The ROP2 GTPase Participates in Nitric Oxide (NO)-Induced Root Shortening in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. The Roots of Arabidopsis rop2-1 Mutant Seedlings Are Less Sensitive to Exogenous NO Than That of the Wild Type
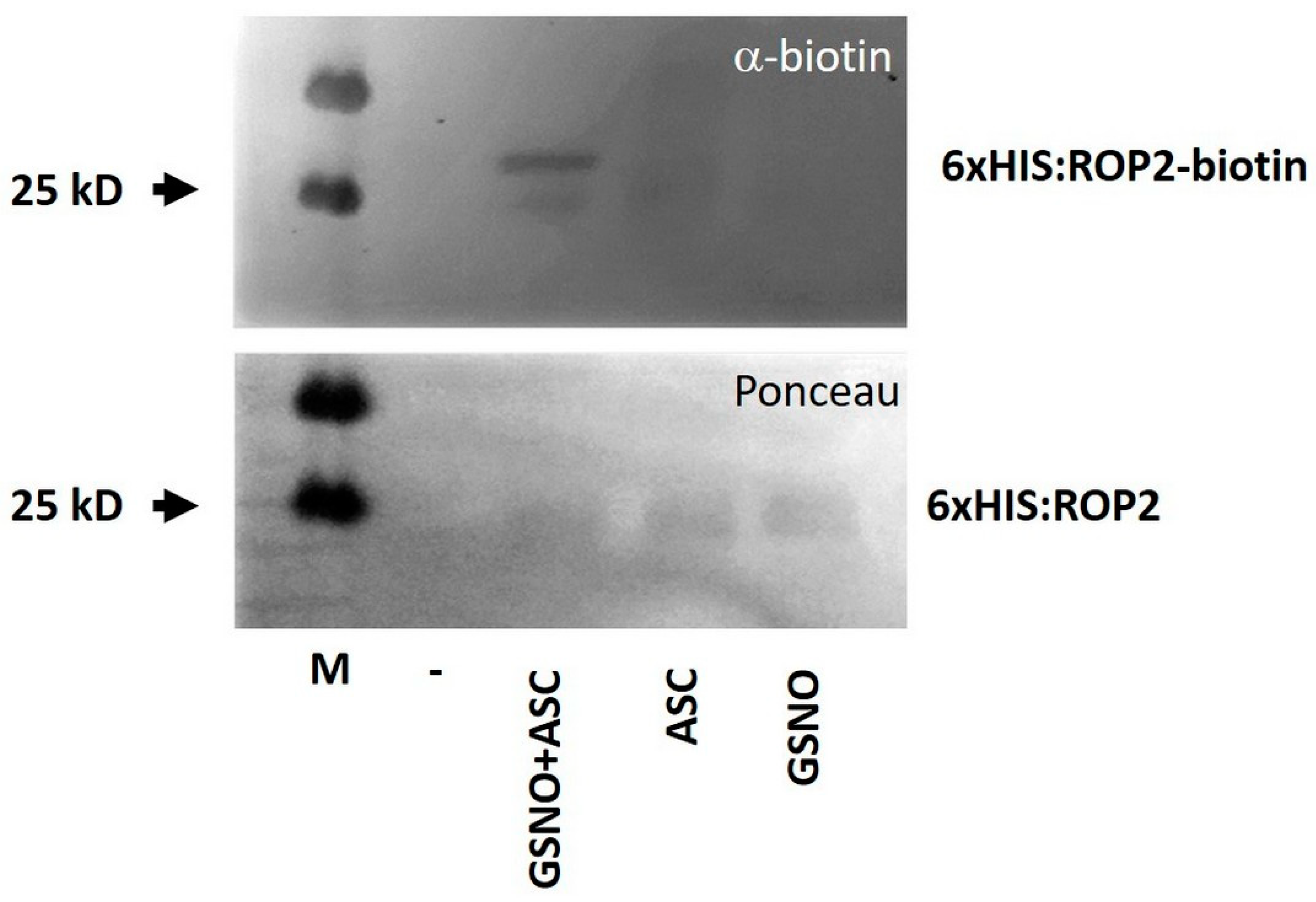
2.2. GSNO Promotes In Vitro S-nitrosation of the ROP2 Protein
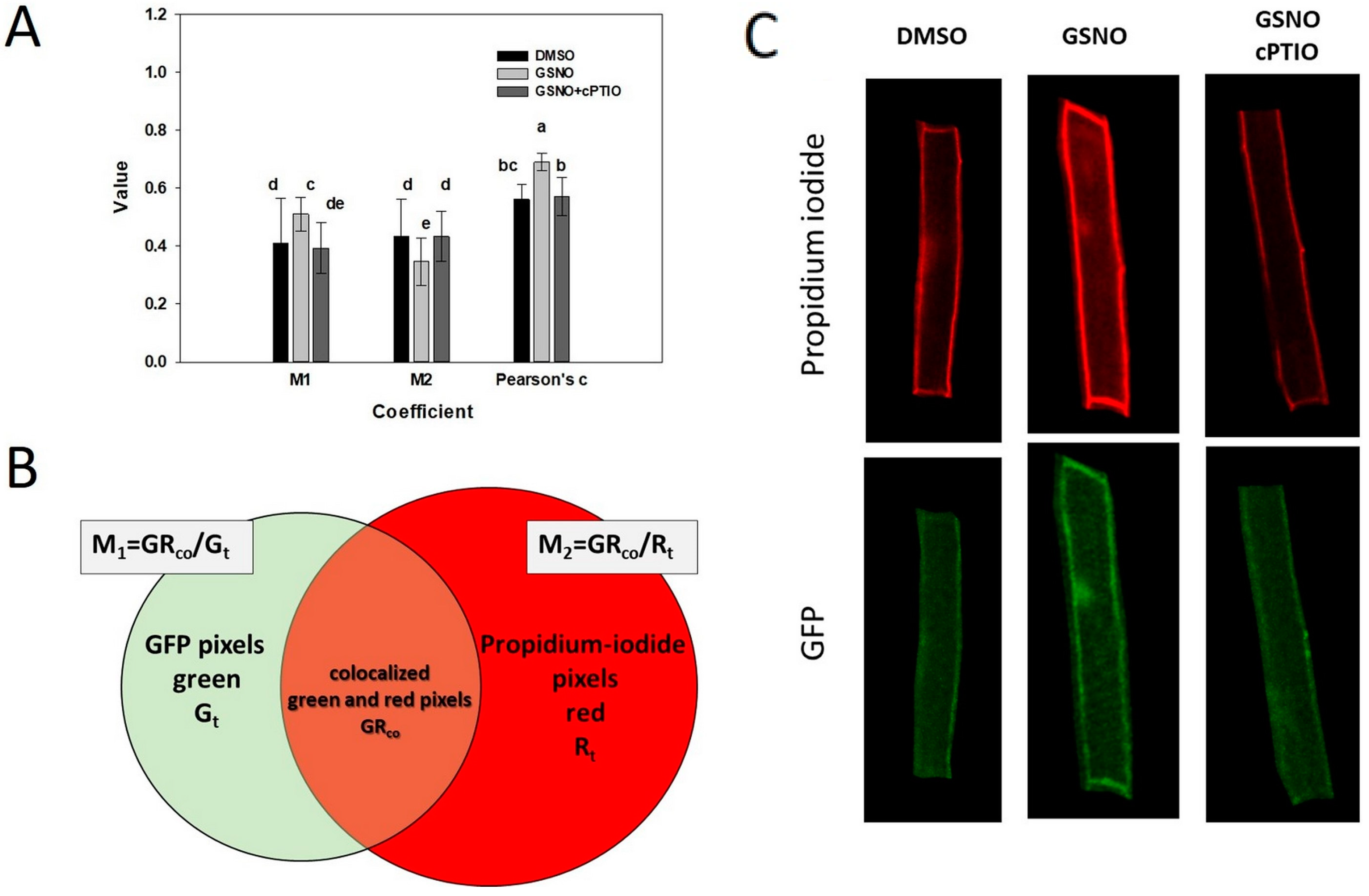
2.3. S-nitrosation Might Alter the Intracellular Localisation of ROP2
2.4. GSNO-Triggered PIN1 Depletion Is Dependent on ROP2 in the Arabidopsis Root Meristem
3. Discussion
3.1. NO Effect on Root Shortening Depends on ROP2
3.2. S-nitrosation of the AtROP2 GTPase Might Control Its Function
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Chemicals and Treatments
4.3. Microscopy-Based Analyses
4.4. Detecting NO in Plant Roots and Its Quantification in Donor Solutions
4.5. qRT-PCR Analysis
4.6. Western Blot Analysis of GFP:ROP2 Protein Abundance
4.7. Molecular Cloning
4.8. Recombinant Protein Expression
4.9. Biotin Switch Assay
4.10. In Vitro S-nitrosation and GTP-Binding Assay
4.11. S-nitrosation Site Prediction and In Silico Gene Expression Analysis
4.12. Whole-Mount Immunolocalization of PIN1
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gruber, B.D.; Giehl, R.F.H.; Friedel, S.; von Wirén, N. Plasticity of the arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, Z.; Amtmann, A. Food for thought: How nutrients regulate root system architecture. Curr. Opin. Plant Biol. 2017, 39, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Giehl, R.F.H.; von Wirén, N. Root Nutrient Foraging. Plant Physiol. 2014, 166, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Burssens, S.; Himanen, K.; Van De Cotte, B.; Beeckman, T.; Van Montagu, M.; Inzé, D.; Verbruggen, N. Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta 2000, 211, 632–640. [Google Scholar] [CrossRef]
- West, G.; Inzé, D.; Beemster, G.T.S. Cell cycle modulation in the response of the primary root of arabidopsis to salt stress. Plant Physiol. 2004, 135, 1050–1058. [Google Scholar] [CrossRef]
- Kochian, L.V. Root architecture. J. Integr. Plant Biol. 2016, 58, 190–192. [Google Scholar] [CrossRef]
- Bishopp, A.; Benková, E.; Helariutta, Y. Sending mixed messages: Auxin-cytokinin crosstalk in roots. Curr. Opin. Plant Biol. 2011, 14, 10–16. [Google Scholar] [CrossRef]
