Metabolite Profiling to Evaluate Metabolic Changes in Genetically Modified Protopanaxadiol-Enriched Rice
Abstract
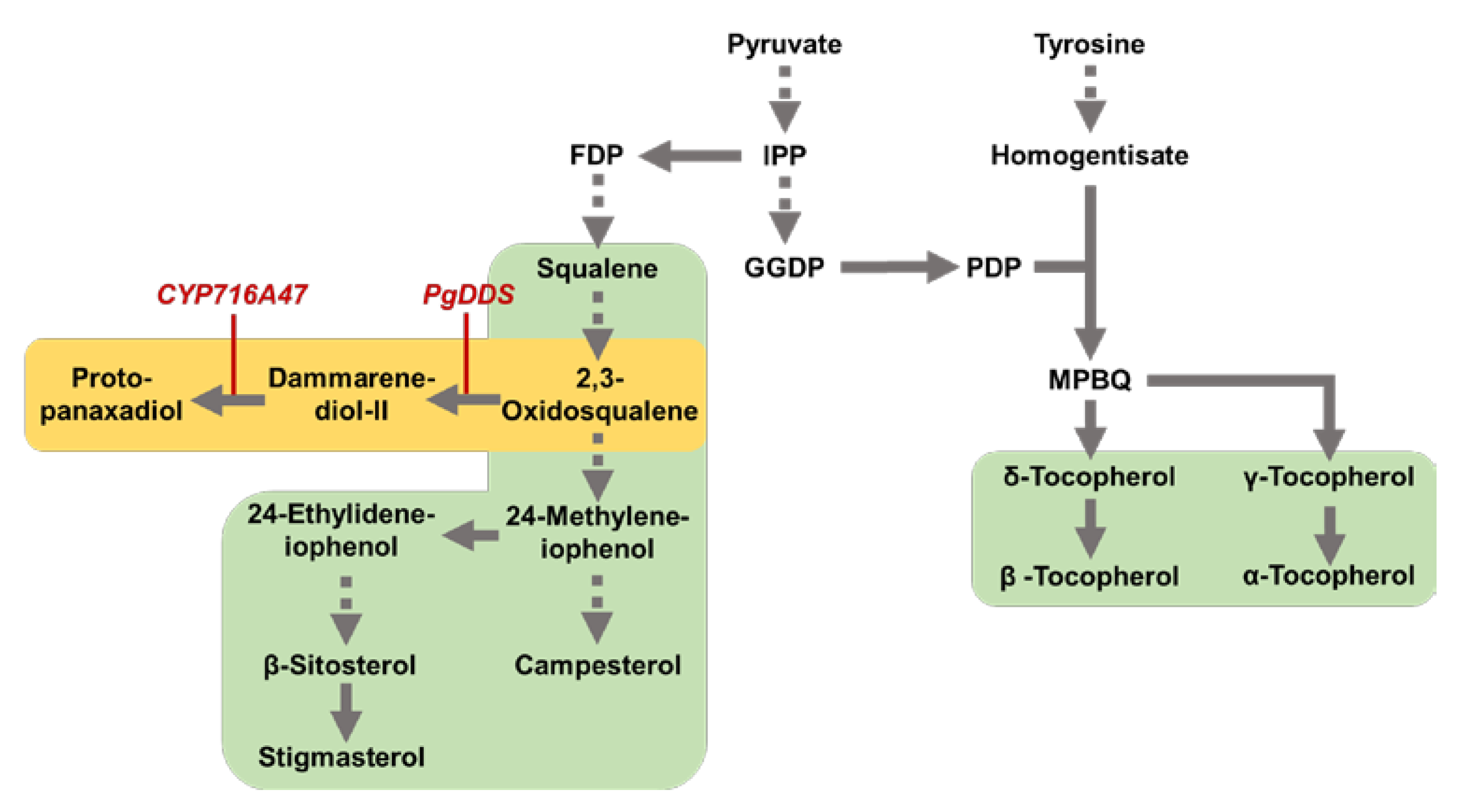
:1. Introduction
2. Results and Discussion
2.1. Multivariate Analysis of Rice Seed
2.2. Tocopherol and Phytosterol Content in Rice Seed
2.3. Multivariate Analysis of Rice Leaves
3. Materials and Methods
3.1. Rice Sample Preparation
3.2. Extraction and Analysis of Hydrophilic Metabolites
3.3. Extraction and Analysis of Lipophilic Metabolites
3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kim, T.J.; Kim, S.Y.; Park, Y.J.; Lim, S.-H.; Ha, S.-H.; Park, S.U.; Lee, B.; Kim, J.K. Metabolite profiling reveals distinct modulation of complex metabolic networks in non-pigmented, black, and red rice (Oryza Sativa L.) cultivars. Metabolites 2021, 11, 367. [Google Scholar]
- Lee, S.Y.; Yeo, Y.S.; Park, S.-Y.; Oh, S.-W.; Yoon, E.K.; Shin, K.-S.; Woo, H.-J.; Lim, M.-H. Composition analysis of herbicide tolerant Ab rice and insect-resistant Bt rice. Korean J. Breed. Sci. 2015, 47, 255–263. [Google Scholar] [CrossRef]
- Shibata, S. Chemistry and cancer preventing activities of ginseng saponins and some related triterpenoid compounds. J. Korean Med. Sci. 2001, 16, S28–S37. [Google Scholar]
- Leung, K.; Wong, A.S.T. Pharmacology of ginsenosides: A literature review. Chin. Med. 2010, 5, 20. [Google Scholar] [CrossRef]
- Chen, X.J.; Zhang, X.J.; Shui, Y.M.; Wan, J.B.; Gao, J.L. Anticancer activities of protopanaxadiol- and protopanaxatriol-type ginsenosides and their metabolites. Evid.-Based Complement. Altern. Med. 2016, 2016, 5738694. [Google Scholar] [CrossRef]
- Han, J.Y.; Baek, S.H.; Jo, H.J.; Yun, D.W.; Choi, Y.E. Genetically modified rice produces ginsenoside aglycone (protopanaxadiol). Planta 2019, 250, 1103–1110. [Google Scholar] [CrossRef]
- Chun, J.H.; Adhikari, P.B.; Park, S.B.; Han, J.Y.; Choi, Y.E. Production of the dammarene sapogenin (protopanaxadiol) in transgenic tobacco plants and cultured cells by heterologous expression of PgDDS and CYP716A47. Plant Cell Rep. 2015, 34, 1551–1560. [Google Scholar] [CrossRef]
- Chung, I.-M.; Lim, J.-J.; Ahn, M.-S.; Jeong, H.-N.; An, T.-J.; Kim, S.-H. Comparative phenolic compound profiles and antioxidative activity of the fruit, leaves, and roots of Korean ginseng (Panax ginseng Meyer) according to cultivation years. J. Ginseng Res. 2016, 40, 68–75. [Google Scholar]
- Jung, W.J.; Oh, S.-D.; Park, S.-Y.; Jang, Y.J.; Lee, S.-K.; Yun, D.-W.; Chang, A.; Park, S.U.; Ha, S.-H.; Kim, J.K. Metabolic profiling and antioxidant properties of hybrid soybeans with different seed coat colors, obtained by crossing β-carotene-enhanced (Glycine max) and wild (Glycine soja) soybeans. Plant Biotechnol. Rep. 2022, 16, 449–463. [Google Scholar]
