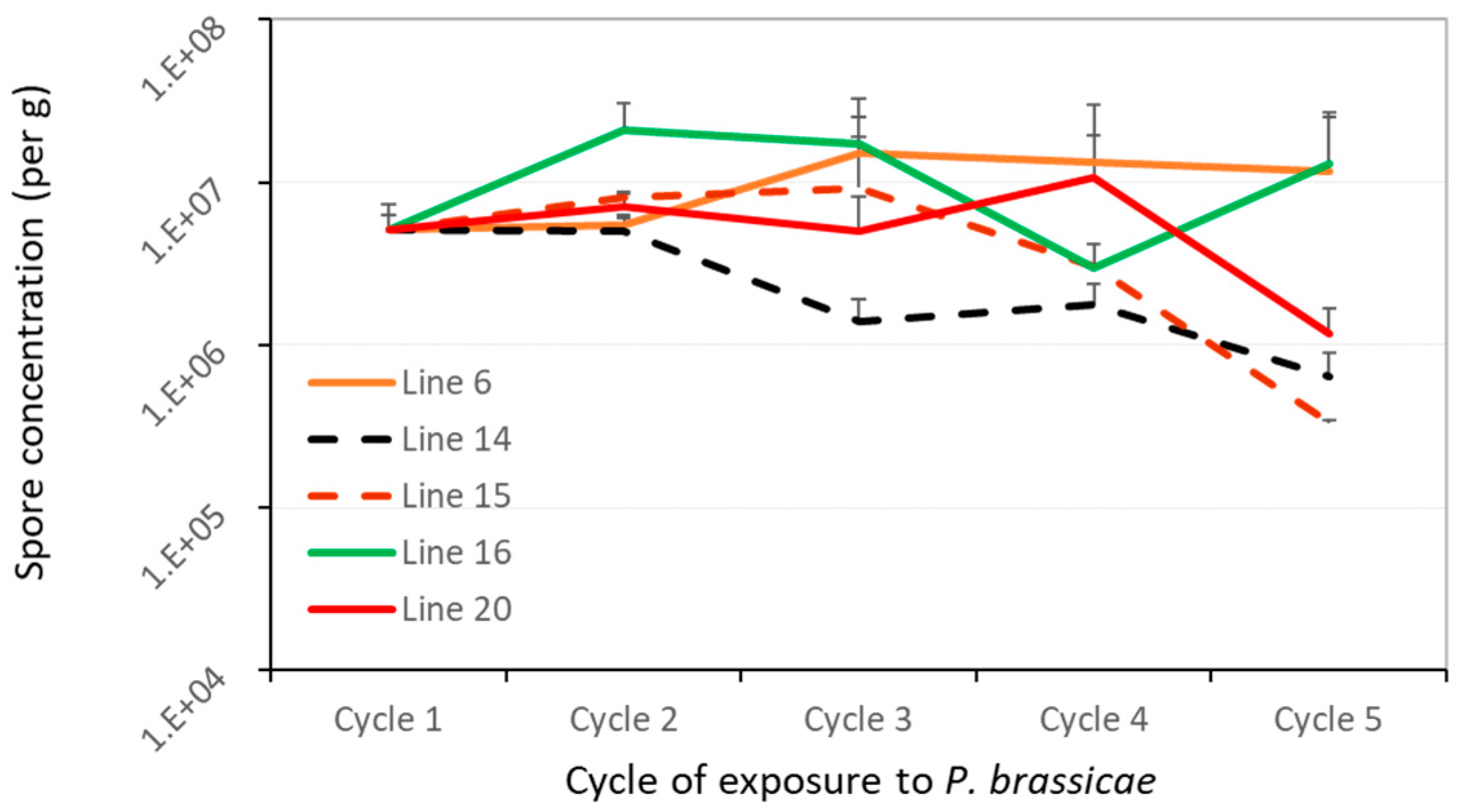
3.3. Quantification of Spores
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rempel, C.B.; Hutton, S.N.; Jurke, C.J. Clubroot and the importance of canola in Canada. Can. J. Plant Pathol. 2014, 36, 19–26. [Google Scholar] [CrossRef]
- Kageyama, K.; Asano, T. Life cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 203–211. [Google Scholar] [CrossRef]
- Peng, G.; Pageau, D.; Strelkov, S.E.; Gossen, B.D.; Hwang, S.E.; Lahlali, R. A >2-year rotation reduces Plasmodiophora brassicae resting spores in soil and the impact of clubroot on canola. Eur. J. Agron. 2015, 70, 78–84. [Google Scholar] [CrossRef]
- Gossen, B.D.; Al-Daoud, F.; Dumonceaux, T.; Dalton, J.A.; Peng, G.; Pageau, D.; McDonald, M.R. Comparison of molecular techniques for estimation of resting spores of Plasmodiophora brassicae in soil. Plant Pathol. 2019, 68, 954–961. [Google Scholar] [CrossRef]
- Hwang, S.F.; Ahmed, H.U.; Zhou, Q.X.; Strelkov, S.E.; Gossen, B.D.; Peng, G.; Turnbullet, G.D. Assessment of the impact of resistant and susceptible canola on Plasmodiophora brassicae inoculum potential. Plant Pathol. 2012, 61, 945–952. [Google Scholar] [CrossRef]
- Hwang, S.F.; Ahmed, H.U.; Zhou, Q.X.; Rashid, A.; Strelkov, S.E.; Gossen, B.D.; Peng, G.; Turnbullet, G.D. Effect of susceptible and resistant canola plants on Plasmodiophora brassicae resting spore populations in the soil. Plant Pathol. 2013, 62, 346–354. [Google Scholar] [CrossRef]
- Hwang, S.F.; Ahmed, H.U.; Zhou, Q.X.; Fu, H.; Fredua-Agyeman, R.; Strelkov, S.E.; Gossen, B.D.; Peng, G. Influence of resistant cultivars and crop intervals on clubroot of canola. Can. J. Plant Sci. 2019, 99, 862–872. [Google Scholar] [CrossRef]
- Fredua-Agyeman, R.; Rahman, H. Mapping of the clubroot disease resistance in spring Brassica napus canola introgressed from European winter canola cv. ‘Mendel’. Euphytica 2016, 211, 201–213. [Google Scholar] [CrossRef]
- Williams, P.H. A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathol. 1966, 56, 624–626. [Google Scholar]
- Strelkov, S.E.; Hwang, S.F. Clubroot in the Canadian canola crop: 10 years into the outbreak. Can. J. Plant Pathol. 2014, 36, 27–36. [Google Scholar] [CrossRef]
- Deora, A.; Gossen, B.D.; McDonald, M.R. Infection and development of Plasmodiophora brassicae in resistant and susceptible canola cultivars. Can. J. Plant Pathol. 2012, 34, 239–247. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Hwang, S.F.; Manolii, V.P.; Cao, T.; Feindel, D. Emergence of new virulence phenotypes of Plasmodiophora brassicae on canola (Brassica napus) in Alberta, Canada. Eur. J. Plant Pathol. 