- Fu, X.; Harberd, N.P. Auxin promotes arabidopsis root growth by modulating gibberellin response. Nature 2003, 421, 740–743. [Google Scholar] [CrossRef]
- Correa-Aragunde, N.; París, R.; Foresi, N.; Terrile, C.; Casalongué, C.; Lamattina, L. The Auxin-Nitric Oxide Highway: A Right Direction in Determining the Plant Root System. In Gasotransmitters in Plants: The Rise of a New Paradigm in Cell Signaling; Lamattina, L., García-Mata, C., Eds.; Signaling and Communication in Plants; Springer International Publishing: Cham, Switzerland, 2016; pp. 117–136. ISBN 978-3-319-40713-5. [Google Scholar]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Grieneisen, V.A.; Xu, J.; Marée, A.F.M.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef] [Green Version]
- Brumos, J.; Robles, L.M.; Yun, J.; Vu, T.C.; Jackson, S.; Alonso, J.M.; Stepanova, A.N. local auxin biosynthesis is a key regulator of plant development. Dev. Cell 2018, 47, 306–318.e5. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Functions of Nitric Oxide (NO) in roots during development and under adverse stress conditions. Plants 2015, 4, 240–252. [Google Scholar] [CrossRef]
- Kolbert, Z. Implication of Nitric Oxide (NO) in excess element-induced morphogenic responses of the root system. Plant Physiol. Biochem. 2016, 101, 149–161. [Google Scholar] [CrossRef]
- Sanz, L.; Albertos, P.; Mateos, I.; Sánchez-Vicente, I.; Lechón, T.; Fernández-Marcos, M.; Lorenzo, O. Nitric Oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015, 66, 2857–2868. [Google Scholar] [CrossRef]
- Fernández-Marcos, M.; Sanz, L.; Lewis, D.R.; Muday, G.K.; Lorenzo, O. Nitric Oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-Dependent acropetal auxin transport. Proc. Natl. Acad. Sci. USA 2011, 108, 18506–18511. [Google Scholar] [CrossRef]
- Correa-Aragunde, N.; Graziano, M.; Lamattina, L. Nitric Oxide plays a central role in determining lateral root development in tomato. Planta 2004, 218, 900–905. [Google Scholar] [CrossRef]
- Pagnussat, G.C.; Simontacchi, M.; Puntarulo, S.; Lamattina, L. Nitric Oxide is required for root organogenesis. Plant Physiol. 2002, 129, 954–956. [Google Scholar] [CrossRef]
- Lombardo, M.C.; Graziano, M.; Polacco, J.C.; Lamattina, L. Nitric Oxide functions as a positive regulator of root hair development. Plant Signal. Behav. 2006, 1, 28–33. [Google Scholar] [CrossRef]