- Kim, J.K.; Kim, E.H.; Park, I.; Yu, B.R.; Lim, J.D.; Lee, Y.S.; Lee, J.H.; Kim, S.H.; Chung, I.M. Isoflavones profiling of soybean [Glycine max (L.) Merrill] germplasms and their correlations with metabolic pathways. Food Chem. 2014, 153, 258–264. [Google Scholar] [CrossRef]
- Kim, M.S.; Baek, S.A.; Park, S.Y.; Baek, S.H.; Lee, S.M.; Ha, S.H.; Lee, Y.T.; Choi, J.; Im, K.H.; Kim, J.K. Comparison of the grain composition in resveratrol-enriched and glufosinate-tolerant rice (Oryza sativa) to conventional rice using univariate and multivariate analysis. J. Food Compos. Anal. 2016, 52, 58–67. [Google Scholar] [CrossRef]
- Kim, Y.J.; Park, Y.J.; Oh, S.-D.; Yoon, J.S.; Kim, J.G.; Seo, J.-S.; Park, J.-H.; Kim, C.-G.; Park, S.-Y.; Choi, M.-S.; et al. Effects of genotype and environment on the nutrient and metabolic profiles of soybeans genetically modified with epidermal growth factor or thioredoxin compared with conventional soybeans. Ind. Crops Prod. 2022, 175, 114229. [Google Scholar]
- Kim, J.K.; Park, S.-Y.; Lee, S.M.; Lim, S.-H.; Kim, H.J.; Oh, S.-D.; Yeo, Y.; Cho, H.S.; Ha, S.-H. Unintended polar metabolite profiling of carotenoid-biofortified transgenic rice reveals substantial equivalence to its non-transgenic counterpart. Plant Biotechnol. Rep. 2013, 7, 121–128. [Google Scholar]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Vriet, C.; Russinova, E.; Reuzeau, C. From squalene to brassinolide: The steroid metabolic and signaling pathways across the plant kingdom. Mol. Plant 2013, 6, 1738–1757. [Google Scholar]
- Lee, H.S.; Hwang, W.H.; Jeong, J.S.H.; Yang, S.Y.; Jeong, N.J.; Lee, C.K.; Choi, M.G. Physiological causes of transplantation shock on rice growth inhibition and delayed heading. Sci. Rep. 2021, 11, 16818. [Google Scholar] [CrossRef]
- Li, X.; Zhong, Q.; Li, Y.; Li, G.-H.; Ding, Y.; Wang, S.; Liu, Z.; Tang, S.; Ding, C.; Chen, L. Triacontanol reduces transplanting shock in machine-transplanted rice by improving the growth and antioxidant systems. Front. Plant Sci. 2016, 7, 501. [Google Scholar] [CrossRef]
- Shi, X.; Chen, S.; Peng, Y.; Wang, Y.; Chen, J.; Hu, Z.; Wang, B.; Li, A.; Chao, D.; Li, Y.; et al. TSC1 enables plastid development under dark conditions, contributing to rice adaptation to transplantation shock. J. Plant Biol. 2018, 60, 112–129. [Google Scholar]
- Kumar, M.S.S.; Ali, K.; Dahuja, A.; Tyagi, A. Role of phytosterols in drought stress tolerance in rice. Plant Physiol. Biochem. 2015, 96, 83–89. [Google Scholar] [CrossRef]
- Song, Y.; Feng, L.; Alyafei, M.A.M.; Jaleel, A.; Ren, M. Function of chloroplasts in plant stress responses. Int. J. Mol. Sci. 2021, 22, 3464. [Google Scholar]