2016, 145, 517–529. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Hwang, S.F.; Manolii, V.P.; Cao, T.; Fredua-Agyeman, R.; Harding, M.; Peng, G.; Gossen, B.D.; Mcdonald, M.R.; Feindel, D. Virulence and pathotype classification of Plasmodiophora brassicae populations collected from clubroot resistant canola (Brassica napus) in Canada. Can. J. Plant Pathol. 2018, 40, 284–298. [Google Scholar] [CrossRef]
- LeBoldus, J.M.; Manolii, V.P.; Turkington, T.K.; Strelkov, S.E. Adaptation to Brassica host genotypes by a single-spore isolate and population of Plasmodiophora brassicae (clubroot). Plant Dis. 2012, 96, 833–838. [Google Scholar] [CrossRef]
- Hirai, M. Genetic analysis of clubroot resistance in Brassica crops. Breed. Sci. 2006, 56, 223–229. [Google Scholar] [CrossRef]
- Li, Z.K.; Sanchez, A.; Angeles, E.; Singh, S.; Domingo, J.; Huang, N.; Khush, G.S. Are the dominant and recessive plant disease resistance genes similar? A case study of rice R genes and Xanthomonas oryzae pv. oryzae races. Genetics 2001, 159, 757–765. [Google Scholar] [CrossRef]
- Balesdent, M.H.; Gautier, A.; Plissonneau, C.; Le Meur, L.; Loiseau, A.; Leflon, M.; Carpezat, J.; Pinochet, X.; Rouxel, T. Twenty years of Leptosphaeria maculans population survey in France suggests pyramiding Rlm3 and Rlm7 in rapeseed is a risky resistance management strategy. Phytopathology 2022, 112. [Google Scholar] [CrossRef]
- Brun, H.; Chevre, A.M.; Fitt, B.D.; Powers, S.; Besnard, A.L.; Ermel, M.; Huteau, V.; Marquer, B.; Eber, F.; Renard, M.; et al. Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 2010, 185, 285–299. [Google Scholar] [CrossRef]
- Matsumoto, E.; Ueno, H.; Aruga, D.; Sakamoto, K.; Hayashida, N. Accumulation of three clubroot resistance genes through marker-assisted selection in Chinese cabbage (Brassica rapa L. ssp. pekinensis). J. Jpn. Soc. Hort. Sci. 2012, 81, 184–190. [Google Scholar] [CrossRef]
- Hasan, M.J.; Rahman, H. Genetics and molecular mapping of resistance to Plasmodiophora brassicae pathotypes 2, 3, 5, 6, and 8 in rutabaga (Brassica napus var. napobrassica). Genome 2016, 59, 805–815. [Google Scholar] [CrossRef]
- Karim, M.M.; Dakouri, A.; Zhang, Y.; Chen, Q.; Peng, G.; Strelkov, S.E.; Gossen, B.D.; Yu, F. Two clubroot-resistance genes, Rcr3 and Rcr9WA mapped in Brassica rapa using bulk segregant RNA sequencing. Int. J. Mol. Sci. 2020, 21, 5033. [Google Scholar] [CrossRef]
- Suwabe, K.; Suzuki, G.; Nunome, T.; Hatakeyama, K.; Mukai, Y.; Fukuoka, H.; Matsumoto, S. Microstructure of a Brassica rapa genome segment homoeologous to the resistance gene cluster on Arabidopsis chromosome 4. Breed. Sci. 2012, 62, 170–177. [Google Scholar] [CrossRef]