- Potters, G.; Pasternak, T.P.; Guisez, Y.; Palme, K.J.; Jansen, M.A.K. Stress-Induced Morphogenic responses: Growing out of trouble? Trends Plant Sci. 2007, 12, 98–105. [Google Scholar] [CrossRef]
- Korver, R.A.; Koevoets, I.T.; Testerink, C. Out of shape during stress: A key role for auxin. Trends Plant Sci. 2018, 23, 783–793. [Google Scholar] [CrossRef]
- Potters, G.; Pasternak, T.P.; Guisez, Y.; Jansen, M.A.K. Different stresses, similar morphogenic responses: Integrating a plethora of pathways. Plant Cell Environ. 2009, 32, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.W.M. Regulatory Role of Nitric Oxide in Alterations of Morphological Features of Plants under Abiotic Stress. In Nitric Oxide Action in Abiotic Stress Responses in Plants; Khan, M.N., Mobin, M., Mohammad, F., Corpas, F.J., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 65–73. ISBN 978-3-319-17804-2. [Google Scholar]
- Correa-Aragunde, N.; Foresi, N.; Lamattina, L. Chapter Three—Auxin and Nitric Oxide: A Counterbalanced Partnership Ensures the Redox Cue Control Required for Determining Root Growth Pattern. In Advances in Botanical Research; Wendehenne, D., Ed.; Nitric Oxide and Signaling in Plants; Academic Press: Cambridge, MA, USA, 2016; Volume 77, pp. 41–54. [Google Scholar]
- Lanteri, M.L.; Graziano, M.; Correa-Aragunde, N.; Lamattina, L. From Cell Division to Organ Shape: Nitric Oxide Is Involved in Auxin-Mediated Root Development. In Communication in Plants: Neuronal Aspects of Plant Life; Baluška, F., Mancuso, S., Volkmann, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 123–136. ISBN 978-3-540-28516-8. [Google Scholar]
- Sanz, L.; Fernández-Marcos, M.; Modrego, A.; Lewis, D.R.; Muday, G.K.; Pollmann, S.; Dueñas, M.; Santos-Buelga, C.; Lorenzo, O. Nitric Oxide Plays a role in stem cell niche homeostasis through its interaction with auxin. Plant Physiol. 2014, 166, 1972–1984. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.; Kleine-Vehn, J. PIN-FORMED and PIN-LIKES auxin transport facilitators. Development 2019, 146, dev168088. [Google Scholar] [CrossRef] [PubMed]
- Kleine-Vehn, J.; Wabnik, K.; Martinière, A.; Łangowski, Ł.; Willig, K.; Naramoto, S.; Leitner, J.; Tanaka, H.; Jakobs, S.; Robert, S.; et al. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol. Syst. Biol. 2011, 7, 540. [Google Scholar] [CrossRef] [PubMed]
- Löfke, C.; Luschnig, C.; Kleine-Vehn, J. Posttranslational modification and trafficking of PIN auxin efflux carriers. Mech. Dev. 2013, 130, 82–94. [Google Scholar] [CrossRef]