- Movahedi, A.; Wei, H.; Pucker, B.; Ghaderi-Zefrehei, M.; Rasouli, F.; Kiani-Pouya, A.; Jiang, T.; Zhuge, Q.; Yang, L.; Zhou, X. Isoprenoid biosynthesis regulation in poplars by methylerythritol phosphate and mevalonic acid pathways. Front. Plant Sci. 2022, 13, 968780. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Zeeman, S.C. Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 2012, 15, 282–292. [Google Scholar] [CrossRef]
- Troncoso-Ponce, M.A.; Cao, X.; Yang, Z.; Ohlrogge, J.B. Lipid turnover during senescence. Plant Sci. 2013, 205–206, 13–19. [Google Scholar]
- Honda, S.; Ohkubo, S.; San, N.S.; Nakkasame, A.; Tomisawa, K.; Katsura, K.; Ookawa, T.; Nagano, A.; Adachi, S. Maintaining higher leaf photosynthesis after heading stage could promote biomass accumulation in rice. Sci. Rep. 2021, 11, 7579. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, J.G.; Lee, W.-K.; So, K.M.; Kim, J.K. Trial data of the anti-obesity potential of a high resistant starch diet for canines using Dodamssal rice and the identification of discriminating markers in feces for metabolic profiling. Metabolomics 2019, 15, 21. [Google Scholar] [CrossRef]
- Kim, T.J.; Lee, K.B.; Baek, S.-A.; Choi, J.; Ha, S.-H.; Lim, S.-H.; Park, S.-Y.; Yeo, Y.; Park, S.U.; Kim, J.K. Determination of lipophilic metabolites for species discrimination and quality assessment of nine leafy vegetables. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 909–918. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sim, J.-E.; Oh, S.-D.; Kang, K.; Shin, Y.-M.; Yun, D.-W.; Baek, S.-H.; Choi, Y.-E.; Park, S.-U.; Kim, J.-K. Metabolite Profiling to Evaluate Metabolic Changes in Genetically Modified Protopanaxadiol-Enriched Rice. Plants 2023, 12, 758. https://doi.org/10.3390/plants12040758
Sim J-E, Oh S-D, Kang K, Shin Y-M, Yun D-W, Baek S-H, Choi Y-E, Park S-U, Kim J-K. Metabolite Profiling to Evaluate Metabolic Changes in Genetically Modified Protopanaxadiol-Enriched Rice. Plants. 2023; 12(4):758. https://doi.org/10.3390/plants12040758
Chicago/Turabian StyleSim, Ji-Eun, Sung-Dug Oh, Kiyoon Kang, Yu-Mi Shin, Doh-Won Yun, So-Hyeon Baek, Yong-Eui Choi, Sang-Un Park, and Jae-Kwang Kim. 2023. "Metabolite Profiling to Evaluate Metabolic Changes in Genetically Modified Protopanaxadiol-Enriched Rice" Plants 12, no. 4: 758. https://doi.org/10.3390/plants12040758
APA StyleSim, J.-E., Oh, S.-D., Kang, K., Shin, Y.-M., Yun, D.-W., Baek, S.-H., Choi, Y.-E., Park, S.-U., & Kim, J.-K. (2023). Metabolite Profiling to Evaluate Metabolic Changes in Genetically Modified Protopanaxadiol-Enriched Rice. Plants, 12(4), 758. https://doi.org/10.3390/plants12040758