- Hatakeyama, K.; Suwabe, K.; Tomita, R.N.; Kato, T.; Nunome, T.; Fukuoka, H.; Matsumoto, S. Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS ONE 2013, 8, e54745. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Song, T.; Falk, K.C.; Zhang, X.; Liu, X.; Chang, A.; Rachid, L.; McGregor, L.; Gossen, B.D.; Yu, F.; et al. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genom. 2014, 15, 1166. [Google Scholar] [CrossRef]
- Diederichsen, E.; Beckmann, J.; Schondelmeier, J.; Dreyer, F. Genetics of clubroot resistance in Brassica napus ‘Mendel’. Acta Hort. 2006, 706, 307–311. [Google Scholar] [CrossRef]
- Matsumoto, E.; Yasui, C.; Ohi, M.; Tsukada, M. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis). Euphytica 1998, 104, 79–86. [Google Scholar] [CrossRef]
- Kato, T.; Hatakeyama, K.; Fukino, N.; Matsumoto, S. Identificaiton of a clubroot resistance locus conferring resistance to a Plasmodiophora brassicae classified into pathotype group 3 in Chinese cabbage (Brassica rapa L.). Breed. Sci. 2012, 62, 282–287. [Google Scholar] [CrossRef]
- Ueno, H.; Matsumoto, E.; Aruga, D.; Kitagawa, S.; Matsumura, H.; Hayashida, N. Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol Biol. 2012, 80, 621–629. [Google Scholar] [CrossRef]
- Kato, T.; Hatakeyama, K.; Fukino, N.; Matsumoto, S. Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa. Breed. Sci. 2013, 63, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Kuginuki, Y.; Yoshikawa, H.; Hirai, M. Variation in virulence of Plasmodiophora brassicae in Japan tested with clubroot-resistant cultivars of Chinese cabbage (Brassica rapa L. ssp. pekinensis). Eur. J. Plant Pathol. 1999, 105, 327–332. [Google Scholar] [CrossRef]
- Peng, G.; Falk, K.C.; Gugel, R.; Franke, C.; Yu, F.Q.; James, B.; Strelkov, S.E.; Hwang, S.F.; McGregor, L. Sources of resistance to Plasmodiophora brassicae (clubroot) pathotypes virulent on canola. Can. J. Plant Pathol. 2014, 36, 89–99. [Google Scholar] [CrossRef]
- Rennie, D.C.; Manolii, V.P.; Cao, T.; Hwang, S.F.; Howard, R.J.; Strelkov, S.E. Direct evidence of seed and tuber infestation by Plasmodiophora brassicae and quantification of inoculum loads. Plant Pathol. 2011, 60, 811–819. [Google Scholar] [CrossRef]