- Ren, H.; Lin, D. ROP GTPase regulation of auxin transport in Arabidopsis. Mol. Plant 2015, 8, 193–195. [Google Scholar] [CrossRef]
- Uhrig, J.F.; Hülskamp, M. Rop GTPases: Polarity and Cell Shape in Plants. In eLS; American Cancer Society: Atlanta, GA, USA, 2014; ISBN 978-0-470-01590-2. [Google Scholar]
- Chen, X.; Friml, J. Rho-GTPase-regulated vesicle trafficking in plant cell polarity. Biochem. Soc. Trans. 2014, 42, 212–218. [Google Scholar] [CrossRef]
- Feiguelman, G.; Fu, Y.; Yalovsky, S. ROP GTPases structure-function and signaling pathways. Plant Physiol. 2018, 176, 57–79. [Google Scholar] [CrossRef]
- Lavy, M.; Bloch, D.; Hazak, O.; Gutman, I.; Poraty, L.; Sorek, N.; Sternberg, H.; Yalovsky, S. A Novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr. Biol. 2007, 17, 947–952. [Google Scholar] [CrossRef]
- Wu, G.; Gu, Y.; Li, S.; Yang, Z. A Genome-wide analysis of arabidopsis rop-interactive CRIB motif–containing proteins that act as rop GTPase targets. Plant Cell 2001, 13, 2841–2856. [Google Scholar] [CrossRef] [Green Version]
- Nagawa, S.; Xu, T.; Lin, D.; Dhonukshe, P.; Zhang, X.; Friml, J.; Scheres, B.; Fu, Y.; Yang, Z. ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLOS Biol. 2012, 10, e1001299. [Google Scholar] [CrossRef]
- Chen, X.; Naramoto, S.; Robert, S.; Tejos, R.; Löfke, C.; Lin, D.; Yang, Z.; Friml, J. ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in arabidopsis roots. Curr. Biol. 2012, 22, 1326–1332. [Google Scholar] [CrossRef]
- Lin, D.; Nagawa, S.; Chen, J.; Cao, L.; Chen, X.; Xu, T.; Li, H.; Dhonukshe, P.; Yamamuro, C.; Friml, J.; et al. A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in arabidopsis roots. Curr. Biol. 2012, 22, 1319–1325. [Google Scholar] [CrossRef]
- Gupta, K.J.; Kolbert, Z.; Durner, J.; Lindermayr, C.; Corpas, F.J.; Brouquisse, R.; Barroso, J.B.; Umbreen, S.; Palma, J.M.; Hancock, J.T.; et al. Regulating the regulator: Nitric Oxide control of post-translational modifications. New Phytol. 2020, 227, 1319–1325. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, D.; Wang, C.; Wang, B.; Fang, H.; Huo, J.; Liao, W. Recent progress in protein S-nitrosylation in phytohormone signaling. Plant Cell Physiol. 2019, 60, 494–502. [Google Scholar] [CrossRef]