- Deora, A.; Gossen, B.D.; Amirsadeghi, S.; McDonald, M.R. A multiplex qPCR assay for detection and quantification of Plasmodiophora brassicae in soil. Plant Dis. 2015, 99, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Lee, J.; Chu, M.; Tonu, N.; Dumonceaux, T.; Gossen, B.D.; Yu, F.; Peng, G. Quantification of Plasmodiophora brassicae resting spores in soils using droplet digital PCR (ddPCR). Plant Dis. 2020, 104, 1188–1194. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhang, Y.; Wang, J.; Chen, Q.; Karim, M.M.; Gossen, B.D.; Peng, G. Identification of two major QTLs in Brassica napus lines with introgressed clubroot resistance from turnip cultivar ECD01. Front. Plant Sci. 2022, 12, 785989. [Google Scholar] [PubMed]
- Holtz, M.D.; Hwang, S.F.; Strelkov, S.E. Genotyping of Plasmodiophora brassicae reveals the presence of distinct populations. BMC Genom. 2018, 19, 254. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Li, Y.; Vossen, J.H.; Visser, R.G.; Jacobsen, E. Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res. 2012, 21, 89–99. [Google Scholar] [CrossRef]
- Fredua-Agyeman, R.; Hwang, S.F.; Strelkov, S.E.; Zhou, Q.X.; Feindel, D. Potential loss of clubroot resistance genes from donor parent Brassica rapa subsp. rapifera (ECD 04) during doubled haploid production. Plant Pathol. 2018, 67, 892–901. [Google Scholar] [CrossRef]
- Sedaghatkish, A.; Gossen, B.D.; Yu, F.; Torkamaneh, D.; McDonald, M.R. Whole genome DNA similarity and population structure of Plasmodiophora brassicae strains in Canada. BMC Genom. 2019, 20, 744. [Google Scholar] [CrossRef]
- Botero-Ramirez, A.; Hwang, S.F.; Strelkov, S.E. Plasmodiophora brassicae inoculum density and spatial patterns at the field level and relation to soil characteristics. Pathogens 2021, 10, 449. [Google Scholar] [CrossRef]
- Al-Daoud, F.; Gossen, B.D.; Mcdonald, M.R. Maturation of resting spores of Plasmodiophora brassicae continues after host cell death. Plant Pathol. 2019, 69, 310–319. [Google Scholar] [CrossRef]
- Drury, S.; Sedaghatkish, A.; Gossen, B.D.; McDonald, M.R. Perennial grasses and common field crops reduce the concentration of resting spores of Plasmodiophora brassicae in soil. Plant Pathol. 2022, 71, 1793–1800. [Google Scholar] [CrossRef]
- Faggian, R.; Strelkov, S.E. Detection and measurement of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 282–288. [Google Scholar] [CrossRef]
- Ernst, T.W.; Kher, S.; Stanton, D.; Rennie, D.C.; Hwang, S.F.; Strelkov, S.E. Plasmodiophora brassicae resting spore dynamics in clubroot resistant canola (Brassica napus) cropping systems. Plant Pathol. 2019, 68, 399–408. [Google Scholar] [CrossRef]
- Hildebrand, P.L.; McRae, K.B. Control of clubroot caused by Plasmodiophora brassicae with nonionic surfactants. Can. J. Plant Pathol. 1998, 20, 1–11. [Google Scholar] [CrossRef]
| Canola Lines | CR Genes Involved | # CR Genes | Location of CR Genes |
|---|---|---|---|
| 1 | CRaM/Crr1rutba | 2 | A03/A08 |
| 2 | CRaM/Crr1rutb | 2 | A03/A08 |
| 3 | CRaM/Crr1rutb | 2 | A03/A08 |
| 4 | CRaM/Crr1rutb | 2 | A03/A08 |
| 5 | CRaM/Crr1rutb | 2 | A03/A08 |
| 6 | CRaM/Crr1rutb | 2 | A03/A08 |
| 7 | CRaM/Crr1rutb | 2 | A03/A08 |
| 8 | CRaM/Crr1rutb | 2 | A03/A08 |
| 9 | Rcr1/CRaM | 2 | A03/A03 |
| 10 | Rcr1//CRaM/Crr1rutbb | 3 | A03//A03/A08 |
| 11 | Rcr1//CRaM/Crr1rutb | 3 | A03//A03/A08 |
| 12 | Rcr1//CRaM/Crr1rutb | 3 | A03//A03/A08 |
| 13 | Rcr1/Rcr1c | 1 | A03/A03 |
| 14 | Crr1rutb/Rcr1 | 2 | A08/A03 |
| 15 | Crr1rutb/CRaM | 2 | A08/A03 |
| 16 | CRaM | 1 | A03 |
| 17 | CRaM//Rcr1/Rcr1 | 2 | A03/A03 |
| 18 | CRaM//CRaM/Crr1rutb | 2 | A03//A03/A08 |
| 19 | CRaM//Crr1rutb/CRaM | 2 | A03//A03/A08 |
| 20 | Crr1rutb | 1 | A08 |
| Canola Lines | CR Genes Involved | # CR Genes | Generation | To Pathotype 3 a | Reaction to L-G3 b |
|---|---|---|---|---|---|
| 6 | CRaM/Crr1rutb | 2 | F1 | Resistant | Susceptible |
| 12 | Rcr1//CRaM/Crr1rutb c | 3 | F1 | Resistant | Susceptible |
| 13 | Rcr1/Rcr1 | 1 | DH d | Resistant | Susceptible |
| 14 | Crr1rutb/Rcr1 | 2 | F1 | Resistant | Moderately resistant |
| 15 | Crr1rutb/CRaM | 2 | F1 | Resistant | Moderately resistant |
| 16 | CRaM | 1 | DH | Resistant | Susceptible |
| 20 | Crr1rutb | 1 | DH | Resistant | Susceptible |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tonu, N.N.; Wen, R.; Song, T.; Guo, X.; Murphy, L.A.; Gossen, B.D.; Yu, F.; Peng, G. Canola with Stacked Genes Shows Moderate Resistance and Resilience against a Field Population of Plasmodiophora brassicae (Clubroot) Pathotype X. Plants 2023, 12, 726. https://doi.org/10.3390/plants12040726
Tonu NN, Wen R, Song T, Guo X, Murphy LA, Gossen BD, Yu F, Peng G. Canola with Stacked Genes Shows Moderate Resistance and Resilience against a Field Population of Plasmodiophora brassicae (Clubroot) Pathotype X. Plants. 2023; 12(4):726. https://doi.org/10.3390/plants12040726
Chicago/Turabian StyleTonu, Nazmoon Naher, Rui Wen, Tao Song, Xiaowei Guo, Lee Anne Murphy, Bruce Dean Gossen, Fengqun Yu, and Gary Peng. 2023. "Canola with Stacked Genes Shows Moderate Resistance and Resilience against a Field Population of Plasmodiophora brassicae (Clubroot) Pathotype X" Plants 12, no. 4: 726. https://doi.org/10.3390/plants12040726
APA StyleTonu, N. N., Wen, R., Song, T., Guo, X., Murphy, L. A., Gossen, B. D., Yu, F., & Peng, G. (2023). Canola with Stacked Genes Shows Moderate Resistance and Resilience against a Field Population of Plasmodiophora brassicae (Clubroot) Pathotype X. Plants, 12(4), 726. https://doi.org/10.3390/plants12040726