- Simontacchi, M.; García-Mata, C.; Bartoli, C.G.; Santa-María, G.E.; Lamattina, L. Nitric Oxide as a key component in hormone-regulated processes. Plant Cell Rep. 2013, 32, 853–866. [Google Scholar] [CrossRef]
- Terrile, M.C.; París, R.; Calderón-Villalobos, L.I.A.; Iglesias, M.J.; Lamattina, L.; Estelle, M.; Casalongué, C.A. Nitric Oxide influences auxin signaling through S-Nitrosylation of the arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 2012, 70, 492–500. [Google Scholar] [CrossRef]
- Raines, K.W.; Bonini, M.G.; Campbell, S.L. Nitric Oxide Cell Signaling: S-Nitrosation of ras superfamily GTPases. Cardiovasc. Res. 2007, 75, 229–239. [Google Scholar] [CrossRef]
- Heo, J.; Campbell, S.L. Mechanism of redox-mediated guanine nucleotide exchange on redox-active Rho GTPases. J. Biol. Chem. 2005, 280, 31003–31010. [Google Scholar] [CrossRef]
- Lin, L.; Xu, C.; Carraway, M.S.; Piantadosi, C.A.; Whorton, A.R.; Li, S. RhoA Inactivation by S-Nitrosylation regulates vascular smooth muscle contractive signaling. Nitric Oxide 2018, 74, 56–64. [Google Scholar] [CrossRef]
- Jeon, B.W.; Hwang, J.-U.; Hwang, Y.; Song, W.-Y.; Fu, Y.; Gu, Y.; Bao, F.; Cho, D.; Kwak, J.M.; Yang, Z.; et al. The arabidopsis small G Protein ROP2 is activated by light in guard cells and inhibits light-induced stomatal opening. Plant Cell 2008, 20, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.; Zheng, M.; Zhang, Y.; Yuan, M.; Yalovsky, S.; Zhu, L.; Fu, Y. The Microtubule-associated Protein MAP18 affects ROP2 GTPase activity during root hair growth1[OPEN]. Plant Physiol. 2017, 174, 202–222. [Google Scholar] [CrossRef] [PubMed]
- Sorek, N.; Henis, Y.I.; Yalovsky, S. How Prenylation and S-acylation regulate subcellular targeting and function of ROP GTPases. Plant Signal Behav. 2011, 6, 1026–1029. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; Cordelières, F.P. A Guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A Practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef]
- Ma, X.; Denyer, T.; Timmermans, M.C.P. PscB: A browser to explore plant single cell RNA-sequencing data sets1[OPEN]. Plant Physiol. 2020, 183, 464–467. [Google Scholar] [CrossRef]
- Omelyanchuk, N.A.; Kovrizhnykh, V.V.; Oshchepkova, E.A.; Pasternak, T.; Palme, K.; Mironova, V.V. A Detailed Expression Map of the PIN1 Auxin Transporter in Arabidopsis thaliana Root. BMC Plant Biol. 2016, 16, 5. [Google Scholar] [CrossRef]
- París, R.; Vazquez, M.M.; Graziano, M.; Terrile, M.C.; Miller, N.D.; Spalding, E.P.; Otegui, M.S.; Casalongué, C.A. Distribution of endogenous NO regulates early gravitropic response and PIN2 localization in arabidopsis roots. Front. Plant Sci. 2018, 9, 495. [Google Scholar] [CrossRef]
- Cséplő, Á.; Zsigmond, L.; Andrási, N.; Baba, A.I.; Labhane, N.M.; Pető, A.; Kolbert, Z.; Kovács, H.E.; Steinbach, G.; Szabados, L. The AtCRK5 protein kinase is required to maintain the ROS NO balance affecting the PIN2-mediated root gravitropic response in arabidopsis. Int. J. Mol. Sci. 2021, 22, 5979. [Google Scholar] [CrossRef]
- Vieten, A.; Vanneste, S.; Wisniewska, J.; Benková, E.; Benjamins, R.; Beeckman, T.; Luschnig, C.; Friml, J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 2005, 132, 4521–4531. [Google Scholar] [CrossRef] [Green Version]
- Hess, D.T.; Matsumoto, A.; Kim, S.-O.; Marshall, H.E.; Stamler, J.S. Protein S -Nitrosylation: Purview and parameters. Nat. Rev. Mol. Cell Biol. 2005, 6, 150–166. [Google Scholar] [CrossRef]
- Lamotte, O.; Bertoldo, J.B.; Besson-Bard, A.; Rosnoblet, C.; Aimé, S.; Hichami, S.; Terenzi, H.; Wendehenne, D. Protein S-Nitrosylation: Specificity and identification strategies in Plants. Front. Chem. 2015, 2, 114. [Google Scholar] [CrossRef]
- Heo, J.; Raines, K.W.; Mocanu, V.; Campbell, S.L. Redox regulation of RhoA. Biochemistry 2006, 45, 14481–14489. [Google Scholar] [CrossRef]
- Ménesi, D.; Klement, É.; Ferenc, G.; Fehér, A. The arabidopsis rho of plants GTPase ROP1 is a potential calcium-dependent protein kinase (CDPK) Substrate. Plants 2021, 10, 2053. [Google Scholar] [CrossRef]
- Sorek, N.; Segev, O.; Gutman, O.; Bar, E.; Richter, S.; Poraty, L.; Hirsch, J.A.; Henis, Y.I.; Lewinsohn, E.; Jürgens, G.; et al. An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Curr. Biol. 2010, 20, 914–920. [Google Scholar] [CrossRef]
- Roberts, P.J.; Mitin, N.; Keller, P.J.; Chenette, E.J.; Madigan, J.P.; Currin, R.O.; Cox, A.D.; Wilson, O.; Kirschmeier, P.; Der, C.J. Rho family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 2008, 283, 25150–25163. [Google Scholar] [CrossRef]
- Wodala, B.; Horváth, F. The Effect of Exogenous NO on PSI photochemistry in intact pea leaves. Acta Biol. Szeged. 2008, 52, 243–245. [Google Scholar]
- Zhang, X. Real Time and in vivo monitoring of Nitric Oxide by electrocehmical sensors- from dream to reality. Front. Biosci. A J. Virtual Libr. 2004, 9, 3434–3446. [Google Scholar] [CrossRef]
- Gallé, Á.; Csiszár, J.; Secenji, M.; Guóth, A.; Cseuz, L.; Tari, I.; Györgyey, J.; Erdei, L. Glutathione transferase activity and expression patterns during grain filling in flag leaves of wheat genotypes differing in drought tolerance: Response to water deficit. J. Plant Physiol. 2009, 166, 1878–1891. [Google Scholar] [CrossRef]
- Jaffrey, S.R.; Snyder, S.H. The biotin switch method for the detection of S-nitrosylated proteins. Sci. Signal. 2001, 2001, pl1. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Liu, Z.; Gao, X.; Jin, C.; Wen, L.; Yao, X.; Ren, J. GPS-SNO: Computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS ONE 2010, 5, e11290. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Luo, X.; Li, Y.; Chen, L.; Ma, W.; Huang, J.; Cui, J.; Zhao, Y.; Xue, Y.; Zuo, Z.; et al. DeepNitro: Prediction of protein nitration and nitrosylation sites by deep learning. Genom. Proteom. Bioinform. 2018, 16, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shao, X.-J.; Wu, L.-Y.; Deng, N.-Y.; Chou, K.-C. ISNO-AAPair: Incorporating amino acid pairwise coupling into PseAAC for predicting cysteine S-nitrosylation sites in proteins. PeerJ. 2013, 1, e171. [Google Scholar] [CrossRef]
- Pasternak, T.; Tietz, O.; Rapp, K.; Begheldo, M.; Nitschke, R.; Ruperti, B.; Palme, K. Protocol: An Improved and Universal Procedure for Whole-Mount Immunolocalization in Plants. Plant Methods 2015, 11, 50. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kenesi, E.; Kolbert, Z.; Kaszler, N.; Klement, É.; Ménesi, D.; Molnár, Á.; Valkai, I.; Feigl, G.; Rigó, G.; Cséplő, Á.; et al. The ROP2 GTPase Participates in Nitric Oxide (NO)-Induced Root Shortening in Arabidopsis. Plants 2023, 12, 750. https://doi.org/10.3390/plants12040750
Kenesi E, Kolbert Z, Kaszler N, Klement É, Ménesi D, Molnár Á, Valkai I, Feigl G, Rigó G, Cséplő Á, et al. The ROP2 GTPase Participates in Nitric Oxide (NO)-Induced Root Shortening in Arabidopsis. Plants. 2023; 12(4):750. https://doi.org/10.3390/plants12040750
Chicago/Turabian StyleKenesi, Erzsébet, Zsuzsanna Kolbert, Nikolett Kaszler, Éva Klement, Dalma Ménesi, Árpád Molnár, Ildikó Valkai, Gábor Feigl, Gábor Rigó, Ágnes Cséplő, and et al. 2023. "The ROP2 GTPase Participates in Nitric Oxide (NO)-Induced Root Shortening in Arabidopsis" Plants 12, no. 4: 750. https://doi.org/10.3390/plants12040750







